Abstract
Objective
To assess the prognostic significance of molecular biomarkers, particularly c-erbB-2 and p53, through study of prospective clinical data and archival breast cancer tissues for women accrued to the Alabama Breast Cancer Project.
Summary Background Data
Defining molecular abnormalities in breast cancer is an important strategy for early detection, assessment of prognosis, and treatment selection. Evidence is strong that selective biomarkers, including c-erbB-2 and p53, have prognostic significance in breast cancer. Few studies have analyzed the prognostic significance of coexpression of biomarkers.
Methods
Study patients were those accrued to the Alabama Breast Cancer Project (1975–1978) who had archival breast cancer tissues available for analysis. Criteria for entrance into the Alabama Breast Cancer Project were T1–3 breast cancer with M0 status. Age, nodal status, and histologic grade were also documented. Patients were randomized to radical versus modified radical mastectomy, and node-positive patients were also randomized to adjuvant chemotherapy (cyclophosphamide, methotrexate, and 5-fluorouracil [CMF]) versus melphalan. Archival breast cancer tissues were studied for c-erbB-2, TGF-α, p53, cathepsin D, bcl-2, and estrogen and progesterone receptor expression using immunohistochemistry. Survival curves were developed using the Kaplan-Meier method. Univariate analysis was performed using the log-rank test, multivariate analysis using a rank regression model.
Results
Three hundred eleven patients were accrued to the Alabama Breast Cancer Project, and paraffin-embedded breast cancer tissues for 90 patients were available for immunohistochemical analysis of molecular biomarkers. Univariate analysis showed nodal status, c-erbB-2 expression, and p53 expression to have prognostic significance. Coexpression of c-erbB-2 and p53 was also found to have prognostic significance by the log-rank test. Multivariate analysis showed T stage, nodal status, c-erbB-2 expression, and p53 expression to have independent prognostic significance.
Conclusions
These data suggest that c-erbB-2 and p53 expression in breast cancer have prognostic significance. After median follow-up of 16 years, coexpression of c-erbB-2 and p53 may have more prognostic significance than traditional prognostic factors such as T stage and nodal status. Prospective study of large numbers of patients with breast cancer is encouraged to validate these findings.
Molecular biomarkers for breast cancer are of several types. Risk biomarkers are those associated with increased cancer risk and include mammographic abnormalities, proliferative breast disease with or without atypia, family clustering, and inherited germ-line abnormalities. Surrogate end-point biomarkers are tissue, cellular, or molecular alterations that occur between cancer initiation and progression. These biomarkers are used as end points in short-term chemoprevention trials. Prognostic biomarkers provide information regarding outcome irrespective of therapy, whereas predictive biomarkers provide information regarding response to therapy. Candidate prognostic biomarkers for breast cancer include elevated proliferation indices such as Ki-67 and proliferating cell nuclear antigen; estrogen receptor (ER) and progesterone receptor (PR) overexpression; markers of oncogene overexpression such as c-erbB-2, transforming growth factor-α (TGF-α), and EGFr; indicators of apoptotic imbalance, including overexpression of bcl-2 and an increased bax/bcl-2 ratio; markers of disordered cell signaling such as p53 nuclear protein accumulation; alteration of differentiation signals, such as overexpression of c-myc and related proteins; loss of differentiation markers, such as TGF-β II receptor and retinoic acid receptor; and alteration of angiogenesis proteins such as vascular endothelial growth factor (VEGF) overexpression. 1 Biomarkers indicative of perturbed molecular processes are often amenable to quantification in small tissue specimens by means of immunohistochemistry.
Review of the pertinent literature suggests that overexpression of c-erbB-2 and p53, in particular, may have prognostic significance in breast cancer, and these molecular biomarkers are the focus of this report. c-erbB-2 is a normal cellular gene that encodes a membrane protein (p185), which is tyrosine phosphorylated after interaction with its ligands. 2,3 Overexpression of c-erbB-2 occurs either through changes in amplification or through mRNA overexpression. 4–6 p53 is involved in regulating cell proliferation, inducing apoptosis, and promoting chromosomal stability. Disruption of these functions appears to have an important role in carcinogenesis. 7 There is evidence that overexpression of c-erbB-2 and p53 is relevant to breast cancer progression. 8 This hypothesis is based on the high frequency of c-erbB-2 and p53 overexpression in invasive breast cancer, 4 in benign breast disease, 9–20 and in animal models of breast cancer, 21,22 which together suggest a role for these genes in the early stages of breast tumorigenesis.
We have studied prospective clinical data and archival breast cancer tissues for women accrued to the Alabama Breast Cancer Project between 1975 and 1978 and in this article report on c-erbB-2 and p53 as prognostic molecular biomarkers after a median follow-up of 16 years.
PATIENTS AND METHODS
Study Population
Between 1975 and 1978, 311 patients with operable breast cancer (T1–3, M0) were accrued to the Alabama Breast Cancer Project and randomized to receive either a Halstead radical mastectomy or a modified radical mastectomy with dissection of at least axillary lymph node levels 1 and 2. 23 Patients with histologically positive metastatic axillary lymph nodes were randomized further to receive either melphalan (L-PAM) or a combination of cyclophosphamide, methotrexate, and fluorouracil (CMF). Clinical and histologic factors such as T stage, nodal status, and histologic grade were recorded on entrance into the Alabama Breast Cancer Project. More recently, all available paraffin-embedded breast cancer specimens for patients accrued to the Alabama Breast Cancer Project were studied for expression of molecular biomarkers. Because of the limited amount of tissue available for analysis, the number of molecular biomarkers studied for each patient varied. All human subject studies were conducted in accordance with the ethical standards of the Helsinki Declaration of 1975.
