Abstract
The eukaryotic translation initiation factor 1 binds to the ribosome during translation initiation. It is instrumental for initiator-tRNA and mRNA binding, and has a function in selection of the authentic start codon. Here, we show that the archaeal homolog aIF1 has analogous functions. The aIF1 protein of the archaeon Sulfolobus solfataricus is bound to the small ribosomal subunit during translation initiation and accelerates binding of initiator-tRNA and mRNA to the ribosome. Accordingly, aIF1 stimulated translation of an mRNA in a S. solfataricus in vitro translation system. Moreover, this study suggested that the C terminus of the factor is of relevance for its function.
Keywords: aIF1, archaea, Sulfolobus solfataricus, translation initiation
INTRODUCTION
When compared to the elongation step of protein synthesis, the process of translation initiation is much less conserved throughout the three kingdoms of life. In bacteria, pairing of complementary RNA motifs, the Shine-Dalgarno (SD) sequence on the mRNA and the anti-Shine-Dalgarno (anti-SD) sequence on the 16S rRNA, are generally required for correct start codon selection (Kozak 1999). Three initiation factors (IF1, IF2, and IF3) are involved in the initiation step (Gualerzi and Pon 1990). IF1 binds to the A-site of the 30S ribosomal subunit (Dahlquist and Puglisi 2000) and most likely prevents premature binding of elongator tRNA. IF2 accelerates the formation of the codon–anticodon interaction by promoting binding of fMet-tRNAfMet to the ribosomal P-site. Moreover, a contribution of IF2-dependent GTP hydrolysis in the final adjustment of fMet-tRNAfMet in the P-site of the 30S subunit has been reported (La Teana et al. 1996; Brandi et al. 2004). IF3 acts as a fidelity factor by destabilizing noncanonical codon–anticodon interactions (Gualerzi and Pon 1990). The discriminatory function exerted by IF3 appears to result from a conformational change in the ribosome rather than from a direct interaction with fMet-tRNAf Met (Petrelli et al. 2001; Pioletti et al. 2001).
Eukaryotic translation initiation requires protein–protein interactions and several factors. The initiator methionyl-tRNA (Met-tRNAi) binds as part of an eIF2·GT·Met-tRNAi ternary complex to the 40S ribosomal subunit, which is in complex with several other factors (eIF1, eIF1A, eIF3, and eIF5), resulting in a 43S complex. The eIF4F complex assembles on the 5′-cap of the mRNA and unwinds structures in the 5′UTR. This is accomplished through the ATP-dependent action of eIF4A assisted by the RNA-binding proteins eIF4B, and in mammals, eIF4H. eIF4F, in conjunction with eIF3 and the poly(A) binding protein (PAB), which is bound to the 3′-poly(A) tail, then facilitate loading of the 43S complex onto the mRNA. The 43S complex begins scanning into the 5′to 3′direction until it encounters an initiation codon. Following this ATP-dependent scanning step, eIF2 bound GTP is hydrolyzed, and the eIF2-GDP complex is released, leaving the Met-tRNAi in the P-site of the 40S subunit (for a recent review, see Kapp and Lorsch 2004).
The two small initiation factors eIF1 and eIF1A have been reported to act together in different aspects of translation initiation. It has been shown that both factors bind to the small ribosomal subunit in a thermodynamically coupled manner (Maag and Lorsch 2003). No other factor is required for binding of eIF1 and eIF1A to yeast ribosomes, while eIF3 is required for binding of both factors to ribosomes of higher eukaryotes (Fletcher et al. 1999; Phan et al. 2001). Although eIF1 is sufficient for eIF2·GTP·Met-tRNAi binding to yeast ribosomes, eIF1A has a stimulatory effect (Algire et al. 2002). The factor eIF1 is thought to be released after start codon recognition through a conformational change in the ribosome (Maag et al. 2005). For eIF1A it was shown that it interacts with eIF2(5B), and it has been suggested that it aids in stabilization of Met-tRNAi on the ribosome after eIF2-GDP release (Marintchev et al. 2003). Both eIF1 and eIF1A are essential for cap-mediated initiation of translation in higher eukaryotes (Pestova et al. 1998). In the absence of these factors, the 43S complex is unable to start scanning and to locate the correct start codon. The factor eIF1 is further important to maintain the accuracy of this process by recognizing and destabilizing aberrant pre-initiation complexes (Yoon and Donahue 1992; Pestova et al. 1998). The solution structure of eIF1 has been resolved by NMR (Fletcher et al. 1999). The smaller N-terminal part is unfolded, while the larger and more conserved C-terminal part forms a tightly folded domain with two α-helices on one side of a five-stranded parallel and anti-parallel β-sheet. It has been reported that both the C-terminal and the N-terminal end of eIF1 are important for its function (Singh et al. 2004). It was shown that C-terminal tagged eIF1 is lethal in yeast and that it has a decreased binding affinity for eIF3c (Asano et al. 1998). In addition, C- and N-terminal tagged eIF1 displayed a reduced binding affinity for eIF2β, eIF5, and for the 40S ribosomal subunit (Singh et al. 2004).
The translation initiation pathway in archaea is poorly understood and little is known about the different components required for this process (Londei 2005). It appears that two different mechanisms for translation initiation exist in archaea (Tolstrup et al. 2000; Benelli et al. 2003). Internal cistrons of polycistronic mRNAs usually have a SD sequence similar to bacteria. In contrast, monocistronic mRNAs as well as proximal cistrons of polycistronic mRNAs have short or no 5′-untranslated regions (5′-UTRs). Such leaderless mRNAs appear to require a mechanism independent of a SD/anti-SD interaction and seem to depend only on codon–anticodon interaction between tRNAi and mRNA (Grill et al. 2000; Benelli et al. 2003). Archaea possess homologs of bacterial and eukaryal translation initiation factors. The trimeric factor a/eIF2 forms a ternary complex with GTP and Met-tRNAi and delivers the Met-tRNAi to the ribosome (Yatime et al. 2004; Pedulla et al. 2005). In contrast to eukaryotes, the αand γsubunits are important for Met-tRNAi binding. The other archaeal initiation factors include aIF2(5B) (homolog to bacterial IF2 and eukaryotic eIF2[5B]), aIF6 (homolog to eIF6), aIF1 (homolog to eIF1) and aIF1A (homolog to bacterial IF1 and eukaryotic eIF1A). S. solfataricus aIF1 shows little sequence homology with eukaryotic eIF1 homologs. Except for a/eIF2, no function has as yet been assigned to any archaeal translation initiation factor.
