Abstract
Recent efforts have been made to unravel the independent roles of monovalent cations in RNA folding, primarily using the Tetrahymena ribozyme as a model. Here we report how monovalent cations impact the folding of the Candida ribozyme. Interestingly, this ribozyme requires an order of magnitude less monovalent cations (Na+ and Tris+) to commit to a new folding starting state in which the J3/4:P6 base triple is partially formed and mispairing in the L2.1 and L6 terminal loops is resolved. When Mg2+-induced ribozyme folding proceeded on the same energy landscape, the altered starting state led to a rapid assembly of the correct ribozyme core and a fivefold to 10-fold increase in the ribozyme activity. Moreover, when the ribozyme folding was started from a misfolding-prone state, high millimolar concentrations of monovalent cations moderately elevated the ribozyme activity by efficiently resolving the misfolding of a peripheral element, P5abc.
Keywords: Candida ribozyme, monovalent cations, RNA folding
INTRODUCTION
The independent roles of monovalent cations in folding of the structurally complex RNA molecules have been appreciated recently. High concentrations of monovalent cations (>1 M) support group I ribozyme folding into compact structures (Heilman-Miller et al. 2001; Takamoto et al. 2002; Perez-Salas et al. 2004). Monovalent cations promote RNA tertiary folding of the Tetrahymena ribozyme using a similar folding principle to that of magnesium (Das et al. 2003; Takamoto et al. 2004), while the folding is faster than that mediated by magnesium (Shcherbakova et al. 2004). It is noteworthy that lower concentrations of Na+ (>250 mM) greatly facilitate the correct and fast folding of the Tetrahymena ribozyme through altering the folding starting state in which P3 and P13 helices are formed (Russell et al. 2002).
Although folding of group I introns shares some common mechanisms in responding to magnesium (Woodson 2005), folding of the Candida ribozyme, a group I ribozyme derived from the 26S rRNA gene of the opportunistic fungal pathogen Candida albicans, behaves quite differently from that of the Tetrahymena ribozyme in many aspects (Sclavi et al. 1998; Zhuang et al. 2000; Xiao et al. 2003; Zhang et al. 2005). In this report, we studied independent functions of monovalent cations in folding of a trans-acting Candida ribozyme. Using the ribozyme activity and T1 ribonuclease footprinting pattern as two readouts, we found that low millimolar monovalent cations were sufficient to promote the correct folding of the Candida ribozyme through changing the folding starting state. Even if the ribozyme folds from the misfolding-prone starting state, monovalent cations are able to relieve the ribozyme misfolding, although via a different mechanism.
RESULT AND DISCUSSION
Candida ribozyme requires more than one order of magnitude less Na+ than does Tetrahymena ribozyme to alter the folding starting state
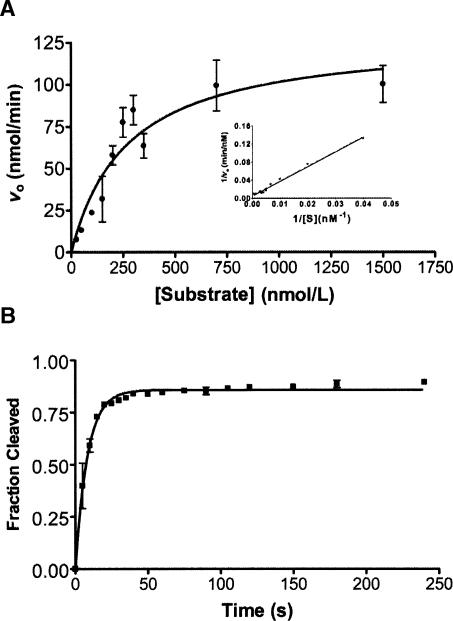
The purified ribozyme RNA molecules were renatured in the presence of varying concentrations of Na+ or Tris+, followed by magnesium-induced folding at exactly the same ionic condition, that is, on the same energetic landscape (Fig. 1A ▶; Russell et al. 2002). The trans-cleavage activity (kobs) of each RNA population folded from a distinct starting state was monitored to reflect the native folding of the Candida ribozyme (Fig. 1B ▶) that followed. However, the observed rate constant for the ribozyme activity (kobs) reflects the rate of ribozyme folding only if the folding process is much slower than the rate constant for the substrate cleavage. We thus determined the rate constant for substrate cleavage by the Candida ribozyme according to the Michaelis-Menten equation, which yielded a k2 of 8.6 min−1 (Fig. 2A ▶). The rate constant for substrate cleavage was also measured for the pre-folded ribozyme, which resulted in a cleavage constant of 7.2 min−1 (Fig. 2B ▶). Apparently, substrate cleavage was a much faster process than the ribozyme folding (2 min−1) (Xiao et al. 2003). We thus validated that the observed ribozyme activity (kobs) in the present study reflects the rate of ribozyme folding.