Follow-Up
The median patient follow-up was 16 (range 1–21) years. Disease-free survival and overall survival were calculated as the time from date of diagnosis to first recurrence of disease or death, respectively. Recurrence of disease was defined as a new manifestation of disease near the site of the original breast cancer, at a distant site, or in the contralateral breast.
Tumor Histology
Original hematoxylin-and-eosin stained microscopic sections from all cases were reviewed by one of the authors for confirmation of diagnosis and for standardization of grading according to the criteria described by Bloom and Richardson. The microscopic sections reviewed included all axillary lymph nodes identified in the axillary dissections.
Immunohistochemistry
Representative blocks of tumor for each case studied were identified and sections were prepared for immunohistochemical staining using standard techniques. Briefly, 5-micron sections were cut from the blocks, deparaffinized in xylene, rehydrated in a series of graded alcohols, and placed in a Tris buffer bath (0.05 mol/L Tris [pH 7.6], 0.15 mol/L NaCl, 0.0002% Triton X-100). Endogenous peroxidase activity was quenched using 3% hydrogen peroxide. Slides were rinsed with deionized water and placed in a Tris buffer bath. After incubation, 1.0% preimmune goat serum was used to block nonspecific staining, and sections were then stained with primary antibodies using an automated immunostaining system (Ventana ES, Ventana Medical Systems, Inc., Tucson, AZ). Appropriate controls were used to ensure uniformity of results (ER and PR staining was done manually). Antibodies used were as follows:
c-erbB-2: rabbit antihuman c-erbB-2 oncoprotein (DAKO, Carpinteria, CA)
TGF-α: mouse monoclonal antihuman TGF-α (Oncogene Science, Uniondale, NY)
p53: mouse monoclonal antihuman p53 (DAKO)
Cathepsin D: rabbit polyclonal antihuman CATH-D (DAKO)
bcl-2: mouse monoclonal antihuman bcl-2 (DAKO)
ER: mouse monoclonal antihuman ER (BioGenix, San Ramon, CA)
PR: mouse monoclonal antihuman PR (BioGenix, San Ramon, CA).
All immunostained sections were independently examined and scored by two of the authors. Any discrepancies were resolved by consensus of the observers before data analysis. Expression of ER and PR, c-erbB-2, TGF-α, cathepsin-D, and bcl-2 was scored 0 to 3 based on the intensity of staining and the percentage of cells staining. For statistical analysis, expression of these molecular biomarkers was described as negative (score 0) or positive (score 1–3). p53 staining was scored and analyzed based on the intensity of nuclear staining (score 0–3) and the percentage of cells staining (score 0–5), the final p53 score being the sum of these two scores (0–8). Figure 1 illustrates c-erbB-2 and p53 immunostaining in breast cancer tissues.
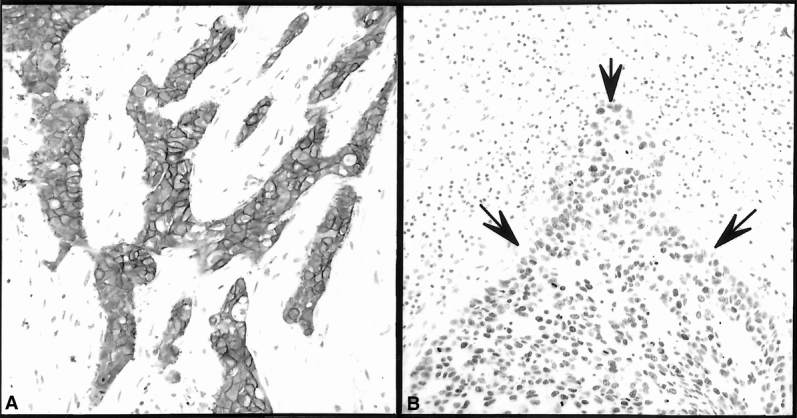
Figure 1. Immunohistochemical determination of c-erbB-2 (A) and p53 (B) expression in breast cancer and adjacent normal-appearing tissue. There is c-erbB-2 (A) membrane immunostaining of the malignant cells shown. This microscopic section was evaluated based on the intensity of staining and the percentage of malignant cells with membrane staining and was scored as 3/3. There is an increase in p53 (B) staining of the malignant cells demarcated by the solid arrows. This microscopic section was evaluated based on the intensity of staining and the percentage of malignant cells staining and was scored as 3/3 and 4/5, respectively (overall score 7).
Statistical Analysis
The patterns of disease-free and overall survival were estimated by the Kaplan-Meier method. Univariate analysis was performed using a log-rank test; multivariate analysis was evaluated using a rank regression model. Coexpression of c-erbB-2 and p53 was analyzed by a log-rank test. The small number of patients studied precluded analysis of the interaction of c-erbB-2 and p53 using a rank regression procedure.
RESULTS
Demographics and Survival Data
Three hundred eleven patients were accrued to the Alabama Breast Cancer Project, and 90 of these patients had paraffin-embedded blocks of breast cancer tissue available for immunohistochemical analysis of molecular biomarkers. Of the 90 patients with blocks available for biomarker analysis, 71% were 50 years of age or older, 80% had T1–2 tumors, 44% were N0, and 45% had high-grade histology (Table 1). Disease-free and overall survival for these patients was 49% and 52%, respectively; for all 311 patients accrued to the Alabama Breast Cancer Project, disease-free and overall survival was 50% and 55%, respectively.
Table 1. PATIENT, DISEASE, AND MOLECULAR VARIABLES AT PRESENTATION
ER, estrogen receptor; PR, progesterone receptor; TGF-α, transforming growth factor-α.