In this work, we have cloned the aIF1 gene and purified the factor from the crenarchaeon S. solfataricus. We show that aIF1 binds to the 30S ribosomal subunit, and that it stimulates translation of a mRNA in a S. solfataricus in vitro translation system by promoting binding of a/eIF2·GTP·Met-tRNAi and mRNA to the ribosome.
RESULTS
Cloning of the S. solfataricus aIF1 gene and purification of the protein
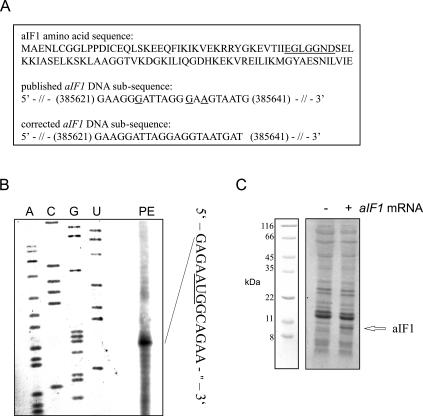
We first compared the sequence of the S. solfataricus aIF1 gene with that of several eukaryotic eIF1 homologs. According to the published DNA sequence of aIF1 (She et al. 2001; http://www-archbac.u-psud.fr/projects/sulfolobus/), the gene would be smaller in size than that of different eukaryotic homologs. We therefore re-sequenced the putative aIF1 gene and found some sequence errors between bp 385 621–385 641 in the annotated S. solfataricus genomic sequence. According to our sequence analysis the S. solfataricus aIF1 gene codes for 100 amino acids. Figure 1A ▶ shows the published and corrected DNA sub-sequence of the aIF1 gene as well as the corrected amino acid sequence of the aIF1 protein.
FIGURE 1.
Assignment of the transcriptional start site, the correct amino acid sequence and expression of the aIF1 gene in vitro. (A) Amino acid sequence of aIF1 (affected amino acids are underlined) and the corrected DNA subsequence (the numbers correspond to the S. solfataricus genome coordinates [Singh et al. 2004]) of the aIF1 gene. Sequence errors in the published aIF1 sequence are underlined. (B) Determination of the 5′-end of aIF1 mRNA by primer extension. The primer extension signal (PE) corresponds to the G nucleotide, 4 nt upstream of the AUG start codon. (C) In vitro translation of aIF1 mRNA using a S. solfataricus in vitro translation system. The position of the aIF1 protein is indicated by an arrow, and the positions of molecular weight markers are shown at the left.
The genes up- and downstream from aiF1 in the S. solfataricus genome are located on the opposite chromosomal strand (She et al. 2001), suggesting that it is transcribed independently as a monocistronic mRNA. To verify this, the 5′-end of aIF1 mRNA was mapped by primer extension (Fig. 1B ▶). This experiment revealed that a short, 4-nt-long 5′UTR precedes the AUG start codon. The DNA sequence of the aIF1 gene including the mini-5′UTR preceded by a T7 promoter was amplified by PCR from genomic DNA. Then, the quasi-leaderless aIF1 RNA was transcribed in vitro, and used to program a cell-free translation system derived from S. solfataricus cell lysates (Condo et al. 1999). As shown in Figure 1C ▶, the in vitro translation yielded a protein of the predicted size of ~10 kDa, corresponding to full-length aIF1.
For purification of the aIF1 protein, the coding sequence of the aIF1 gene was PCR-amplified and inserted into the Escherichia coli “His-tag vectors” pRSET (for N-terminal His-tagging [NH-aIF1]) and pET28b (for C-terminal His-tagging [CH-aIF1]). The recombinant proteins were purified by a two-step procedure, including heating the E. coli lysates at 70°C to remove most of the host proteins, followed by selective capture of the recombinant polypeptides by affinity chromatography. The proteins were purified to homogeneity and displayed the expected molecular weights in SDS-polyacrylamide gels (data not shown).
Localization of aIF1 in S. solfataricus cell extracts
To analyze the function of aIF1, we first determined its localization in both cell lysates and in lysates programmed for protein synthesis. First, S. solfataricus cell lysates were fractionated on a 10%–30% sucrose gradient, and the ribosome profile was determined by measuring the A260 (Fig. 2A ▶). The fractions were then subjected to Western blot analysis and probed with anti-aIF1 antibodies. This analysis revealed that aIF1 was exclusively associated with 30S ribosomal subunits. No aIF1 was associated with the large ribosomal subunits or found in the fractions containing low-molecular-weight components. Since S. solfataricus ribosomes are known to dissociate during ribosome preparation (Londei et al. 1986), 70S monosomes or polysomes were not detectable (Fig. 2A ▶).
FIGURE 2.
aIF1 is localized on the 30S ribosomal subunit. (A) Sedimentation profile (10%–30% sucrose gradient) of a S. solfataricus cell lysate, determined by A260 measurement in the absence of added mRNA. (B) Sedimentation profile (10%–30% sucrose gradient) of a S. solfataricus cell lysate programmed with 104 mRNA upon treatment with formaldehyde. The top and the bottom of the gradient are indicated. The gradients were fractionated and the fractions were probed for the presence of aIF1 with polyclonal antibodies raised against recombinant S. solfataricus aIF1 as described in Materials and Methods. The corresponding Western blots are shown below the sedimentation profiles.
