Abstract
In Saccharomyces cerevisiae, the cleavage/polyadenylation factor Pcf11 is an important regulatory factor required for recruiting the polyadenylation machinery to the elongating RNA polymerase II (RNAPII) and is necessary for correct transcriptional termination. The interaction with RNAPII is mediated by a CTD-interacting domain (CID) located in the N-terminal region of Pcf11 that binds in a phospho-dependent manner the heptad repeats in the RNAPII CTD. We have previously investigated this protein–protein interaction. We examine here the interaction of the CID with different RNA sequences and look at the effect of phosphopeptides derived from the CTD heptad repeats on the RNA–protein interaction. Our findings demonstrate that the CID displays weak RNA-binding activity, but with some degree of sequence preference, the RNA–protein and peptide–protein interfaces overlap and the CTD-derived phosphopeptides and RNA compete for the binding site. We propose that competition between the protein–peptide and the protein–RNA interaction is important mechanistically and required for the disengagement of polyadenylation factors from RNAPII.
Keywords: transcription termination, mRNA cleavage, protein–RNA interactions, protein–protein interactions, NMR, surface plasmon resonance
INTRODUCTION
Coupling of transcription, 3′-end processing and polyadenylation is achieved by the direct association of several effector complexes with RNA polymerase II (RNAPII) (Howe 2002; Proudfoot et al. 2002) and provides a means to coordinate transcriptional and post-transcriptional events (Zorio and Bentley 2004). The assembly of the different complexes on the C-terminal domain (CTD) of RNAPII can function by increasing the local concentration of processing factors, by coupling the rate of transcription with the assembly of specific RNA–protein complexes, and by allosterically activating and inhibiting the same complexes (Bentley 2005). While a number of recent studies have significantly improved our knowledge of the interactions that lie at the basis of the assembly of these complexes (Fabrega et al. 2003; Meinhart and Cramer 2004; Zorio and Bentley 2004; Meinhart et al. 2005), the physical basis for the disassembly of the elongation and mRNA-processing complexes are still largely uncharted.
The Saccharomyces cerevisiae cleavage factor IA (CFIA) is an essential component of the mRNA 3′-end processing machinery. During transcription, this multisubunit complex associates with the RNAPII elongation complex (Licatalosi et al. 2002; Kim et al. 2004a) and is required for cleavage-site selection. Subsequently, CFIA recruits other protein complexes, resulting in transcript cleavage and polyadenylation (Chen and Moore 1992). CFIA consists of the proteins Rna14, Rna15, Pcf11, and Clp1 (Gross and Moore 2001a). Rna15 and a CFIA-associated factor (Hrp1) interact with sequence recognition elements present in the 3′ UTR (Gross and Moore 2001b) and tether the CFIA complex at the 3′ end of nascent mRNAs. Rna14 is tightly associated with Rna15, and this interaction enhances Rna15 binding to the nascent mRNA (Noble et al. 2004). In turn, the Rna14–Rna15 complex interacts with sequences located within the central region of Pcf11 (Amrani et al. 1997a,b; Sadowski et al. 2003). Pcf11 then couples CFIA to RNAPII by the interaction of its N-terminal CTD interaction domain (CID) with serine 2-phosphorylated heptapeptide repeats of the RNAPII CTD (Barilla et al. 2001; Licatalosi et al. 2002; Sadowski et al. 2003).
In a recent study, Zhang and coworkers (Zhang et al. 2005) identified a new RNA-binding activity in the Pcf11 CID and related this activity to CTD binding, proposing a new model for the dismantling of the RNA polymerase II elongation complex. We have previously investigated the interaction between the Pcf11 CID and a serine 2-phosphorylated peptide (PTSPSYSpPTSPSY) derived from the CTD of RNAPII (Noble et al. 2005). Here, we explore the RNA-binding capability of the CID and examine the relationship between binding to the CTD and binding to RNA. Using NMR and SPR, we have analyzed this three-component system in vitro and show that RNA and RNAPII CTD are competitive ligands of Pcf11 CID. We propose that competition for the CID-binding site is an important contributory factor to CFIA release from RNAPII after initial deployment of CFIA onto the polyadenylation signals.