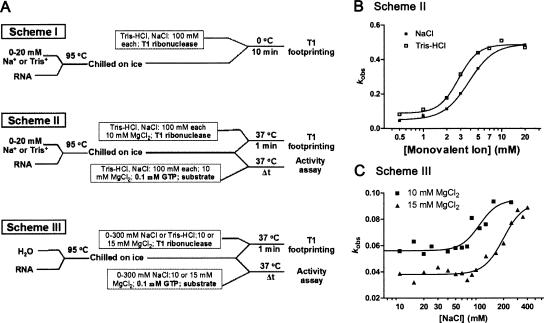
FIGURE 1.
(A) Experimental schemes. (Scheme I) Analyzing the structure of different folding starting states renatured at 0–20 mM monovalent cations. The Candida ribozyme Ca.L-11 was denatured at 95°C for 1.5 min and renatured by a rapid annealing (on ice). Tris-HCl (pH 7.5) and NaCl were added on ice to reach the final concentration of 100 mM for each cation. Ribonuclease T1 at a final concentration of 0.4 U/μL was then added to initiate the cleavage reaction on ice for 10 min. (Scheme II) Analyzing how the monovalent cation-promoted folding starting states impact the Mg2+-induced ribozyme folding by exploring the altered ribozyme structure (T1 footprinting) and ribozyme activity (kobs). The ribozyme was denatured and renatured as in Scheme I. However, in addition to the same concentrations of Tris-HCl and NaCl, a final concentration of 10 mM MgCl2 was added to initiate tertiary folding. In order to probe the rapidly compacted ribozyme structure, a final concentration of 0.1 U/μL T1 ribonuclease was added, and the reactions proceeded at 37°C for 1 min. In order to probe the ribozyme activity, GTP and the 5′-end-labeled substrate RNA at a final concentration of 0.1 mM and 20 nM, respectively, were added, and the reactions proceeded at 37°C from 0 to 60 min to obtain the kobs for each ribozyme sample (10 nM) renatured at a distinct concentration of Tris-HCl or NaCl. (Scheme III) Analyzing if monovalent cations present during the Mg2+-induced RNA folding could relieve ribozyme misfolding. The ribozyme RNA was denatured and renatured in water following the same procedure as in Scheme I. Then 0–400 mM NaCl was added together with 10 or 15 mM MgCl2 to the ribozyme RNA on ice. T1 footprinting analysis and activity analysis were then performed as in Scheme II. (B) The observed catalytic activity (kobs) of the Candida ribozyme following Scheme II. The kobs of each reaction was obtained and plotted against the concentrations of Na+ and Tris+. The half-maximal activity for Na+ and Tris+ was 3.81 and 2.82 mM, respectively. The Hill coefficients were 2.69 ± 0.262 and 3.50 ± 0.613 for Na+ and Tris+, respectively. (C) The observed catalytic activity (kobs) of the Candida ribozyme following Scheme III. The half-maximal activity was achieved at 99.25 mM and 199.3 mM Na+ in the presence of 10 mM and 15 mM Mg2+, respectively. The corresponding Hill coefficients were 3.73 ± 1.705 and 3.516 ± 0.9829.
FIGURE 2.