Second, the same experiment was performed using cell lysates programmed for translation with S. solfataricus 104 mRNA encoding ribosomal protein L30 (Condo et al. 1999). The samples were incubated for 15 min at 70°C and then treated with 6% formaldehyde for 30 min at 4°C. This was done to stabilize 70S ribosomes, which would otherwise dissociate during sample centrifugation (Londei et al. 1986). As shown in Figure 2B ▶, aIF1 was only detected in ribosome free fractions or bound to 30S ribosomes, whereas no aIF1 was associated with 50S subunits or 70S monosomes. Since free aIF1 was not detected in translationally silent extracts (Fig. 2A ▶), these results suggested that aIF1 recycles between the 30S subunit and the soluble fraction upon entering the elongation phase.
aIF1 stimulates translation
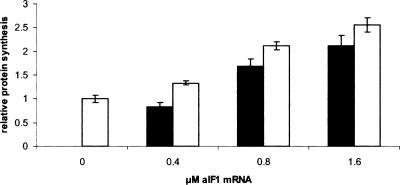
Next, we tested whether aIF1 exerts a general effect on translation. Initially, increasing amounts of the recombinant purified aIF1 were added to an in vitro translation system programmed with S. solfataricus 104 mRNA. However, as the high-salt storage buffer of recombinant aIF1 nonspecifically inhibited in vitro translation, these attempts were unsuccessful. Therefore, the in vitro translation system programmed with 104 mRNA was supplemented with increasing amounts of aIF1 quasi-leaderless mRNA to overproduce aIF1 in the translation assay. The samples were incubated in the presence of [35S]-methionine for 40 min at 70°C, and were then loaded on a SDS-polyacrylamide gel to quantify the amount of synthesized proteins. As shown in Figure 3 ▶, the production of the L30 protein increased up to ~2.5-fold in the presence of increasing concentrations of aIF1, suggesting that the factor stimulated some step during translation initiation. No stimulation of 104 mRNA translation was observed in a control sample supplemented with increasing amounts of a S. solfataricus mRNA encoding a transcription factor, demonstrating that the observed effect was attributable to aIF1 (not shown). The fractionation by density gradients of the translation mixtures programmed only with aIF1 mRNA revealed that the newly-synthesized radioactively labeled aIF1 protein was exclusively located on 30S ribosomal subunits (not shown), suggesting that under these conditions aIF1 was present in substoichiometric amounts to ribosomes. Next, we studied the specific step(s) affected by aIF1 during translation initiation.
FIGURE 3.
Cosynthesis of aIF1 increases in vitro translation of S. solfataricus 104 mRNA. The 104 mRNA (0.4 μM) was translated in a S. solfataricus in vitro translation system in the presence of increasing concentrations of aIF1 mRNA (0, 0.4, 0.8, and 1.6 μM). The white and the black bars represent the relative protein synthesis rate of 104 mRNA and aIF1 mRNA, respectively. The samples were separated on a 15% SDS-polyacrylamide gel and the radioactive bands were visualized using a PhosphorImager. A graphical representation of two independent experiments is shown. The amount of 104 protein in the absence of aIF1 mRNA was set to 1. The relative signal intensities have been corrected for the number of methionine residues in each protein. The error bars represent standard deviations.
aIF1 stimulates binding of a/eIF2·GTP·Met-tRNAi to the ribosome
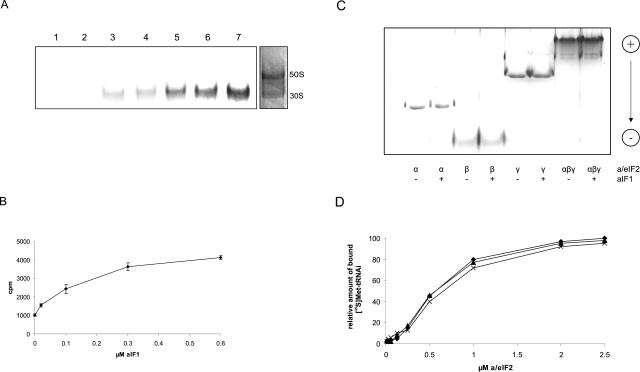
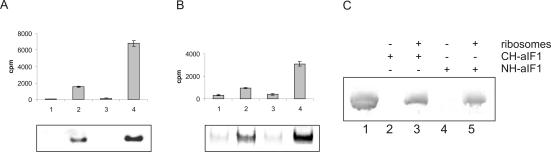
Recently, we have shown that Met-tRNAi binding to S. solfataricus ribosomes requires a/eIF2 (Pedulla et al. 2005). To examine whether aIF1, like its eukaryal counterpart, can stimulate the formation of a pre-initiation complex containing the small ribosomal subunit and a/eIF2·GTP·Met-tRNAi the following experiment was performed: Dissociated S. solfataricus ribosomes were incubated at 70°C with recombinant N-terminally His-tagged aIF1 (NH-aIF1) and with pre-formed a/eIF2·GTP· [35S]Met-tRNAi complex. Binding of the radioactively labeled [35S]-Met-tRNAi to the ribosome was monitored through separation of the ribosomal subunits on native gels. The position of the ribosomal subunits was determined by Coomassie staining of the gel (Fig. 4A ▶, right panel). Per se, · [35S]Met-tRNAi did not bind to ribosomes (Fig. 4A ▶, lane 1) and aIF1 had no stimulatory effect on Met-tRNAi binding to ribosomes in the absence of a/eIF2 (Fig. 4A ▶, lane 2). As shown previously (Pedulla et al. 2005), the incubation of the a/eIF2·GTP· [35S]Met-tRNAi complex with ribosomes resulted in binding of the tRNAi-complex to the 30S subunit (Fig. 4A ▶, lane 3). The addition of increasing amounts of aIF1 stimulated binding of a/eIF2·GTP· [35S]-Met-tRNAi to the ribosome up to fourfold over that observed in the absence of aIF1 (Fig. 4A ▶, lanes 4–7, 4B).