RESULTS AND DISCUSSION
RNA binding of Pcf11 CID
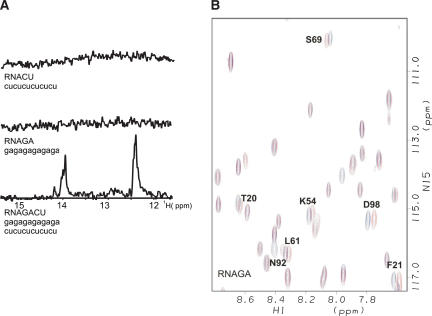
In order to investigate the interaction between RNA and the CID from Pcf11, two ribo-oligonucleotides, GA-RNA (GAGAGAGAGAGA) and CU-RNA (CUCUCUCUCUCU), were prepared (Fig. 1A ▶). These oligos were chosen to both maximize sequence diversity and minimize the possibility of the RNA to form secondary structures. The absence of imino resonances in 1D NMR experiments recorded at physiological pH and salt concentration confirmed that the ribo-oligonucleotides do not form stable secondary structures (Fig. 1A ▶). However, when the RNAs were mixed with a 1:1 ratio, two closely grouped sets of peaks were observed, corresponding to the quasidegenerate resonances of the alternate AU and GC base pairs in the RNA duplex (Fig. 1A ▶).
FIGURE 1.
(A) RNA oligonucleotides used in this study and their 1D 1H NMR spectra (imino region). (B) Superimposition of a representative region of the HSQC spectra of Pcf11-CID free (blue) and bound to the GA-rich oligonucleotide (red).
The interaction of the RNAs with the Pcf11 CID was examined by recording 15N-1H TROSY spectra of 15N 2H-labeled Pcf11-CID (Pcf11; residues 1–142) at increasing concentrations of (1) GA-RNA, (2) CU-RNA, and (3) GA/CU-RNA duplex. Addition of GA-RNA up to an RNA:protein ratio of 1:1 resulted in selective chemical-shift changes in the protein spectrum (Fig. 1B ▶). The residues affected cluster in four groups (Fig. 2 ▶). The first and by far largest of these clusters comprises T20, F21, N22, and S23 in the loop between helices 1 and 2, and K54, Q59, and L61 between helices 3 and 4. An isolated residue, S69, located in the phosphopeptide binding site is also significantly affected by RNA binding. Further changes are observed in three residues located within the first two turns of helix 7 (D98, T101, and R102) and in the last two residues of the protein. Globally, these changes define a binding site that spans the top third of the concave face of the protein and includes the ridge separating it from the convex surface. Similar, but more limited changes were observed upon addition of CU-RNA and GA/ CU-RNA. The relatively small changes observed in all three titrations indicate that no major structural rearrangement (e.g., significant movement or distortion of the helices or intercalation of aromatic rings) accompanies the binding and the (very fast) exchange regime of the resonances suggests that the binding is weak.
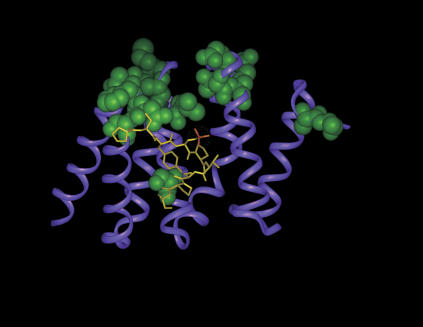
FIGURE 2.
Ribbon representation of Pcf11 CID (magenta) in complex with the CTD-derived peptide (Meinhart and Cramer 2004). The peptide residues are shown in yellow and phosphoserine 2 is highlighted in orange. The CID residues whose resonances are most affected during the titration with GA-RNA are displayed using a CPK render (green). The binding sites of the CTD-peptide and of the RNA partially overlap.