Measuring the rate constant for substrate cleavage by the Candida ribozyme Ca.L-11. (A) Kinetics for the ribozyme catalysis. The initial velocity (v0 ) for each substrate concentration ranging from 25 nM to 1500 nM was determined from a single exponential plot of the fraction of reaction as a function of time: Y = Ymax × [1 − exp(−kobs × X)], in which v0 = kobs[S]. v0 was then plotted as a function of substrate concentration. The graph shown is a nonlinear least-squares fit to the Michaelis-Menten equation; the inserted graph is also a linear least-squares fit to a plot of 1/v0 against 1/[substrate]. The resulting kinetic parameters are KM = 268.3 ± 86.76 nM, Vmax = 128.8 ± 17.04 nM/min, and kcat = 8.6 ± 1.1 min−1, kcat/KM = 3.4316 ± 0.9895 × 107 mol−1 min−1. (B) Rate constant for substrate cleavage (kc) measured by the prefolded ribozyme. After the ribozyme renatured in Tris-HCl and prefolded in the presence of 10 mM Mg2+ and substrate for 10 min, GTP was added to initiate the substrate cleavage for 0–240 sec. The rate constant kc was 0.1195 ± 0.005986 sec−1.
As shown in Figure 1B ▶ k,obs at each monovalent ion concentration was obtained and plotted against [Na+] or [Tris+]. Renaturation of the ribozyme RNA in both monovalent cations significantly increased the ribozyme activity in a dose-dependent manner; the half-maximal increase was achieved at 2.82 mM Tris+ and 3.81 mM Na+. The maximal increase was fivefold to 10-fold of the control sample renatured in H2O, which was achieved at ≥7 mM Tris+ and 20mM Na+. These results lead us to conclude that monovalent cations increase the native folding of the Candida ribozyme through altering the folding starting state, which is similar to that of the Tetrahymena ribozyme. However, the Tetrahymena ribozyme requires >400 mM Na+ to render a folding initial state that leads to the maximal increase (fourfold) of the correctly folded ribozyme. Such a dramatic difference in the Na+ requirement to alter the starting states of these two ribozymes suggests that the Na+-changed structure identities in these starting states are different. Nevertheless, monovalent cations are capable of drastically facilitating native folding of both the Tetrahymena and the Candida ribozymes through altering the folding starting states, suggesting a general mechanism of monovalent cations in supporting RNA folding.
Monovalent cation-promoted folding starting state: Mispairing in L2.1 and L6 is resolved and a J3/4:P6 base triple is partially formed
RNase T1 footprinting was then used to probe the structure of the Candida ribozyme formed after renaturation in the presence and absence of monovalent cations. Because ionic strength strongly affected the T1 ribonuclease activity at low concentrations of cations (data not shown), compensating concentrations of Na+ and Tris+ were added during T1 cleavage. To avoid changing the ribozyme folds by the later added monovalent cations, T1 cleavage was run on ice. As shown in Figure 3B ▶, Na+ and Tris+ in renaturation dose-dependently protect G121/122 from T1 cleavage, indicating an improved J3/4:P6 base triple structure in the folding starting state of the ribozyme. However, protection at two Gs of the P3 helix was very marginal, consistent with our recent report that formation of P3 requires magnesium (Zhang et al. 2005). Surprisingly, G220/222 in L6 and G86/G89 and G95/G97/98/99 in L2.1 were not well accessible to T1 ribonuclease cleavage if the ribozyme was renatured in water, suggesting that these two loops are mispaired with some other structures of the ribozyme in the folding starting state. The presence of either Na+ or Tris+ greatly enhanced the accessibility of those loop Gs to T1 cleavage, indicating that mispairing in L2.1 and in L6 was resolved by the presence of monovalent ions (Fig. 3B ▶). Therefore, renaturation in monovalent ions converts the Candida ribozyme to the folding starting state in which the J3/4:P6 base triple is partially formed and mispairing in the terminal loops of P2.1 and P6 is resolved.