FIGURE 4.
NH-aIF1 stimulates Met-tRNAi binding to ribosomes but not to purified a/eIF2. (A) S. solfataricus ribosomes (0.1 μM) were incubated with a/eIF2·GTP· [35S]Met-tRNAi (lanes 3–7) and increasing amounts of aIF1 (0.02, 0.1, 0.3 and 0.6 μM, respectively) (lanes 4–7) at 70°C. (Lane 1) [35S]Met-tRNAi (1 μM) was incubated with ribosomes (0.1 μM). (Lane 2) aIF1 (1 μM) was incubated together with [35S]Met-tRNAi (1 μM) and ribosomes (0.1 μM). Samples were loaded on a native 4% polyacrylamide gel, and the radioactive bands were visualized using a PhosphorImager. One representative experiment is shown. The positions of the 30S and 50S ribosomal subunits in the gel were revealed in a parallel experiment by Coomassie blue staining. The positions of the 50S and 30S ribosomal subunits are indicated. (B) Graphical representation of two independent experiments of [35S]Met-tRNAi binding to ribosomes as a function of the aIF1 concentration. The error bars represent standard deviations. (C) The α-, β-, and γ-polypeptides or reconstituted a/eIF2 trimer (50 pmol each) were incubated at 65°C for 15 min in the presence (+) or absence (−) F1 (100 pmol). The samples were electrophoresed on a nondenaturing polyacylamide gel as described in Materials and Methods. The pI of aIF1 is ~7; therefore, free protein does not enter the gel per se. Migration of the proteins from the cathode toward the anode is indicated on the right. (D) Increasing amounts of S. solfataricus a/eIF2 were incubated with GTP (1 mM) and [35S]Met-tRNAi (1μM) at 70°C in the absence of aIF1 (diamonds) or in the presence of 2 (triangles) and 5 (crosses) μM aIF1, respectively. The amount of [35S]Met-tRNAi bound by 2.5 μM a/eIF2 in the absence of aIF1 was set to 100%.
In eukaryotes, eIF1 binds directly to the eIF2 β-subunit (Singh et al. 2004). The domain required for this interaction is absent in the archaeal homolog a/eIF2β(Pedulla et al. 2005). To test whether this function is transferred to another subunit of a/eIF2, we used acidic, native polyacrylamide gels to assess whether aIF1 binds to intact a/eIF2 or to any of the a/eIF2 subunits. As shown in Figure 4C ▶ these experiments did not reveal any interactions. This finding could be reconciled with the observation that the affinity of purified a/eIF2 for Met-tRNAi was not affected in the presence of aIF1 (Fig. 4D ▶).
aIF1 stimulates binding of a model RNA to the ribosome
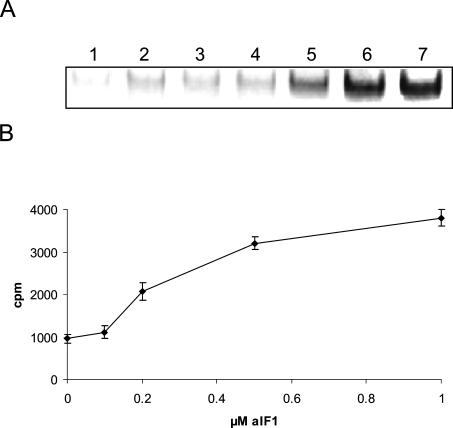
As a minimal mRNA template the 5′radioactively labeled RNAo [(U)3 GAGGUGACUCUCUCAUG(U)10] containing a SD-motif (bold) and an AUG start codon (underlined) was used to study whether aIF1 stimulates binding of a mRNA to the ribosome. Dissociated S. solfataricus ribosomes were incubated at 70°C with NH-aIF1, the pre-formed unlabeled a/eIF2·GTP·Met-tRNAi complex and labeled RNAo. Binding of the radioactively labeled RNAo to the ribosome was assessed by native gel electrophoresis. The 30S ribosomal subunits alone bound RNAo with low efficiency (Fig. 5 ▶, lane 1), and binding was only slightly stimulated by addition of a/eIF2·GTP·Met-tRNAi (Fig. 5 ▶, lane 2) or in the presence of aIF1 alone (Fig. 5 ▶, lane 3). However, the addition of increasing amounts of aIF1 to ribosomes in the presence of the ternary complex a/eIF2·GTP·Met-tRNAi stimulated binding of RNAo to 30S ribosomes significantly (Fig. 5A ▶, lane 4–7, 5B).
FIGURE 5.
NH-aIF1 stimulates binding of RNAo to S. solfataricus 30S subunits. (A) S. solfataricus ribosomes (0.1 μM) were incubated with a/eIF2·GTP·Met-tRNAi (lanes 2,4–7), radioactively labeled RNAo (0.01 μM) (lanes 1–7) and increasing amounts of aIF1 (0.1, 0.2, 0.5 and 1 μM, respectively) (lanes 4–7) at 70°C. The samples were loaded on a native 4% polyacrylamide gel and the radioactive bands were visualized using a PhosphorImager. (Lane 1) Only RNAo (0.01 μM) was incubated with ribosomes (0.1 μM). (Lanes 2,3) Ribosomes were incubated with RNAo in the presence of a/eIF2·GTP·Met-tRNAi or aIF1 (1μM), respectively. The position of the ribosomal subunits was determined by Coomassie staining of the gel as described in the legend to Figure 4 ▶. (B) A graphical representation of two independent experiments of RNAo binding to ribosomes as a function of the aIF1 concentration is shown. The error bars represent standard deviations.