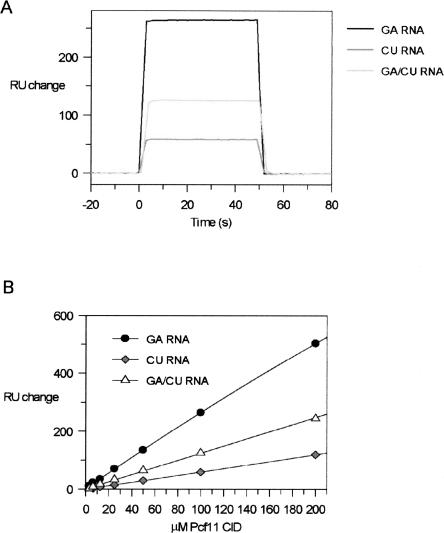
In a complementary set of experiments, the CID–RNA interaction was examined using surface plasmon resonance (SPR). Here, 3′-biotinylated RNA molecules (GA-RNA, CU-RNA, and GA/CU-RNA) were immobilized onto the surface of streptavidin-coated sensor chips, and increasing concentrations of Pcf11–CID were then applied to each RNA in order to assess the binding. A signal change was observed for all three RNAs, although at the same analyte concentration, GA-RNA retained twice as much Pcf11-CID as the GA/CU-RNA or CU-RNA (Fig. 3A ▶). Thus, the rank order of the RNA-binding preference of the Pcf11-CID is consistent between the NMR and the SPR experiments. Inspection of the associative and dissociative phases of the SPR response indicates that the CID–RNA interaction is characterized by a fast on-and-off rate, in agreement with the transience indicated by the fast exchange seen by NMR. Weak binding is also evident from the trend of the SPR signal obtained at increasing protein concentrations (Fig. 3B ▶). Although the data were fitted to a single-site binding equation, the plots are close to linear over the concentration range used, with no indication of a breakpoint. Taken together, these data confirm that the CID only weakly interacts with RNA with an estimated Kd in the range of 10−4 to 10−3M. Interestingly, this places the affinity of the CID for RNA in the same range as the affinity reported for the CID–phosphopeptide interaction (Noble et al. 2005).
FIGURE 3.
Binding of the RNA oligos to Pcf11 monitored by SPR. (A) Overlap of three SPR curves obtained by the introduction of Pcf11-CID to immobilized GA-RNA, CU-RNA, and GA/CU-RNA. (B) The increase in response units resulting from each addition of Pcf11-CID titration plotted against protein concentration. The curve is the line of best fit using an expression for a 1:1 binding model.
Our NMR and SPR data are in agreement with a recent report that the Pcf11 CID is capable of RNA binding (Zhang et al. 2005). Although some sequence specificity is apparent, the affinity is weak and it is unlikely that the CID–RNA interaction provides a stable association in the absence of other protein–RNA interactions. Interestingly, the CID does not show a priori preference for single-stranded or double-stranded RNA; the interaction of the protein with the CU-RNA results in smaller chemical-shift changes and SPR signal than the interaction with the duplex. However, the differences observed between the binding to the polypyrimidine and polypurine RNAs indicate that a degree of sequence preference does exist. Although it is possible that a specific high-affinity CID target sequence is still unidentified, our data show that Pcf11 would bind to purine-rich sequences within an mRNA molecule already associated with Rna15, Hrp1, and other 3′-end processing factors.