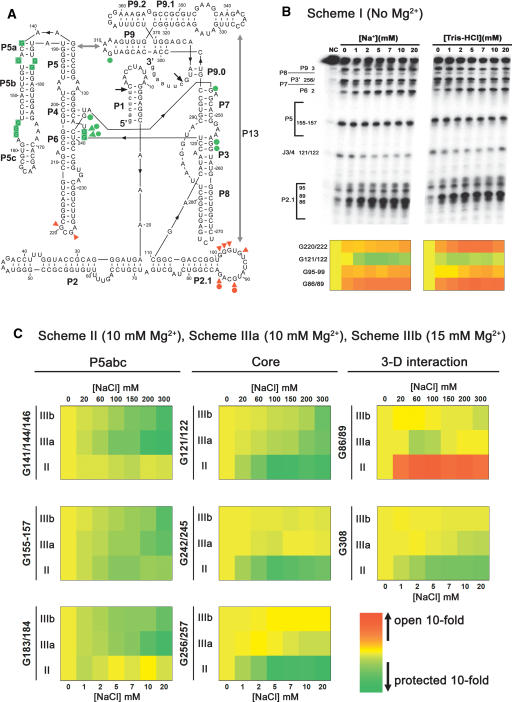
FIGURE 3.
(A) Illustration of the secondary structure of the Candida ribozyme. T1 footprinting results are incorporated into the structure. The Gs whose T1 sensitivity was changed in Scheme I, Scheme II, and Scheme III are indicated as filled triangles, filled circles, and shaded, respectively. The protected and opened Gs are indicated in green and red, respectively. (B) T1 footprinting analysis of the monovalent cation-altered folding starting state. Experiments were done following Scheme I illustrated in Figure 1A ▶, and reactions were fractionated on 8% PAGE/7 M urea gels. Two representative gels (top) for renaturation in NaCl and Tris-HCl (pH 7.5) are shown, with the cation concentrations present in renaturation indicated. False-color maps of G sites showing the altered T1 sensitivity in response to the concentrations of Na+ and Tris+ are shown (bottom). The band intensity at the indicated G sites on the denatured gels was analyzed as previously described (Xiao et al. 2003). For each site, the G-band intensity of the sample lacking monovalent cations (0 mM) was set as the reference, and the G-band intensity for the sample renatured in various concentrations of monovalent cations was normalized to the reference to obtain the ratio. A base 10 logarithm (log10) of each ratio was then calculated and plotted with Kyplot 2.0 software to illustrate the change of T1 cleavage pattern; the decrease (protected) and increase (opened) of the T1 cleavage are plotted in green and red, respectively. (C) False-color maps showing the results of T1 footprinting of the ribozyme structure folded in the presence of magnesium. Only the Gs displaying an altered T1 sensitivity in response to Na+ are shown. Experiments were done following Schemes II and III as illustrated in Figure 1A ▶, and reactions were fractionated on 8% PAGE/7 M urea gels. The band intensity was analyzed and plotted as shown in B. In each map, row II indicates the results from Scheme II, and rows IIIa and IIIb indicate data from Scheme III in the presence of 10 mM and 15 mM magnesium, respectively. The Na+ concentrations labeled at the bottom and on the top of each map indicate those present in Scheme II and Scheme III, respectively.
The improved local structures in the folding starting state ensure rapid assembly of the ribozyme core helices
All three local structures improved by low concentrations of monovalent cations are proposed to be important for the ribozyme tertiary folding. The J3/4:P6 base triple is conserved among all group I introns and is important for assembly of P4–P6 and P3–P9 domains (Michel and Westhof 1990; Adams et al. 2004; Guo et al. 2004; Golden et al. 2005). Hydroxyl radical footprinting analysis suggests that the L6 GNRA tetraloop interacts with a receptor in the P8 stem, providing a driving force for assembly of the P4–P6 and P3–P7 domains (Xiao et al. 2005). L2.1 is known to pair with L9.1 to establish the P13 helix, a structure conserved among all group I introns of the IE subgroup, and plays an important role in stabilizing the core structure of the Candida ribozyme (Li and Zhang 2005; Xiao et al. 2005). It is conceivable that partial formation of the J3/4:P6 base triple and relieving of mispairing in L2.1 and L6 encode a folding starting state committed to the correct folding of the Candida ribozyme upon the addition of magnesium.