C-terminally modified aIF1 is nonfunctional
The C-terminal end of eIF1 is important for its function (Singh et al. 2004). To obtain a hint whether the same holds true for aIF1, we compared the C-terminal His-tagged aIF1 (CH-aIF1) with the N-terminal His-tagged factor (NH-aIF1) regarding its function to stimulate Met-tRNAi and mRNA binding to ribosomes. As shown in Figure 6A ▶, lane 3, CH-aIF1 abolished binding of a/eIF2·GTP·Met-tRNAi to the ribosome, which contrasts with the stimulating effect of NH-aIF1 on binding of a/eIF2·GTP·Met-tRNAi to ribosomes (Fig. 4 ▶, 6A ▶, lane 4). The amount of bound [35S]Met-tRNAi was strongly reduced in the presence of CH-aIF1 (Fig. 6A ▶, lane 3) when compared to the experiment when only a/eIF2·GTP·Met-tRNAi was added to the ribosomes (Fig. 6A ▶, lane 2). Similar results were obtained when the effect of CH-aIF1 on binding of RNAo to ribosomes was tested. When compared to the reaction mixture where only a/eIF2·GTP·Met-tRNAi was added (Fig. 6B ▶, lane 2), the amount of RNAo bound to the ribosome was threefold reduced in the presence of the a/eIF2·GTP·Met-tRNAi complex and CH-aIF1 (Fig. 6B ▶, lane 3). In contrast NH-aIF1 stimulated binding of a/eIF2·GTP·Met-tRNAi (Fig. 6B ▶, lane 4) threefold when compared to the experiment conducted in the absence of aIF1 (Fig. 6B ▶, lane 2). It is worth noting that the observed effects of NH-aIF1 and CH-aIF1 are not attributable to a lack of binding of CH-aIF1 to ribosomes. As shown in Figure 6C ▶, both the N- and C-terminal His-tagged aIF1 bound to ribosomes. Taken together, these observations indicated that the C terminus of aIF1 is of functional importance.
FIGURE 6.
C-terminal His-tagged aIF1 inhibits binding of tRNAi-Met and RNAo to ribosomes. (A) S. solfataricus ribosomes (0.1 μM) were incubated with RNAo (0.01 μM), a/eIF2·GTP· [35S]Met-tRNAi (lanes 2–4) and 0.3 μM CH-aIF1 (lane 3) or 0.3 μM NH-aIF1 (lane 4) at 70°C. (Lane 1) [35S]Met-tRNAi (1 μM) was incubated with ribosomes (0.1 μM). Samples were loaded on a native 4% polyacrylamide gel and the radioactive bands were visualized using a PhosphorImager (bottom). A graphical representation of two independent experiments is shown (top). The error bars represent standard deviations. (B) S. solfataricus ribosomes (0.1 μM) were incubated with a/eIF2·GTP·Met-tRNAi (lanes 2–4), the radioactively labeled RNAo (0.01 μM) and 1 μM CH-aIF1 (lane 3) or 1 μM NH-aIF1 (lane 4) at 70°C. (Lane 1) Only RNAo (0.01 μM) was incubated with ribosomes (0.1 μM). The samples were loaded on native 4% polyacrylamide gels and radioactive bands were visualized using a PhosphorImager (bottom). A graphical representation of two independent experiments is shown (top). The error bars represent standard deviations. (C) Both NH-aIF1 and CH-aIF1 bind to S. solfataricus ribosomes. S. solfataricus ribosomes (400 pmol) were incubated in the presence or the absence of either CH-aIF1 or NH-aIF1 and loaded on 0%–17% sucrose density gradients. The pellet fractions were resuspended in protein sample buffer and resolved on a 12% SDS-polyacrylamide gel using standard procedures. The proteins were blotted on nitrocellulose and the Western blots were probed for the presence of aIF1 as described above. (Lane 1) 50 pmol of purified CH-aIF1 was loaded on the gel as a positive control and marker for the Western blot. (Lanes 2,4) 200 pmol CH-aIF1 and 200 pmol NH-aIF1, respectively, were loaded on the gradients in the absence of ribosomes. (Lanes 3,5) 200 pmol CH-aIF1 and 200 pmol NH-aIF1, respectively, were loaded on the gradients upon incubation with 400 pmol of ribosomes.
DISCUSSION
In eukaryotes, eIF1 has been shown to act as a general fidelity factor, affecting the accuracy of translation initiation and probably also of elongation. Furthermore, it has been demonstrated that the process of translation initiation is faster in the presence of eIF1 (Algire et al. 2002) and that eIF1 is essential for the precise location of the correct start codon (Pestova et al. 1998). These functions of eIF1 are expected to accelerate the forward kinetics of translation initiation. The stimulation of the translation rate of a S. solfataricus mRNA by addition of aIF1 mRNA (Fig. 3 ▶) together with its other functions discussed below lends support to the hypothesis that the function of aIF1 is analogous to that of eIF1.
Like its eukaryal homolog (Schreier et al. 1977; Trachsel et al. 1977), aIF1 was shown to bind to the small ribosomal subunit (Fig. 2 ▶), and to stimulate Met-tRNAi (Fig. 4 ▶) and mRNA (Fig. 5 ▶) binding to ribosomes. The factor did not stimulate binding of Met-tRNAi to a/eIF2 in the absence of ribosomes, which can be reconciled with a lack of direct interaction between aIF1 and a/eIF2 (Fig. 4 ▶). Hence, only ribosome-bound aIF1 appears to stimulate the interaction of the a/eIF2·GTP·Met-tRNAi ternary complex with the ribosome. The NMR structure of eIF1 suggested a RNA binding domain in the C-terminal part of the protein, but no interaction between eIF1 and a RNA oligonucleotide could be demonstrated (Fletcher et al. 1999). Bandshift experiments with NH-aIF1 and labeled RNAo revealed that the factor binds to the RNA with a Kd of ~0.2 μM, whereas CH-aIF1 was unable to bind to RNAo (D. Hasenöhrl, unpubl.). The putative RNA binding domain of eIF1 includes the last amino acid of the protein. Given the lack of binding of CH-aIF1 to RNAo, the C-terminal domain of aIF1 seems thus to be likewise of functional importance. As both CH-aIF1 and NH-aIF1 bind to S. solfataricus ribosomes (Fig. 6C ▶), it is further possible that CH-aIF1 interferes with binding of a/eIF2·GTP·Met-tRNAi.