Univariate Analysis
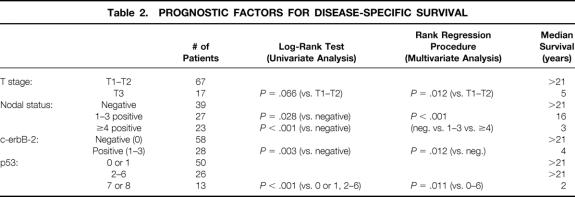
Kaplan-Meier survival curves were calculated and compared using a log-rank test for clinical factors in 90 patients and for histologic factors and molecular biomarker expression in the corresponding paraffin-embedded breast cancer tissues. Nodal status (P = .028 for one to three nodes positive, P < .001 for four or more nodes positive), c-erbB-2 expression (P = .003), and p53 expression (P < .001) were shown to have prognostic significance (Table 2). Coexpression of c-erbB-2 and p53 was also shown to be significant by a log-rank test (Table 3). Survival was better in patients with no expression of c-erbB-2 (negative, score 0) and low expression of p53 (score 0–6) versus patients with expression of c-erbB-2 (positive, score 1–3) and low expression of p53 (score 0–6) (P = .011); versus patients with no expression of c-erbB-2 (negative, score 0) and high expression of p53 (score 7 or 8) (P = .0002); and versus patients with expression of both c-erbB-2 (positive, score 1–3) and high expression of p53 (score 7 or 8) (P = .0001). Survival was also better for patients with expression of c-erbB-2 (positive, score 1–3) and low expression of p53 (score 0–6) versus patients with expression of c-erbB-2 (positive, score 1–3) and high expression of p53 (score 7 or 8) (P = .0009). Figures 2, 3, and 4 show the Kaplan-Meier disease-specific survival curves for c-erbB-2 expression, p53 expression, and coexpression of c-erbB-2 and p53, respectively.
Table 2. PROGNOSTIC FACTORS FOR DISEASE-SPECIFIC SURVIVAL
Table 3. PROGNOSTIC SIGNIFICANCE OF c-erbB-2 AND p53 COEXPRESSION BY UNIVARIATE ANALYSIS
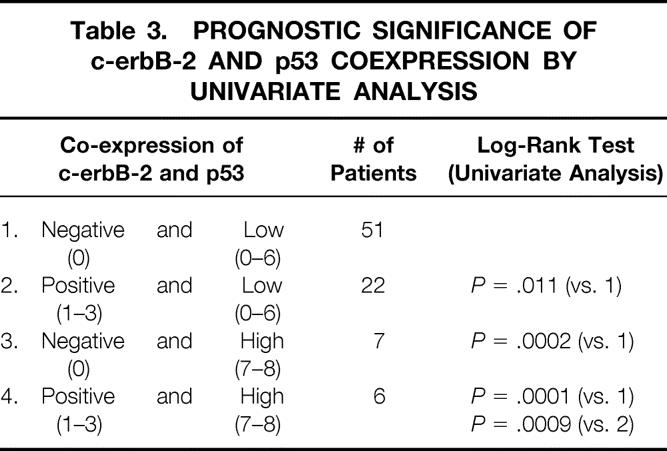
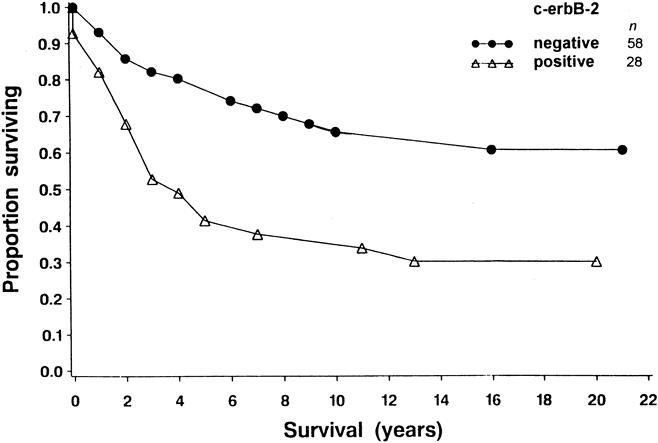
Figure 2. Cause-specific survival curves for 86 patients with breast cancer. For each time period (years, x-axis), the probability of not dying from breast carcinoma is shown (y-axis). Survival was better in patients with no expression of c-erbB-2 (negative, score 0) than in patients with expression of that gene (positive, score 1–3) (P = .003).

Figure 3. Cause-specific survival curves for 89 patients with breast cancer. For each time period (years, x-axis), the probability of not dying from breast carcinoma is shown (y-axis). Survival was better in patients with low expression of p53 (score 0–6) than in patients with high expression of that gene (score 7 or 8) (P < .001).
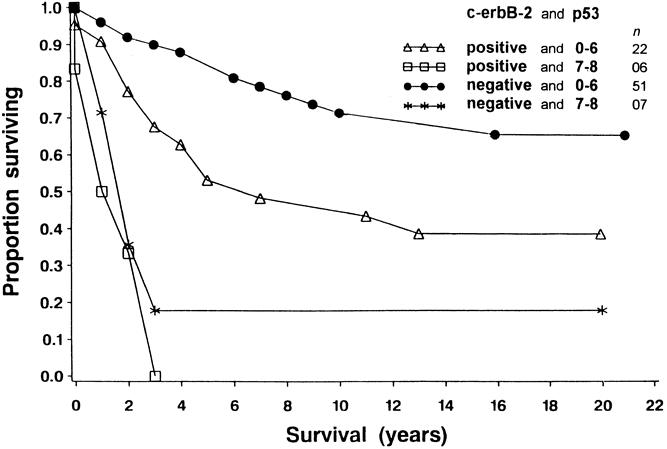
Figure 4. Cause-specific survival curves for 86 patients with breast cancer. For each time point (years, x-axis), the probability of not dying from breast cancer is shown (y-axis). Survival was better in patients with no expression of c-erbB-2 (negative, score 0) and low expression of p53 (score 0–6) versus patients with expression of c-erbB-2 (positive, score 1–3) and low expression of p53 (score 0–6) (P = .011); versus patients with no expression of c-erbB-2 (negative, score 0) and high expression of p53 (score 7 or 8) (P = .0002); and versus patients with expression of both c-erbB-2 (positive, score 1–3) and high expression of p53 (score 7 or 8) (P = .0001). Survival was also better for patients with expression of c-erbB-2 (positive, score 1–3) and low expression of p53 (score 0–6) than for patients with expression of c-erbB-2 (positive, score 1–3) and high expression of p53 (score 7 or 8) (P = .0009).