T1 ribonuclease footprinting was then conducted to discover whether the Na+-improved local structures benefited the tertiary folding of the ribozyme when magnesium was added. T1 ribonuclease footprinting could not distinguish the correctly formed tertiary base-pairing from mispairing, because either case resulted in reduced T1 cleavage signals. Fortunately, correct formation of the P13 structure accompanies the opening of G86 and G89 in L2.1 (Zhang et al. 2005). Consistent with the prediction that mispairing in L2.1 prevents the formation of the P13 helix during Mg2+-induced folding, G86 and G89 in L2.1 were not sensitive to T1 cleavage in the H2O-renatured ribozyme (Fig. 3C ▶). A deficiency in formation of the J3/4:P6 base triple was also observed for the H2O-renatured ribozyme, as reflected by the moderate T1 cleavage at G121/122 (Fig. 3C ▶). Strikingly, a strong cleavage at G308 was evident, indicating that the L9/P5 tertiary interaction could not form for the H2O-renatured ribozyme (Fig. 3C ▶). Lacking of such an interaction indicated that the ribozyme core was not assembled. The strong cleavages at G256/257 and G245/247 in the P3–P7 core helices, as well as G242 in J6/7:P4 base triples, further proved that the H2O-renatured ribozyme was deficient in folding of the native ribozyme core (Fig. 3C ▶), consistent with the very low ribozyme activity shown in Figure 1B ▶.
When the ribozyme was renatured in Na+ before being subjected to Mg2+-induced folding, low millimolar concentrations of Na+ dose-dependently protected the accessible Gs in L2.1, L9, and P3–P7 core helices and J3/4:P6, J6/7:P4 base triples from T1 cleavage, indicating the formation of P13 and L9/P5 tertiary interactions as well as a correctly assembled ribozyme core (Fig. 3C ▶). The maximal protection was achieved at 5 mM or higher concentrations of Na+, consistent with the dose-dependent activity increase shown in Figure 1B ▶. Similar results were obtained using Tris+ (data not shown). In addition, the presence of Na+ in renaturation also resulted in the protection of G155/156/157 in P5abc from T1 cleavage in a dose-dependent manner, suggesting that this peripheral element tends to misfold in the H2O-renatured ribozyme.
Taken together, the Na+-altered starting state characterized by opening of L2.1 and L6 terminal loops and a partial formation of the J3/4:P6 triple is essential for the rapid assembly of the core helices of the Candida ribozyme. These observations suggest that formation of some long-range tertiary interactions is required for the ribozyme core assembly.
Na+ present in magnesium-induced RNA folding resolves misfolding of the P5abc structure
As described above, renaturation of the Candida ribozyme in H2O resulted in ribozyme misfolding upon the addition of magnesium. We further addressed whether monovalent cations could rescue the ribozyme from the misfolding fate.
Varying concentrations of Na+ were added in the Mg2+- induced folding of the H2O-renatured ribozyme, and kobs was measured for each monovalent cation. We noted that high concentrations of Na+ did not alter the rate constant for substrate cleavage (data not shown); thus the difference in kobs should reflect the effect of monovalent cations on ribozyme folding. As shown in Figure 1C ▶, in reactions containing 10 mM or 15 mM Mg2+, high millimolar concentrations of Na+ dose-dependently increased the ribozyme activity, with the half-maximal increase being achieved at 99 mM and 199 mM Na+ for the presence of 10 mM and 15 mM Mg2+, respectively, indicating that Na+ antagonizes the negative effect of Mg2+ on ribozyme folding. In both cases, the maximal increment of the ribozyme activity was about twofold, smaller than the increase from alteration of the folding starting state.
