Abstract
Some species introduced into new geographical areas from their native ranges wreak ecological and economic havoc in their new environment. Although many studies have searched for either species or habitat characteristics that predict invasiveness of exotic species, the match between characteristics of the invader and those of members of the existing native community may be essential to understanding invasiveness. Here, we find that one metric, the phylogenetic relatedness of an invader to the native community, provides a predictive tool for invasiveness. Using a phylogenetic supertree of all grass species in California, we show that highly invasive grass species are, on average, significantly less related to native grasses than are introduced but noninvasive grasses. The match between the invader and the existing native community may explain why exotic pest species are not uniformly noxious in all novel habitats. Relatedness of invaders to the native biota may be one useful criterion for prioritizing management efforts of exotic species.
Keywords: biological invasions, community phylogenetics, grass, novel weapons, supertree
Because increasingly large numbers of species are being imported by humans to areas far from their native geographic ranges, and because some of these species have huge ecological and economic impacts on the novel communities to which they are introduced (1), the ability to predict which species are likely to have large impacts becomes paramount (2). Many studies have searched for either species or habitat characteristics that predict invasiveness (e.g., refs. 3–8); however, identifying such attributes has proven challenging (7). Rather than considering traits of invaders and invaded communities in isolation, we support the view that the match between the invader and the invaded community is key to understanding invasiveness (9).
With respect to the establishment of introduced species, several studies have argued that phenotypic similarity between native communities and invaders reduces the success of invading species (10–13). Darwin’s naturalization hypothesis (14) suggested that novel genera would be more successful in naturalizing in new ranges than genera with native representatives. Darwin proposed that species closely related to native species should not succeed because they overlap in resource use (i.e., are unsuccessful because of limiting similarity). Another mechanism that predicts the same pattern is that release from enemies allows exotic species to establish. Natural enemies are expected to shift onto close relatives of their host more easily than onto distantly related taxa (15); thus, release from enemies should be greater in taxa more distantly related to natives. In both Darwin’s naturalization and the enemy-release hypotheses, phylogenetic relatedness is implicitly expected to correlate with net ecological similarity and inversely with the probability of sharing natural enemies. This expectation is based on the common ancestry of species, is fundamental to evolutionary biology theory (16), and is generally borne out by observation (17–19). In support of the proposed taxonomic pattern, species introduced to California from the plant families Poaceae, Brassicaceae, and Asteraceae were more likely to belong to novel genera than would be expected by chance (20, 21). The pattern, however, was not supported in the Hawaiian flora (22), where no association was found. The approach used by these authors depends very much on knowing the appropriate source pool of all potential invaders, and assessing potential source pools is not trivial (23).
An alternative hypothesis to Darwin’s naturalization hypothesis predicts a diametrically opposing pattern: invaders more closely related to natives should be more likely to succeed in novel environments, owing to similarities based on common ancestry. Species more closely related to natives in New Zealand were more likely to establish, perhaps because they share traits with their native relatives that allow them to thrive in their new environment (24). Again, testing this approach depends on specifying the appropriate source pool for invaders. The view that similarity predisposes invaders to success is also supported by work showing that native plants inhabiting particular microhabitats are more closely related to one another than would be expected by chance alone (17, 18, 25). Importantly, both phylogenetic repulsion (17) (predicted by Darwin’s naturalization hypothesis) and phylogenetic attraction (predicted from sharing conserved characters) suggest that whether an exotic species will be successful in establishing depends on how its traits match with the traits of native species in the invaded community.
Although the cases cited above have focused on establishment of invaders, only a few studies have tried to understand not just establishment, but the impact of exotics as a function of the match between recipient community and invader. There is a fundamental difference between species that are introduced and persist at relatively low numbers in a new habitat and those that dominate and become ecosystem engineers, thereby profoundly affecting ecological communities (26, 27). For example, spotted knapweed has swept through grasslands in western North America (11, 28, 29), and many introduced diseases have virulent effects that decimate naïve populations of native hosts (30–32). Only two studies, one on aquatic communities and the other on plant communities, have considered whether high-impact invaders differ in their relatedness to the native community from low-impact invaders. Both studies find that invaders with large impacts are more likely to come from genera not represented in the native fauna or flora than from genera with native representatives (9, 33).