As shown in Figure 5 ▶, aIF1 is important for efficient mRNA binding to the ribosome. The ribosome by itself displayed a low intrinsic binding affinity for the SD-sequence containing RNAo. An efficient binding of RNAo was only achieved in the presence of both aIF1 and a/eIF2·GTP·Met-tRNAi. As aIF1 requires the presence of a/eIF2·GTP·Met-tRNAi to stimulate mRNA binding, its effect on mRNA binding is most likely indirect. Possibly, the presence of aIF1 enhances the binding affinity of the ternary a/eIF2·GTP·Met-tRNAi complex for the ribosome, which in turn stabilizes the mRNA on the ribosome by means of the additional interaction between the start codon and Met-tRNAi. Taken together, these results revealed that aIF1 is an important factor in archaeal translation initiation with diverse functions in the initiation process.
MATERIALS AND METHODS
Cloning of aIF1 and purification of the protein
The aIF1 gene was amplified from S. solfataricus genomic DNA by PCR, sequenced (VBC-Genomics, Austria) and compared to the published sequenced (She et al. 2001). The gene encoding aIF1 was cloned by means of PCR amplification from Sulfolobus solfataricus genomic DNA. The forward primers CH-aIF1_FP (5′-GTGTCAAACCATGGCAGAAAATCTGTGTGGTGGTCTTCC-3′) and NH-aIF1_FP (5′-GTGTCAAAGGATCCGGCAGAAAA TCTGTGTGGTGGTCTTCC-3′) and the reverse primers CH-aIF1_RP (5′-CGCGCCGCGCTCGAGCTACTCAATAACTAGAA TATTGGATTCTGC-3′) and NH-aIF1_RP (5′-GCGCCGCGGAAT TCCTACTCAATAACTAGAATATTGGATTCTGC-3′), respectively, were used for PCR. They contained restriction sites for insertion into the expression vectors pRSETB (Invitrogen) for NH-aIF1 (containing a N-terminal His-tag sequence) and pET28b (Novagen) for CH-aIF1 (containing a C-terminal His-tag sequence). The recombinant plasmids were sequenced and transformed into E. coli BL21(DE3) (Stratagene). The expression of the two aIF1 derivatives was induced for 3 h with 1 mM IPTG at an OD600 of 0.5. The cells were then lysed and the cell extracts were heated for 10 min at 70°C and centrifuged at 10,000 g for another 10 min to precipitate mesophilic E. coli proteins. The recombinant proteins, which contained either a C- or N-terminal tag of six histidines, were purified to homogeneity by affinity chromatography on Ni-NTA agarose following standard protocols (Qiagen). The purified proteins were dialyzed against storage buffer (10 mM MOPS, 200 mM KCl, 10 mM β-mercaptoethanol and 10% glycerol) and stored at −80°C in aliquots. Antibodies against NH-aIF1 were raised by Eurogentec, Belgium.
Determination of the 5′-end of the aIF1 mRNA
The 5′-end of the aIF1 mRNA was determined using primer extension reactions (Hartz et al. 1988). S. solfataricus total RNA (20 μg) was annealed to the [32P]5′-end labeled oligonucleotide (5′-GTTCTTCCTTAGAAAGTTGCTC-3′) by heating to 85°C for 5 min and slowly cooling to 37°C. For the primer extension reaction, 10 μL of the annealing reaction were incubated in the presence of 5 μL of 5 × RT buffer (250 mM Tris/HCl at pH 8.3, 300 mM NaCl, 30 mM Mg[OAc]2, 50 mM DTT), 2 μL of 2.5 mM dNTPs, 1 μL of RNAse Inhibitor (40 U/μL; Fermentas), and 1μL of M-MuLV RT (200 U/μL; Promega) in a final volume of 25 μL for 1 h at 42°C. The reaction was stopped by heat inactivation of the M-MuLV RT at 70°C for 10 min. After phenol extraction, the cDNA was precipitated and analyzed on a 6% polyacrylamide-8M urea gel after heating to 95°C for 5 min in parallel with sequencing reactions.
Preparation of ribosomes, Met-tRNAi, a/eIF2 and mRNAo
S. solfataricus ribosomes were obtained from frozen cells as described in Londei et al. (1986). Initiator tRNA was transcribed and charged with cold methionine or [35S]-methionine (Amersham Pharmacia Biotech) and the three subunits of a/eIF2 were prepared as recently described (Pedulla et al. 2005). The model mRNA-Oligo [(U)3GAGGUGACUCUCUCAUG(U)10], containing a SD sequence (bold) and a start codon (underlined) was ordered from Dharmacon. The RNA oligonucleotide was 5′-end labeled with [γ-32P]-ATP (Amersham Pharmacia Biotech) and purified on 6%-polyacrylamide–8 M urea gels following standard procedures.