RNA vs. CTD binding
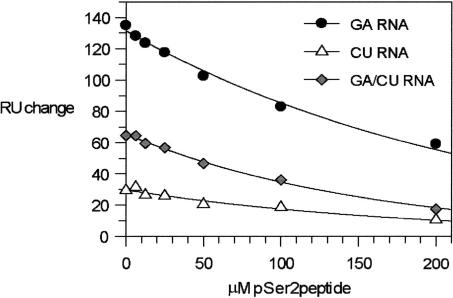
Comparison of the (PTSPSYSpPTSPSY)–CID and RNA–CID interaction surfaces reveals a significant overlap (Fig. 2 ▶). Such an overlap could facilitate a cooperative RNA–CTD interaction when both ligands are bound to the CID. Alternatively, competition between the two ligands for the same site could take place. To investigate these two possibilities, we evaluated the effect of increasing concentrations of the phosphorylated CTD peptide on CID–RNA binding by SPR. The experiments show that addition of the CTD phosphopeptide reduces the binding of the CID to the immobilized RNAs (Fig. 4 ▶), and that this reduction in RNA binding is titratable with respect to the phosphopeptide concentration. At the equivalence point, 50 μM (PTSPSYSpPTSPSY) and 50 μM CID, the SPR signal monitoring the CID–RNA interaction is reduced by ~30%. The reduction in binding demonstrates that RNA and the CTD compete for the CID interface rather than interact in an additive or cooperative manner, and the moderate but significant competition between the two ligands indicates that the affinity of the CID for RNA and the CTD are of comparable magnitude. Control experiments demonstrate that the observed competitive effect is not due to a peptide–RNA interaction resulting in displacement of the CID from the RNA, as no peptide–RNA interaction was detectable when a high concentration (200 μM) of the peptide alone was introduced as the SPR analyte. Further, a shorter CTD-derived peptide (YSpPTSPS) (Fig. 1 ▶) also phosphorylated on Serine 2, which does not bind to the CID (Noble et al. 2005), does not compete with CID–RNA binding (data not shown).
FIGURE 4.
RNA and CTD peptide compete for binding to Pcf11-CID. Maximal intensity of the SPR-binding sensograms recorded for Pcf11-CID interaction with GA-RNA, CU-RNA, and GA/CU-RNA are plotted against increasing concentration of pSer2 peptide.
What is the function of the peptide–RNA competition? Recent ChIp analyses have demonstrated that Pcf11 and the other CFIA factors are mostly associated with transcribing RNAPII that is located toward the 3′ ends of genes and around the polyadenylation site, concomitant with high levels of CTD serine-2 phosphorylation (Ahn et al. 2004; Kim et al. 2004a). Given that the interaction of Pcf11 with the CTD is strongly phosphoserine 2 dependent (Licatalosi et al. 2002; Ahn et al. 2004; Meinhart and Cramer 2004; Noble et al. 2005), the likelihood is that, upon transcription through the polyadenylation signals, CFIA is coupled to RNAPII through this CID–phosphoCTD interaction. The competition we observe between the CTD peptide and RNA suggests that the in vivo association between the CTD and CFIA could be disrupted by RNA binding to the Pcf11 CID, resulting in the disengagement of CFIA from the CTD. Of course, RNA has to be available in order to compete for the CID-binding site. This situation specifically arises at polyadenylation signals, when the nascent RNA is bound by CFIA at the positioning element by Rna15, and at the efficiency element by Hrp1 (Gross and Moore 2001b). The association of the nascent RNA with CFIA has the potential to bring the 3′ end of the transcript into proximity with the CID and switch Pcf11 from CTD-bound to RNA-bound and to release CFIA from RNAPII. The effectiveness of the competition by RNA may be further enhanced as the polymerase moves downstream of the polyadenylation site and the density of serine-2 phosphorylation in the CTD decreases.