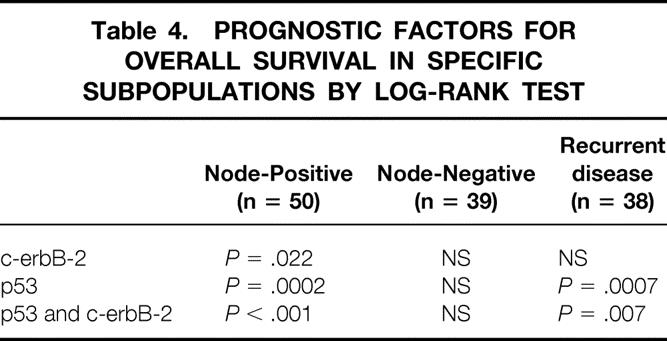
c-erbB-2 expression, p53 expression, and coexpression of c-erbB-2 and p53 were also studied in specific subpopulations (Table 4). In node-positive patients (n = 50), c-erbB-2 expression (P = .022), p53 expression (P = .0002), and coexpression of c-erbB-2 and p53 (P < .001) had prognostic significance; in node-negative patients (n = 39), they were not significant. In patients with recurrent disease (n = 38), both p53 expression (P = .0007) and coexpression of c-erbB-2 and p53 (P = .007) were significant prognostic factors.
Table 4. PROGNOSTIC FACTORS FOR OVERALL SURVIVAL IN SPECIFIC SUBPOPULATIONS BY LOG-RANK TEST
Multivariate Analysis
By rank regression analysis of clinical factors in 90 patients and of histologic factors and molecular biomarker expression in the corresponding paraffin-embedded breast cancer tissues, T stage (P = .012), nodal status (P < .001), c-erbB-2 expression (P = .012), and p53 expression (P = .011) had prognostic significance (see Table 2). The number of patients coexpressing c-erbB-2 and p53 was too small to study that interaction using a rank regression procedure.
DISCUSSION
The results of this study suggest that p53 expression, c-erbB-2 expression, and coexpression of c-erbB-2 and p53 have prognostic significance in breast cancer. Overexpression of c-erbB-2 correlated strongly with poor patient survival. In this regard, our results are similar to those in several other studies 24–27 but contrast with studies that suggest c-erbB-2 overexpression has no 28 or only limited prognostic value. 29 These conflicting results may be due to differences in the immunohistochemical techniques applied in the various studies and to the scoring systems used. 26,27 Whereas some studies reported that cytoplasmic immunostaining of c-erbB-2 may have prognostic significance, we confined our analysis of c-erbB-2 expression to membrane immunostaining because that has been found to correlate with c-erbB-2 gene amplification. 29 We scored c-erbB-2 membrane staining as 0 to 3 based on both the intensity of staining and the percentage of cells staining.
p53 overexpression correlated strongly with poor patient survival in our study. It has also been associated with a poor prognosis in other studies of patients with breast cancer. 30,31 These studies suggested both a prognostic and a predictive role for this molecular biomarker. 32 Immunohistochemistry is a practical and relatively inexpensive method of detecting p53 alterations in cancer tissues and allows precise localization of the cells that exhibit p53 alterations; however, it cannot detect the complete loss of p53. 33 In addition, because viral and cellular oncoproteins and DNA-damaging agents can alter the stability of the p53 protein and affect its detection by immunohistochemistry, p53 immunostaining does not necessarily equate to a p53 gene mutation. 34 We scored p53 nuclear staining as 0 to 8 based on both the intensity of staining (0–3) and the percentage of cells staining (0–5).
Coexpression of c-erbB-2 and p53 has been reported in several studies, with a frequency of coexpression as high as 42%. 27,35–49 Patients whose breast cancer tissues coexpress c-erbB-2 and p53 have been found to have a poor prognosis in several studies. 27,37,39,40,44,45,47,49 It has been postulated that breast cancers coexpressing c-erbB-2 and p53 (both genes map to chromosome 17) have lost a key mechanism for control of cell proliferation and have gained an activator of malignant cell potential, resulting in a highly malignant tumor phenotype. 36,50 However, other studies have shown c-erbB2 and p53 coexpression to have no impact on prognosis, 38,43 and some have shown a better prognosis for breast cancers coexpressing c-erbB-2 and p53. 41 These differences may reflect the effect of varying therapeutic regimens on the interaction between c-erbB-2 and p53. 50 Patients with breast cancer tumors coexpress c-erbB-2 and p53 have an improved 10-year survival when treated with a high-dose FAC regimen (fluorouracil, Adriamycin [doxorubicin], cyclophosphamide). 48 In the Alabama Breast Cancer Project, node-positive patients were randomized to CMF or L-PAM. The small number of node-positive patients with archival breast tissues available for analysis prevented us from determining differences in molecular biomarker expression between the two treatment groups.