Interaction of aIF1 with the 30S subunit
A S. solfataricus in vitro translation extract (25 mg/mL) was programmed with 2 μg in vitro transcribed 104 mRNA (Condo et al. 1999) per 10 μL cell lysate. The samples were incubated for 15 min at 70°C and afterward treated with formaldehyde (6% final concentration) at 4°C for 30 min. S. solfataricus cell lysate (10 μL, 25 mg/mL) or 10 μL of S. solfataricus cell lysate programmed with 104 mRNA and treated with formaldehyde were loaded on a 10%–30% sucrose density gradient prepared in 30 mM KCl, 10 mM MgCl2, 20 mM Tris/HCl (pH 7). The gradient was centrifuged at 36,000 rpm for 4 h in a Beckman SW41 rotor and analyzed by measuring the A260 using an ISCO UA-6 spectrophotometer. The gradient was fractionated and the fractions were then probed for the presence of aIF1 with anti-aIF1 polyclonal mouse antibodies raised against recombinant S. solfataricus aIF1. The fractions were separated on a 12% SDS-polyacrylamide gel, and transferred to nitrocellulose membranes (Schleicher & Schuell) by electroblotting. The blots were blocked with 5% dry milk in TBS (140 mM NaCl, 2.7 mM KCl, and 25 mM Tris/HCl at pH 7.5), and then probed with anti-aIF1 antibodies. The antibody–antigen complex was visualized with goat anti-mouse immunoglobulin alkaline-phosphatase-conjugated antibody (Sigma Immuno Chemicals) using NBT (Nitroblue-tetrazolium-chloride, BIOMOL) and BCIP (5-Bromo-4-chloro-3-indolyl phosphate toluidine salt, BIOMOL) in alkaline phosphatase-buffer (10 mM NaCl, 5 mM MgCl2, 100 mM Tris/HCl at pH 9.5) as a chromogenic substrate.
Binding of C- and N-terminal His-tagged aIF1 to S. solfataricus ribosomes was determined as follows: S. solfataricus ribosomes (400 pmol) were incubated with 200 pmol of either CH- or NH-aIF1 in 20 mM KCl, 10 mM MgCl2, 20 mM Tris/HCl (pH 7) for 10 min at 70°C. As a control 200 pmol of either CH- or NH-aIF1 were incubated in buffer in the absence of ribosomes. Samples were chilled on ice and loaded on 0%–17% sucrose density gradients prepared with incubation buffer. The gradients were centrifuged at 45,000 rpm for 5 h in a Beckman Ti50 rotor. The pellets were resuspended in protein sample buffer and probed for the presence of aIF1 with anti-aIF1 antibodies as described above.
mRNA preparation and in vitro translation assay
104 mRNA was prepared as described before (Condo et al. 1999). The sequence of the aIF1 gene together with its 4-nt-long 5′-UTR was amplified from chromosomal S. solfataricus DNA and placed under transcriptional control of the T7 Φ 10 promoter by means of PCR using the forward primer aIF1_FP (5′-CTGCAGAACAC TACGTGTAATACGACTCACTATAGGGAGAATGGCAGAAAAT CTG-3′) and the reverse primer aIF1_RP (5′-ATAAGAATGC GGCCGCCTACTCAATAACTAGAATATTGG-3′). The obtained DNA was used as template for in vitro transcription with T7 RNA Polymerase (Fermentas). The run-off transcripts were purified on 6% polyacrylamide-8 M urea gels following standard procedures. The mRNA concentration was determined by measuring the A260.
The in vitro translation reactions were performed as described before (Condo et al. 1999). The samples contained in a final volume of 25 μL: 10 mM KCl, 20 mM Tris/HCl (pH 7), 20 mM MgCl2, 7 mM β-mercaptoethanol, 3 mM ATP, 1 mM GTP, 5 μg of S. solfataricus tRNA, 1 μL (10 μCi) of [35S]-methionine (Amersham Bioscience Biotech), 5 μL of S. solfataricus S30 extract, 0.4 μM of 104 mRNA and 0.4–1.6 μM of aIF1 mRNA. The samples were incubated for 40 min at 70°C, then resolved on a 15% SDS-polyacrylamide gel, and the radioactive bands were visualized using a PhosphorImager.
Interaction of [35S]Met-tRNAi with the ribosome
The stimulatory effect of aIF1 on binding of [35S]Met-tRNAi to the ribosome was determined as follows: One micromole (1 μM) each of α, β, and γ a/eIF2 subunits were mixed and incubated at 70°C for 10 min in 20 mM KCl, 20 mM Tris/HCl (pH 7), 10 mM MgCl2 (incubation buffer) with 1 mM GTP. Then 1 μM of [35S]Met-tRNAi was added and incubation was continued for 5 min to allow formation of the ternary a/eIF2·GTP· [35S]Met-tRNAi complex. Next, 0.1 μM of ribosomes and 0.02–0.6 μM of aIF1 were added and incubation was continued for 15 min. For comparison of the effects of CH-aIF1, 0.3 μM of the factor was added to the reaction, which was carried out as described above. The samples were immediately electrophoresed on running native 4% polyacrylamide gels prepared in 20 mM potassium acetate, 40 mM Tris/HCl (pH 6) and 2.5 mM MgCl2. Electrophoresis was continued at 4°C and 30 mA for 4 h. Then, the gels were dried and the radioactive bands were visualized using a PhosphorImager. The amount of radioactivity bound to the ribosome was determined using the ImageQuant software. A part of the gel was stained with Coomassie blue to determine the position of 30S and 50S ribosomal subunits.
Binding of Met-tRNAi to a/eIF2 in the presence of aIF1
To test whether purified aIF1 stimulates binding of [35S]Met-tRNAi to a/eIF2, 0.015–2.5 μM of reconstituted a/eIF2 were incubated at 70°C in incubation buffer with 1 mM GTP and 1 μM of [35S]Met-tRNAi in the absence of aIF1 or in the presence of 2 and 5 μM aIF1, respectively. The incubation was carried out for 15 min. Samples were withdrawn and filtered through 0.22 μm nitrocellulose disks, which were washed with incubation buffer, dried, and then subjected to scintillation counting.
Interaction of aIF1 with a/eIF2 and the a/eIF2 subunits
To test whether aIF1 interacts with a/eIF2 or with the individual a/eIF2 subunits, 50 pmol of the α-, β-, and γ-subunits were mixed or incubated alone in incubation buffer in the presence or absence of 100 pmol aIF1. The proteins/complexes were visualized by nondenaturing electrophoresis on 12% polyacrylamide gels prepared in acetate buffer (120 mM potassium-acetate, 72 mM acetic acid at pH 4.3). The gels included a stacking overlay of 4% polyacrylamide in acetate buffer (120 mM potassium-acetate, 12 mM acetic acid at pH 6.8). The running buffer was 133 mM acetic acid, 350 mM β-alanine (pH 4.4). The gels were stained with Coomassie brilliant blue.