As well as its involvement in the cleavage and polyadenylation reaction, Pcf11 is also required for correct transcriptional termination to occur (Birse et al. 1998; Sadowski et al. 2003). Interestingly, mutations in the CID affect termination, while mutations elsewhere in the protein affect cleavage/polyadenylation (Sadowski et al. 2003). In a recent report, it has been suggested that a ternary Pcf11-CID/ CTD/RNA interaction is directly responsible for Pcf11 termination activity by dismantling transcription elongation complexes (Zhang et al. 2005). Our data now reveal that RNA competes effectively with the serine-2 phosphorylated CTD for the Pcf11-binding site. Intriguingly, an important component of the “torpedo” model for transcriptional termination is that transcript cleavage and disengagement of the polyadenylation factors occurs prior to recruitment of another CID containing protein, Rtt103, and the associated Rat1/Rai1 ribonuclease complex (Kim et al. 2004b). One possibility is that the switch in Pcf11 from CTD-bound to RNA-bound is a prerequisite for the recruitment of Rtt103/ Rat1/Rai1, and perhaps the association of Pcf11 with transcriptional termination is related to the subtleties of this disengagement and exchange process.
Interestingly, a recent study has unveiled a specific role for protein–RNA interactions in human (Kaneko and Manley 2005). The investigators have shown that human, but not yeast, CTD has an RNA-binding activity, and that this activity is localized in its noncanonical repeats. These repeats show a loose specificity for ACCCACACC sequences, and the newly identified RNA–protein interaction has been proposed to compete with essential protein–protein ones (Kaneko and Manley 2005). The RNA-binding activity of the Pcf11 CID and of the noncanonical repeats of polymerase II CTD highlights the potential role of competitive protein–protein and protein–RNA interactions in the regulation of transcriptional elongation and mRNA processing.
MATERIALS AND METHODS
Protein expression and purification
Pcf11-CID was prepared as previously described (Noble et al. 2005). In brief, the protein was expressed in the Escherichia coli strain BL21 (DE3) as an N-terminal glutathione-S-transferase fusion and purified from clarified crude cell extracts using immobilized glutathione-affinity chromatography. This was combined with on-column cleavage by PreScission protease (Amersham Biosciences) to release Pcf11-CID and then gel-filtration chromatography on Superdex 75 (Amersham Biosciences). The purity and the monodispersity of preparations were monitored by ESI-MS and SDS-PAGE. Protein concentration was determined from the absorbance at 280 nm using a molar extinction coefficient of 15,010 m−1 cm−1.
Peptides
PTSPSYSpPTSPSY and YSpPTSPS peptides were prepared using standard solid-phase peptide synthesis, purified by reverse-phase HPLC and analyzed using ESI-MS (Dept of Biochemistry, University of Bristol). The concentration of the peptides was determined using UV absorbance spectroscopy exploiting the near-UV absorbance of the tyrosine residues.
RNA
3′-Biotinylated and nonbiotinylated versions of the GAGAGAGAGAGA and CUCUCUCUCUCU ribo-oligonucleotides were chemically synthesized and gel purified (CureVac, GmbH). Concentrations were determined from the calculated molar extinction coefficients.
Surface plasmon resonance
Experiments were performed at 25°C on a Biacore 2000 instrument (Biacore). A GA/CU-RNA duplex, biotinylated on the GA ribo-oligonucleotide, was prepared by annealing the GA oligonucleotide with a 1.5 molar excess of nonbiotinylated CU-RNA. The GA, CU, and GA/CURNA ribo-oligonucleotides were attached to flow cells 2–4 of a SA sensor chip (Biacore) as described previously (Noble et al. 2004). Pcf11-CID was dialyzed into 40 mM Tris-HCl (pH 7.8), 50 mM NaCl, 5 mM MgCl2, 3 mM DTT, and 0.002% Tween 20 (v/v). Typically, 25 μL of 6.25–200 μM Pcf11-CID samples were injected across the chip in the same buffer at 30 μL/min. Signal increases as a function of protein concentration were fitted to a single-site binding equation. For competition experiments, 50 μM Pcf11-CID was injected with or without increasing concentrations of PTSPSYSpPTSPSY or YSpPTSPS peptides. The effectiveness of peptide competition was determined from a plot of the signal decrease as a function of peptide competitor concentration.