The results of this study must be carefully applied because of biases in patient selection and data collection, which cannot be quantified. Our findings may reflect the selection bias resulting from a study of only 90 archival breast cancer specimens when 311 patients were accrued to the Alabama Breast Cancer Project. However, because there was little difference with respect to disease-free and overall survival in these two populations, it is unlikely that the bias introduced invalidates our findings. In addition, with the relatively small populations studied, conclusions must be formulated with caution. Study population size can influence the results of both univariate and multivariate analysis, affecting the number of factors found to have prognostic significance. Prospective study of a much larger number of patients is encouraged to validate the prognostic significance of c-erbB-2 expression, p53 expression, and coexpression of c-erbB-2 and p53.
As our knowledge regarding molecular biomarkers for breast cancer increases, prognostic indices will be developed that combine the predictive power of individual molecular biomarkers with specific clinical and pathologic factors. In laboratories at Brown University, the molecular biomarkers c-fos, c-myc, Ha-ras, and p53 were studied in patients with stage I, IIa, and IIb breast cancer. Although no single biomarker had independent prognostic significance, coexpression of three or more biomarkers was confirmed to endow breast cancers with an aggressive phenotype, significantly affecting both disease-free and overall survival. 51 The results of the study presented here suggest that c-erbB-2 and p53 overexpression may provide additional criteria for assessing prognosis in breast cancer. Our results suggest that coexpression of c-erbB-2 and p53 may have more prognostic significance than traditional prognostic factors such as T stage and nodal status.
Discussion
DR. MICHAEL J. EDWARDS (Louisville, Kentucky): I’d like to begin by commending my mentor Dr. Bland and my colleague Dr. Beenken on their initiative to determine the specific role of certain molecular biomarkers on breast cancer prognosis. It’s a tough job and one that has been problematic over time, but I do believe that we are now entering an era when some of these molecular markers will become clinically important, perhaps even more important than some of our historical parameters used in defining biological behavior. The real problem we have had in the recent past is translating some of these early biological findings to clinical practice and clinical relevance. To date, what we have seen that might pertain to a given population in a tertiary medical center with the molecular expertise of Drs. Bland and Beenken, has not necessarily apply to community practice. These markers have not yet penetrated our daily clinical practice. A lot of work has been done in this area. If you do a literature search on the two topics of prognostic factors and breast cancer, you will retrieve over 2,000 publications. Yet we still rely upon the TNM system and ER/PR testing in spite of these some 2,000 publications. I am encouraged by this work because to date we have not had anyone comment on a database with this degree of follow-up. I think that through this smoke there really is fire, and that we will discover something important. I suspect that these two markers are as important as anything else that we have ever studied. But I want to ask Dr. Beenken to comment on how easy it is to quantitate these parameters. We have had a nightmare of a problem trying to figure out what it means to be node positive and node negative with the advent of immunohistochemical staining for nodal disease. How are these molecular markers going to translate? Are these molecular techniques and procedures easily applicable in the community? There may be a problem here, because there are already conflicting studies showing no value with these same markers, while there are more studies showing value. I would like for you to comment about the potential clinical applicability and what barriers you see that might need to be overcome to achieve clinical relevance. And, finally, I note that this study stems from very old data, and that is wonderful. It is available to us from some of our forefathers who had the foresight to save tissue so that people in next generations could come by and determine the biological behavior of the disease. I think we are indebted to Bill Mattox as the principal investigator of the Alabama Breast Cancer Project in terms of making this available to us today. Thank you.
DR. WILLIAM C. WOOD (Atlanta, Georgia): For molecular markers of prognosis to enter actual clinical use and begin to replace the parameters that we have used for the past 50 years, two steps are necessary, as you all know. The first step is that a large number of patients with similarly treated tumors are analyzed in order to develop hypotheses that, first, make biological sense and, secondly, segregate the data sets statistically more successfully than the standard parameters of prognosis that we have used in the past. Step two is that the hypothesis must then be tested by prospective application to a different population to see if a new principle has been found or to see if the principle merely reflected the play of chance in the first data set. Many a fine hypothesis has been wrecked by brutish facts in step two! And the authors have called for just such a prospective study. This is, as they said, a step one study. It illustrates the difficulty of finding a population even with a common tumor where women have been treated similarly and the much tougher problem of then obtaining sufficient tissue specimens to ask biological and molecular biological questions. A 4-year collection of tissues from Alabama women with breast cancer yielded only 90 paraffin blocks for analysis in this series.
Your conclusion or hypothesis that any membrane overexpression of c-erbB-2 protein on breast cancer cells coupled with the highest levels of mutated p53 expression portends a grim prognosis rests on only six patients. Now, it is made more credible than this number behind it because it fits with our present thoughts about the role of these two markers biologically.
My first question is this: we would also have expected bcl-2 and TGF-α to relate to prognosis. Why do you think they didn’t in this series? Was the data set too small to detect it? Do you think the reagents were inadequate? Or are you challenging our understanding of their involvement in breast cancer?
In our analysis of 1,500 women in the Intergroup Study C7581 with randomization to three levels of FAC chemotherapy dose intensity, we were only able to collect blocks from two sets of about 300 women. We found overexpression of c-erbB-2 and p53 protein elevation to indicate a group of women that greatly benefited from increased levels of dose intensity with our chemotherapy. My second question: do you see those results as conflicting with or compatible with your own data? In other words, do you see a predictive possibility for the results that you reported today?
I really wish to join in congratulating the authors for bringing a study of mature clinical data set, well-followed patients for many years, together with the difficult task of obtaining blocks and doing these biochemical assays to fruition and presenting it today. I also want to thank you for sending me your manuscript. I enjoyed reading it and recommend its very thoughtful discussion to all the membership.