Interaction of RNAo with the ribosome
The influence of aIF1 on RNAo binding to the ribosome was determined by first forming the ternary a/eIF2·GTP·Met-tRNAi complex as described above. Then, 0.1 μM of ribosomes and 0.1–1.0 μM of NH-aIF1 and 0.01 μM of the [32P]-RNAo-Oligo were added and incubation was continued for 15 min. For comparison of the effects of CH-aIF1, one micromole (1 μM) of the factor was added to reactions (see Fig. 6 ▶), which were carried out as described above. Gel preparation and running conditions were as described above.
Acknowledgments
P.L. was supported by funds from the MIUR PRIN 2000 and 2002 projects. The work in U.B.’s laboratory was supported by grant P15334 from the Austrian Science Fund.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2289306.
REFERENCES
- Algire, M.A., Maag, D., Savio, P., Acker, M.G., Tarun Jr., S.Z., Sachs, A.B., Asano, K., Nielsen, K.H., Olsen, D.S., Phan, L., et al. 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, K., Phan, L., Anderson, J., and Hinnebusch, A.G. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 18573–18585. [DOI] [PubMed] [Google Scholar]
- Benelli, D., Maone, E., and Londei, P. 2003. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol. Microbiol. 50: 635–643. [DOI] [PubMed] [Google Scholar]
- Brandi, L., Marzi, S., Fabbretti, A., Fleischer, C., Hill, W.E., Gualerzi, C.O., and Stephen Lodmell, J. 2004. The translation initiation functions of IF2: Targets for thiostrepton inhibition. J. Mol. Biol. 335: 881–894. [DOI] [PubMed] [Google Scholar]
- Condo, I., Ciammaruconi, A., Benelli, D., Ruggero, D., and Londei, P. 1999. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 34: 377–384. [DOI] [PubMed] [Google Scholar]
- Dahlquist, K.D. and Puglisi, J.D. 2000. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol. 299: 1–15. [DOI] [PubMed] [Google Scholar]
- Fletcher, C.M., Pestova, T.V., Hellen, C.U., and Wagner, G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18: 2631–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, S., Gualerzi, C.O., Londei, P., and Bläsi, U. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J. 19: 4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, C.O. and Pon, C.L. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29: 5881–5889. [DOI] [PubMed] [Google Scholar]
- Hartz, D., McPheeters, D.S., Traut, R., and Gold, L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164: 419–425. [DOI] [PubMed] [Google Scholar]
- Kapp, L.D. and Lorsch, J.R. 2004. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73: 657–704. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- La Teana, A., Pon, C.L., and Gualerzi, C.O. 1996. Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol. 256: 667–675. [DOI] [PubMed] [Google Scholar]
- Londei, P. 2005. Evolution of translational initiation: New insights from the archaea. FEMS Microbiol. Rev. 29: 185–200. [DOI] [PubMed] [Google Scholar]
- Londei, P., Altamura, S., Cammarano, P., and Petrucci, L. 1986. Differential features of ribosomes and of poly(U)-programmed cell-free systems derived from sulphur-dependent archaebacterial species. Eur. J. Biochem. 157: 455–462. [DOI] [PubMed] [Google Scholar]
- Maag, D. and Lorsch, J.R. 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330: 917–924. [DOI] [PubMed] [Google Scholar]
- Maag, D., Fekete, C.A., Gryczynski, Z., and Lorsch, J.R. 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17: 265–275. [DOI] [PubMed] [Google Scholar]
- Marintchev, A., Kolupaeva, V.G., Pestova, T.V., and Wagner, G. 2003. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: A new interaction between old partners. Proc. Natl. Acad. Sci. 100: 1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla, N., Palermo, R., Hasenöhrl, D., Bläsi, U., Cammarano, P., and Londei, P. 2005. The archaeal eIF2 homologue: Functional properties of an ancient translation initiation factor. Nucleic Acids Res. 33: 1804–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Borukhov, S.I., and Hellen, C.U. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Petrelli, D., LaTeana, A., Garofalo, C., Spurio, R., Pon, C.L., and Gualerzi, C.O. 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism EMBO J. 20: 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, L., Schoenfeld, L.W., Valasek, L., Nielsen, K.H., and Hinnebusch, A.G. 2001. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 20: 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioletti, M., Schlunzen, F., Harms, J., Zarivach, R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T., Jacobi, C., et al. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20: 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier, M.H., Erni, B., and Staehelin, T. 1977. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J. Mol. Biol. 116: 727–753. [DOI] [PubMed] [Google Scholar]
- She, Q., Singh, R.K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M.J., Chan-Weiher, C.C., Clausen, I.G., Curtis, B.A., De Moors, A., et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. 98: 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C.R., He, H., Ii, M., Yamamoto, Y., and Asano, K. 2004. Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J. Biol. Chem. 279: 31910–31920. [DOI] [PubMed] [Google Scholar]
- Tolstrup, N., Sensen, C.W., Garrett, R.A., and Clausen, I.G. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4: 175–179. [DOI] [PubMed] [Google Scholar]
- Trachsel, H., Erni, B., Schreier, M.H., and Staehelin, T. 1977. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol. 116: 755–767. [DOI] [PubMed] [Google Scholar]
- Yatime, L., Schmitt, E., Blanquet, S., and Mechulam, Y. 2004. Functional molecular mapping of archaeal translation initiation factor 2.J. Biol. Chem. 279: 15984–15993. [DOI] [PubMed] [Google Scholar]
- Yoon, H.J. and Donahue, T.F. 1992. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol. Cell. Biol. 12: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]