T1 footprinting was then performed to elucidate the mechanism by which monovalent cations rescue the Candida ribozyme from the misfolding fate in the presence of magnesium. Figure 3C ▶ shows that during Mg2+-induced folding of the H2O-renatured ribozyme, Na+ dose-dependently protected three regions of the P5abc peripheral element including G183/184, G155/156157, and G141/143/146 from T1 cleavage, suggesting that misfolding of the P5abc peripheral element was relieved. However, the deficiency in formation of the L9/P5 interaction (G308) and P13 long-range base-pairing (G86/89) was not recovered. In the ribozyme core, folding of the J3/4:P6 base triple (G121/122) but not the P3–P7 core helices or the J6/7:P4 base triple was slightly improved. The moderately elevated ribozyme activity caused by Na+ presence during ribozyme folding was thus attributed to preventing the misfolding of the P5abc peripheral element. We recently found that P5abc facilitates the native folding of the Candida ribozyme through a couple of tertiary interactions (X. Yuan, M. Xiao, X. Dai, and Y. Zhang, unpubl.).
The role of monovalent cations in resolving RNA misfolding
Both monovalent and divalent metal ions have a large general effect on folding of RNA molecules that form complex structures (DeRose 2003; Draper 2004; Klein et al. 2004; Woodson 2005), but it seems that they play distinct roles. Magnesium is about two orders of magnitude more efficient than sodium in inducing RNA tertiary folding and stabilizing RNA structures (Heilman-Miller et al. 2001; Takamoto et al. 2002; Perez-Salas et al. 2004). However, the nonspecific RNA collapse induced by magnesium results in an ensemble of native and non-native folding intermediates, which eventually leads to both correctly and incorrectly folded RNA structures (Buchmueller et al. 2000; Thirumalai et al. 2001). Interestingly, Russell et al. (2002) and this work demonstrated that monovalent cations prevent ribozyme misfolding by altering the folding starting states of two different group I ribozymes, although the altered folding starting states contain different structure identities. More than 250 mM Na+ is required to promote the formation of P3 and P13 helices, leading to the correct and fast folding of the Tetrahymena ribozyme (Russell et al. 2002). However, several millimolar Na+ promotes a partially formed J3/4:P6 base triple and resolves mispairing in L2.1 and L6 terminal loops of the Candida ribozyme, correlating with the fast formation of the catalytically active ribozyme. We have also demonstrated that monovalent cations are capable of preventing misfolding of a peripheral structure of the Candida ribozyme, and enhance the ribozyme activity. Therefore, monovalent cations play an essential role in folding of the native structure of large complex RNAs by resolving the misfolding problem caused by magnesium-induced nonspecific RNA collapse, similar to the function of an RNA chaperone (Schroeder et al. 2004). It is noteworthy that monovalent cations achieve the function of preventing RNA misfolding at physiological concentrations, indicating that monovalent cations could display such a function in living cells.
MATERIALS AND METHODS
RNA preparation
The Ca.L-11 ribozyme and the 5′-end-radiolabeled RNA substrate Ca/sub-11 were prepared as previously described (Xiao et al. 2003). The ribozyme purified from the denatured PAGE gels was dissolved in ion-free water.
Catalytic activity analysis
The Ca.L-11 ribozyme in all experiments was denatured at 95°C for 1.5 min and chilled on ice for at least 3 min. Ions and buffer were adjusted to the same levels on ice. The substrate RNA at the indicated concentrations and 100 μM GTP were added to initiate the reaction. Reactions were run at 37°C, and an aliquot of 5 μL was removed at each time point and put into an equal volume of loading buffer containing 100 mM EDTA to stop the reaction. The denatured PAGE electrophoresis, data analysis, and kobs values were obtained as previously described (Xiao et al. 2003).
Ribonuclease T1 footprinting
The Ca.L-11 ribozyme was labeled at the 5′-end and purified as previously described (Xiao et al. 2003). The purified ribozyme was folded as illustrated in Figure 1A ▶. T1 cleavage reactions were stopped by phenol-chloroform extraction and analyzed as previously described (Xiao et al. 2003).