DR. SAMUEL W. BEENKEN (Birmingham, Alabama): I would like to begin my reply by thanking Dr. Wood and Dr. Edwards for their willingness to review this paper and for their very thoughtful comments this afternoon. Biomarker research is in an early stage of development. As new information becomes available about the molecular events responsible for the development of breast cancer, there will undoubtedly be changes in the emphasis that we place on the current clinical and prognostic biomarkers. A long-term goal of biomarker research is to decipher local growth characteristics, metastatic potential, and ultimately prognosis based on genetic analysis of the primary tumor. c-erbB-2 and p53 coexpression may or may not be of importance when we have a more complete understanding of the molecular events underlying breast cancer, but the current literature suggests it to be very significant.
Clinical application of our findings awaits validation in a much larger breast cancer population than we have studied to date. At our institution, we are actively engaged in developing the infrastructure necessary to coordinate such a study. Dr. William Grizzle, the pathologist primarily involved with us in the current study, has been appointed to direct a biomarker validation laboratory in the Early Detection Research Network of the National Cancer Institute. This laboratory will focus on the use of immunohistochemistry for the analysis of early detection and prognostic biomarkers. Most pathology laboratories around the country are already familiar with immunohistochemistry; in fact it is the preferred methodology for ER/PR receptor analysis. With standardization and validation of immunohistochemical techniques and with adequate education of potential users, these techniques can be successfully applied to new clinical situations.
I believe these comments address the very important points that Dr. Edwards brought forward about the ease of quantification and utilization. Let me add that the recent award of a National Cancer Institute Breast SPORE to our institution will enable us to gather clinical data and tissue specimens from all of the 500+ women undergoing surgery at our institution yearly for breast cancer. In addition, we are upgrading our current biostatistics capabilities to enable us to analyze the interaction of multiple biomarkers in tissues from large numbers of patients.
As Dr Wood pointed out, bcl-2 and TGF-α expression were not shown to have prognostic significance in our analysis. In breast cancer, overexpression of bcl-2 has been shown to correlate with high histologic grade, the presence of axillary lymph node metastases, and reduced disease-free and overall survival. bcl-2-family proteins appear to regulate a step in the apoptosis pathway that can be triggered by p53-dependent or -independent factors. Some bcl-2 proteins function as inhibitors of apoptosis and others as promoters of apoptosis, which may account for some of the inconsistencies seen in the breast cancer literature. In addition, the bax/bcl-2 ratio is now thought to provide better prognostic information in breast cancer. The death signal protein, bax, is induced by a variety of factors including growth factor deprivation in the presence of wild-type (normal) p53. The bax/bcl-2 ratio and the resulting formation of either bax-bax homodimers, which stimulate apoptosis, or bax-bcl-2 heterodimers, which inhibit apoptosis, represent an intra-cellular regulatory mechanism that appears to have prognostic importance in breast cancer.
TGF-α shares 33% homology with the epidermal growth factor (EGF) and binds to its receptor (EGFr). In breast cancer, TGF-α overexpression has been associated with high histologic grade, tumor recurrence and poor prognosis, although there is controversy regarding its prognostic significance. Recently, an association between the expression of TGF-α and EGFr and angiogenesis has been shown. It is postulated that TGF-α and EGFr coexpression may predict blood-borne metastases in breast carcinoma. In the study presented today, bcl-2 expression was determined in only 40 tissue specimens because of the limited amount of tissue available for study in some patients, while TGF-α expression was only studied in 50 tissue specimens. The relatively small number of specimens studied may have prevented us from demonstrating prognostic significance in our patient population.
Finally, the literature does show some variability regarding the prognostic significance of c-erbB-2 and p53 coexpression, although most papers confirm its prognostic significance. The variability seen may reflect the impact that differing chemotherapeutic regimes have on survival. As Dr. Wood indicated, patients whose breast cancers coexpress c-erbB-2 and p53 respond favorably to higher doses of FAC chemotherapy, which is in agreement with the finding that breast cancers which overexpress c-erbB-2 respond better to chemotherapeutic regimes that include Adriamycin. Because of the small number of patients randomized to L-PAM or CMF chemotherapy in our study, we were not able to demonstrate any effect of chemotherapy on outcome in our patient population.
Footnotes
Presented at the 112th Annual Meeting of the Southern Surgical Association, December 4–6, 2000, Palm Beach, Florida.
Correspondence: Samuel W. Beenken, MD, Dept. of Surgery, University of Alabama at Birmingham, Suite 321, 1922 7th Ave. South, Birmingham, AL 35294-0016.
E-mail: samuel.beenken@ccc.uab.edu
Accepted for publication December 2000.
References
- 1.Fabian CJ, Kimler BF, Beenken SW, et al. Models for early chemoprevention trials in breast cancer. Hemat Oncol Clinic North Am 1998; 12: 993–1017. [DOI] [PubMed] [Google Scholar]
- 2.Stancovski I, Sela M, Yardin Y. Molecular and clinical aspects of the neu/erbB-2 receptor tyrosine kinase. In: Dickson R, Lippman M, eds. Mammary tumorigenesis and malignant progression. Boston: Kluwer Academic Publishers; 1994: 161–191.