Nuclear magnetic resonance
NMR spectra were recorded at 27°C on a Varian INOVA spectrometer operating at 800 MHz (1H frequency). All experiments described below were recorded in 10 mM Tris-HCl (pH 7.4) and 120 mM NaCl in a 90% H2O/10% D2O mixture. Water suppression was achieved by the WATERGATE pulse-sequence (Piotto et al. 1992). 1D 1H NMR experiments were recorded on 0.2 mM samples of GA-RNA, CU-RNA, and GA/CU-RNA. 1H–15N TROSY spectra (Pervushin et al. 1997) were recorded at each step of titrations of 15N–2H-labeled 0.15 mM samples of Pcf11-CID with the three RNAs, up to a concentration ratio of 1:1.
All spectra were processed and zero filled to the next power of two using the NMRPIPE program (Delaglio et al. 1995). Baseline correction was applied when necessary. The spectral analysis was performed using the Felix (Accelerys) and XEASY programs (Bartels et al. 1995).
Acknowledgments
NMR spectra were recorded at the MRC Biomedical NMR Centre, Mill Hill. We thank Dr. G. Kelly and Dr. T.A. Frenkiel for assistance in data recording.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2304506.
REFERENCES
- Ahn, S.H., Kim, M., and Buratowski, S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67–76. [DOI] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Le Gouar, M., Lacroute, F., and Wyers, F. 1997a. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17: 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Wyers, F., Dufour, M.E., Aggerbeck, L.P., and Lacroute, F. 1997b. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 17: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla, D., Lee, B.A., and Proudfoot, N.J. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Nat. Acad. Sci. 98: 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, C., Xia, S.H., Billeter, M., Guntert, P., and Wuthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6: 1–10. [DOI] [PubMed] [Google Scholar]
- Bentley, D.L. 2005. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17: 251–256. [DOI] [PubMed] [Google Scholar]
- Birse, C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Chen, J. and Moore, C. 1992. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 12: 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Fabrega, C., Shen, V., Shuman, S., and Lima, C.D. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 11: 1549–1561. [DOI] [PubMed] [Google Scholar]
- Gross, S. and Moore, C. 2001a. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Nat. Acad. Sci. 98: 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S. and Moore, C.L. 2001b. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21: 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, K.J. 2002. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta 1577: 308–324. [DOI] [PubMed] [Google Scholar]
- Kaneko, S. and Manley, J.L. 2005. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol. Cell 20: 91–103. [DOI] [PubMed] [Google Scholar]
- Kim, M., Ahn, S.H., Krogan, N.J., Greenblatt, J.F., and Buratowski, S. 2004a. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Krogan, N.J., Vasiljeva, L., Rando, O.J., Nedea, E., Greenblatt, J.F., and Buratowski, S. 2004b. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522. [DOI] [PubMed] [Google Scholar]
- Licatalosi, D.D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., McNeil, J.B., and Bentley, D.L. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9: 1101–1111. [DOI] [PubMed] [Google Scholar]
- Meinhart, A. and Cramer, P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226. [DOI] [PubMed] [Google Scholar]
- Meinhart, A., Kamenski, T., Hoeppner, S., Baumli, S., and Cramer, P. 2005. A structural perspective of CTD function. Genes & Dev. 19: 1401–1415. [DOI] [PubMed] [Google Scholar]
- Noble, C.G., Walker, P.A., Calder, L.J., and Taylor, I.A. 2004. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 32: 3364–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, C.G., Hollingworth, D., Martin, S.R., Ennis-Adeniran, V., Smerdon, S.J., Kelly, G., Taylor, I.A., and Ramos, A. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 12: 144–151. [DOI] [PubMed] [Google Scholar]
- Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. 1997. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Nat. Acad. Sci. 94: 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Sadowski, M., Dichtl, B., Hubner, W., and Keller, W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22: 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Fu, J., and Gilmour, D.S. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 19: 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio, D.A. and Bentley, D.L. 2004. The link between mRNA processing and transcription: Communication works both ways. Exp. Cell Res. 296: 91–97. [DOI] [PubMed] [Google Scholar]