Acknowledgments
We thank L. Guo for her comments on the manuscript. This work is supported by the NSFC funds (30170213 and 30330170) and the Wuhan University Fund (0000028) granted to Y.Z.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2188306.
REFERENCES
- Adams, P.L., Stahley, M.R., Gill, M.L., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a group I intron splicing intermediate. RNA 10: 1867–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmueller, K.L., Webb, A.E., Richardson, D.A., and Weeks, K.M. 2000. A collapsed non-native RNA folding state. Nat. Struct. Biol. 7: 362–366. [DOI] [PubMed] [Google Scholar]
- Das, R., Kwok, L.W., Millett, I.S., Bai, Y., Mills, T.T., Jacob, J., Maskel, G.S., Seifert, S., Mochrie, S.G., Thiyagarajan, P., et al. 2003. The fastest global events in RNA folding: Electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 332: 311–319. [DOI] [PubMed] [Google Scholar]
- DeRose, V.J. 2003. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 13: 317–324. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. 2004. A guide to ions and RNA structure. RNA 10: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, B.L., Kim, H., and Chase, E. 2005. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 12: 82–89. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gooding, A.R., and Cech, T.R. 2004. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell 16: 351–362. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller, S.L., Thirumalai, D., and Woodson, S.A. 2001. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol. 306: 1157–1166. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., and Steitz, T.A. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. and Zhang, Y. 2005. Predicting the secondary structures and tertiary interactions of 211 group I introns in IE subgroup. Nucleic Acids Res. 33: 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F. and Westhof, F. 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216: 585–610. [DOI] [PubMed] [Google Scholar]
- Perez-Salas, U.A., Rangan, P., Krueger, S., Briber, R.M., Thirumalai, D., and Woodson, S.A. 2004. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry 43: 1746–1753. [DOI] [PubMed] [Google Scholar]
- Russell, R., Zhuang, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S., and Herschlag, D. 2002. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. 99: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, R., Barta, A., and Semrad, K. 2004. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 5: 908–919. [DOI] [PubMed] [Google Scholar]
- Sclavi, B., Sullivan, M., Chance, M.R., Brenowitz, M., and Woodson, S.A. 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–1943. [DOI] [PubMed] [Google Scholar]
- Shcherbakova, I., Gupta, S., Chance, M.R., and Brenowitz, M. 2004. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J. Mol. Biol. 342: 1431–1442. [DOI] [PubMed] [Google Scholar]
- Takamoto, K., He, Q., Morris, S., Chance, M.R., and Brenowitz, M. 2002. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat. Struct. Biol. 9: 928–933. [DOI] [PubMed] [Google Scholar]
- Takamoto, K., Das, R., He, Q., Doniach, S., Brenowitz, M., Herschlag, D., and Chance M.R. 2004. Principles of RNA compaction: Insights from the equilibrium folding pathway of the P4–P6 RNA domain in monovalent cations. J. Mol. Biol. 343: 1195–1206. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., Lee, N., Woodson, S.A., and Klimov, D. 2001. Early events in RNA folding. Annu. Rev. Phys. Chem. 52: 751–762. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. 2005. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 92: 104–109. [DOI] [PubMed] [Google Scholar]
- Xiao, M., Leibowitz, M.J., and Zhang, Y. 2003. Concerted folding of a Candida ribozyme into the catalytically active structure posterior to a rapid RNA compaction. Nucleic Acids Res. 31: 3901–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, M., Li, T., Yuan, X., Shang, Y., Wang, F., Chen, S., and Zhang, Y. 2005. A peripheral element assembles the compact core structure essential for group I intron self-splicing. Nucleic Acids Res. 33: 4602–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Xiao, M., Lu, C., and Zhang, Y. 2005. Fast formation of the P3P7 pseudoknot: A strategy for efficient folding of the catalytically active ribozyme. RNA 111: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X.W., Bartley, L.E., Babcock, H.P., Russell, R., Ha, T., Herschlag, D., and Chu, S. 2000. A single-molecule study of RNA catalysis and folding. Science 288: 2048–2051. [DOI] [PubMed] [Google Scholar]