- 3.Singleton TP, Strickler JG. Clinical and pathological significance of the c-erbB-2 (HER-2/neu) oncogene. Pathol Ann 1992; 27: 165–190. [PubMed] [Google Scholar]
- 4.Thiberville L, Payne P, Vielkinds J, et al. Evidence of cumulative gene losses with progression of premalignant epithelial lesions to carcinoma of the bronchus. Cancer Res 1995; 55: 5133–5139. [PubMed] [Google Scholar]
- 5.Slamon DJ, Clark GM. Amplification of c-erbB-2 and aggressive human breast tumors? Science 1988; 240: 1795–1798. [DOI] [PubMed] [Google Scholar]
- 6.Reese DM, Slamon DJ. HER-2/neu signal transduction in human breast and ovarian cancer. Stem Cells 1997; 15: 1–8. [DOI] [PubMed] [Google Scholar]
- 7.Levine AJ. P53, the cellular gatekeeper for growth and division. Cell 1997; 88: 323–331. [DOI] [PubMed] [Google Scholar]
- 8.Rohan TE, Hartwick W, Miller AB, et al. Immunohistochemical detection of c-erbB-2 and p53 in benign breast disease and breast cancer risk. J Natl Cancer Inst 1998; 90: 1262–1269. [DOI] [PubMed] [Google Scholar]
- 9.Tauchi K, Hori S, Itoh H, et al. Immunohistochemical studies on oncogene products (c-erbB-2, EGFR, c-myc) and estrogen receptor in benign and malignant breast lesions. Virchows Arc A Pathol Anat Histopathol 1989; 416: 65–73. [DOI] [PubMed] [Google Scholar]
- 10.Lodato RF, Maguire HC Jr, Greene MI, et al. Immunohistochemical evaluation of c-erbB-2 oncogene expression in ductal carcinoma in situ and atypical ductal hyperplasia of the breast. Mod Pathol 1990; 3: 449–454. [PubMed] [Google Scholar]
- 11.Pechoux C, Chardonnet Y, Noel P. Immunohistochemical studies on c-erbB-2 oncoprotein expression in paraffin-embedded tissues in invasive and non-invasive human breast lesions. Anticancer Res 1994; 14: 1343–1360. [PubMed] [Google Scholar]
- 12.Kalogeraki A, Tzardi M, Datseris G, et al. c-erbB-2 expression in patients with breast carcinoma in comparison to patients with benign breast disease. Anticancer Res 1996; 16: 765–771. [PubMed] [Google Scholar]
- 13.Bartek J, Bartkova J, Vojtesek B, et al. Pattern of expression of the p53 tumour suppressor in human breast tissues and tumors in situ and in vitro. Int J Cancer 1990; 46: 839–844. [DOI] [PubMed] [Google Scholar]
- 14.Heyderman E, Dagg B. p53 immunostaining in benign breast disease. Lancet 1991; 338: 1532. [DOI] [PubMed] [Google Scholar]
- 15.Barbareschi M, Leonardi E, Mauri FA, et al. p53 and c-erbB-2 protein expression in breast carcinomas. An immunohistochemical study including correlations with receptor status, proliferation markers, and clinical stage in human breast cancer. Am J Clin Pathol 1992; 98: 408–418. [DOI] [PubMed] [Google Scholar]
- 16.Thor AD, Moore DH II, Edgerton SM, et al. Accumulation of p53 tumor suppressor gene protein: an independent marker of prognosis in breast cancers. J Natl Cancer Inst 1992; 84: 845–855. [DOI] [PubMed] [Google Scholar]
- 17.Qi FY, Zuo LF, Zhen YJ. Quantitative study of p53 gene protein expression in the benign disease and cancer of the breast. Chung-hua Ping Li Hsueh Tsa Chi 1994; 23: 330–333. [PubMed] [Google Scholar]
- 18.Schmitt FC, Leal C, Lopes C. p53 protein expression and nuclear DNA content in breast intraductal proliferations. J Pathol 1995; 176: 233–241. [DOI] [PubMed] [Google Scholar]
- 19.Younes M, Lebovitz RM, Bommer KE, et al. p53 protein accumulation in benign breast biopsy specimens. Hum Pathol 1995; 26: 155–158. [DOI] [PubMed] [Google Scholar]
- 20.Millikan R, Hulka B, Thor A, et al. p53 mutations in benign breast tissue. J Clin Oncol 1995; 13: 2293–2300. [DOI] [PubMed] [Google Scholar]
- 21.Muller WJ, Sinn E, Pattengale PK, et al. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 1988; 54: 105–115. [DOI] [PubMed] [Google Scholar]
- 22.Jerry DJ, Ozbun MA, Kittrell FS, et al. Mutations in p53 are frequent in the preneoplastic stage of mouse mammary tumor development. Cancer Res 1993; 53: 3374–3381. [PubMed] [Google Scholar]
- 23.Maddox MA, Carpenter JT, Laws HL, et al. A randomized prospective trial of radical (Halstead) mastectomy versus modified radical mastectomy in 311 breast cancer patients. Ann Surg 1983; 198: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker RA, Gullick WJ, Varley JM. An evaluation of immunoreactivity for c-erbB-2 protein as a marker of poor short-term prognosis in breast cancer. Br J Cancer 1989; 60: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallioniemi O-P, Holli K, Visakorpi T, et al. Association of c-erbB-2 protein over-expression with high rate of cell proliferation, increased risk of visceral metastasis and poor long-term survival in breast cancer. Int J Cancer 1991; 49: 650–655. [DOI] [PubMed] [Google Scholar]
- 26.Winstanley J, Cooke T, Murray GD, et al. The long-term prognostic significance of c-erbB-2 in primary breast cancer. Br J Cancer 1991; 63: 447–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjogren S, Inganas M, Lindgren A, et al. Prognostic and predictive value of c-erbB-2 over-expression in primary breast cancer, alone and in combination with other prognostic markers. J Clin Oncol 1998; 16: 462–469. [DOI] [PubMed] [Google Scholar]
- 28.Barnes DM, Lammie GA, Millis RR, et al. An immunohistochemical evaluation of c-erbB-2 expression in human breast carcinoma. Br J Cancer 1988; 58: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van de Vijver MJ, Peterse JL, Mooi WJ, et al. Neu-protein over-expression in breast cancer. Association with comedo-type ductal carcinoma in situ and limited prognostic value in stage II breast cancer. N Engl J Med 1988; 319: 1239–1245. [DOI] [PubMed] [Google Scholar]
- 30.Thor AD, Moore DH II, Edgerton SM, et al. Accumulation of p53 tumor suppressor gene protein: an independent marker of prognosis in breast cancers. J Natl Cancer Inst 1992; 84: 845–855. [DOI] [PubMed] [Google Scholar]
- 31.Allred DC, Clark GM, Elledge R, et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst 1993; 85: 200–206. [DOI] [PubMed] [Google Scholar]
- 32.Allred DC, Harvey JM, Berardo M, et al. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Modern Pathol 1998; 11: 155–168. [PubMed] [Google Scholar]
- 33.Chang F, Syranen S, Syrjanen K. Implications of the p53 tumor-suppressor gene in clinical oncology. J Clin Oncol 1995; 13: 1009–1022. [DOI] [PubMed] [Google Scholar]
- 34.Wynford-Thomas D. p53 in tumor pathology: can we trust immunohistochemistry? J Pathol 1992; 166: 329–330. [DOI] [PubMed] [Google Scholar]
- 35.Chang K, Ding I, Kern FG, et al. Immunohistochemical analysis of p53 and HER-2/neu proteins in human tumors. J Histochem Cytochem 1991; 39: 1281–1287. [DOI] [PubMed] [Google Scholar]
- 36.Barbareschi M, Leonardi E, Mauri FA, et al. p53 and c-erbB-2 protein expression in breast carcinomas. An immunohistochemical study including correlations with receptor status, proliferation markers, and clinical stage in human breast cancer. Am J Clin Pathol 1992; 98: 408–418. [DOI] [PubMed] [Google Scholar]
- 37.Isola J, Visakorpi T, Holli K, et al. Association of over-expression of tumor suppressor protein p53 with rapid cell proliferation and poor prognosis in node-negative breast cancer patients. J Natl Cancer Inst 1992; 84: 1109–1114. [DOI] [PubMed] [Google Scholar]
- 38.Jacquemier J, Penault-Llorca F, Viens P, et al. Breast cancer response to adjuvant chemotherapy correlates with erbB2 and p53 expression. Anticancer Res 1994; 14: 2773–2778. [PubMed] [Google Scholar]
- 39.Marks JR, Humphrey PA, Wu K, et al. Over-expression of p53 and HER-2/neu proteins as prognostic markers in early-stage breast cancer. Ann Surg 1994; 219: 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiltschke C, Kindas-Muegge I, Steininger A, et al. Co-expression of HER-2/neu and p53 is associated with a shorter disease-free survival in node-positive breast cancer patients. J Cancer Res Clin Oncol 1994; 120: 737–742. [DOI] [PubMed] [Google Scholar]
- 41.Rosen PP, Lesser ML, Arroyo CD, et al. p53 in node-negative breast carcinoma: an immunohistochemical study of epidemiologic risk factors, histologic features and prognosis. J Clin Oncol 1995; 13: 821–830. [DOI] [PubMed] [Google Scholar]
- 42.Schneider J, Rubio MP, Barbazan MJ, et al. P-glycoprotein, HER-2/neu, and mutant p53 expression in human gynecologic tumors. J Natl Cancer Inst 1994; 86: 850–855. [DOI] [PubMed] [Google Scholar]
- 43.Menard S, Casalini P, Pilotti S, et al. No additive impact on patient survival of the double alteration of p53 and c-erbB-2 in breast carcinomas. J Natl Cancer Inst 1996; 88: 1002–1003. [DOI] [PubMed] [Google Scholar]
- 44.Nakopoulou LL, Alexiadou A, Theodoropoulos GE, et al. Prognostic significance of the co-expression of p53 and c-erbB-2 proteins in breast cancer. J Pathol 1996; 179: 31–38. [DOI] [PubMed] [Google Scholar]
- 45.Barbati A, Cosmi EV, Sidoni A, et al. Value of c-erbB-2 and p53 oncoprotein co-over-expression in human breast cancer. Anticancer Res 1997; 17: 401–405. [PubMed] [Google Scholar]
- 46.Rudas M, Neumayer R, Gnant MFX, et al. p53 protein expression, cell proliferation and steroid hormone receptors in ductal and lobular in situ carcinomas of the breast. Eur J Cancer 1997; 33: 39–44. [DOI] [PubMed] [Google Scholar]
- 47.Bebenek M, Bar JK, Harlozinska A, et al. Prospective studies of p53 and c-erbB-2 expression in relation to clinicopathological parameters of human ductal breast cancer in the second stage of clinical advancement. Anticancer Res 1998; 18: 619–623. [PubMed] [Google Scholar]
- 48.Thor AD, Berry DA, Budman DR, et al. erbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J Natl Cancer Inst 1998; 90: 1346–1360. [DOI] [PubMed] [Google Scholar]
- 49.Tsuda H, Sakamaki C, Tsugane S, et al. A prospective study of the significance of gene and chromosome alterations as prognostic indicators of breast cancer patients with lymph node metastases. Breast Cancer Res Treat 1998; 48: 21–32. [DOI] [PubMed] [Google Scholar]
- 50.Pich A, Margaria E, Chiusa L. Oncogenes and male breast carcinoma: c-erbB-2 and p53 co-expression predicts a poor survival. J Clin Oncol 2000; 18: 2948–2957. [DOI] [PubMed] [Google Scholar]
- 51.Bland KI, Konstadoulakis MM, Vezeridis MP, et al. Oncogene protein co-expression: value of Ha-ras, c-myc, c-fos and p53 as prognostic discriminants for breast carcinoma. Ann Surg 1995; 221: 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]