Here, we expand on these studies by asking whether already established introduced species with lesser impacts differ in their phylogenetic relatedness to native species than do high-impact, invasive species. Instead of using comparisons at the generic level, as all other prior studies have done, we use phylogenetic supertrees as a rigorous framework with which to estimate phylogenetic distinctiveness of invasive pests. We classify introduced species into two categories: introduced-pest species (as defined by state and federal authorities) and naturalized, nonnoxious species (introduced-nonpest). By comparing two different classes of already established introduced species (pest and nonpest) to natives, we do not require knowledge of the source pool of all possible colonizers, nor do we need to account for species that have failed to establish (because we compare only species that have already naturalized). Our general approach is to compare the relatedness of native species to introduced species of varying impact. We use two metrics to compare the degree to which pest and nonpest species are related to natives: (i) the mean phylogenetic distance of each introduced species to the whole native community and (ii) the mean phylogenetic distance of each introduced species to its nearest native relative in the native community. The first metric provides a community-wide perspective on overall ecological novelty, whereas an analysis of nearest taxon more closely addresses whether limiting similarity to the closest relative, as proposed by Darwin and others, is important in limiting invasiveness.
Results
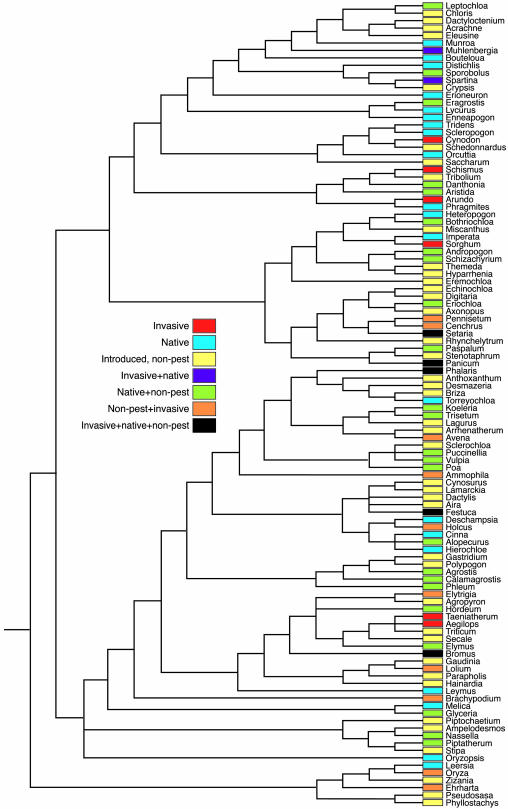
The distribution of species status (native, introduced-nonpest, introduced-pest) on the main supertree is indicated in Fig. 1. Both introduced-nonpest and pest character states were significantly conserved on the tree by using the Maddison–Slatkin test (34) implemented in macclade (35) (i.e., taxa with these states were nonrandomly clustered: nonpest, 54 steps; P < 0.004 random values and pest, 22 steps; P < 0.001, respectively). Despite being nonrandomly distributed, the occurrence of pest status occurs many independent times on the tree (red, orange, purple, and black genera in Fig. 1 have at least one pest member). Of the 42 pest species belonging to 13 genera, there are at least 12 independent origins of “pestiness.” Thus, although there might be shared traits among pests, these traits would not all have arisen from shared common descent.
Fig. 1.
Supertree of the grass genera of California based on Salamin et al. (38) grass supertree. Genera are color coded to represent the types of species contained within that genus (native, introduced-nonpest, introduced-pest, and combinations of these groups).
Pest species were significantly more distantly related to the community of native species than were nonpest introduced species (tree B, pest = 104.03 relative time units; SE = 5.76, n = 42; nonpest = 91.95 units; SE = 2.67, n = 165; t = 2.01, df = 205; P = 0.046). Similarly, in tree A, when branch lengths were not adjusted for time from divergence, we still find that the mean phylogenetic distance between invasive pest species and native grasses was significantly greater than that between nonpest, introduced species and natives (pest, mean branch length from natives = 15.31; SE = 0.28, n = 42; whereas nonpest species were, on average, 14.46 mean branch length from natives; SE = 0.15, n = 165; t = 2.39, df = 205; P = 0.018).
We subsequently conducted a few supplementary tests to determine how robust this result was to tree topology. To consider alternative tree topologies, we used a random sample of 100 trees (tree set C1–100) of the 985 most parsimonious trees that were the basis of the consensus tree to form a set of alternative trees against which to test the hypothesis. The result that invasive species were more distantly related to natives than nonpest introduced species held (P ≤ 0.02 in all 100 cases). Thus, our results are robust to some variation in tree topology.
These results support the idea that, at the community level, phylogenetic distinctiveness, presumably accompanied by ecological difference, is associated with increased invasiveness.
The results are similar but not as strong when we consider phylogenetic distance of invaders to the nearest native taxon. Using the time-adjusted tree B, we find that pest species are still less related to natives than nonpest species, but this difference is no longer significant (mean phylogenetic distance for pest = 10.20, SE = 1.83, n = 42; mean distance for nonpest = 8.55, SE = 1.09, n = 165; t-ratio = 0.694, df = 205, P = 0.49). For the tree in which topology, but not divergence time among taxa, was taken into account (tree A) we do find that pest species are significantly less related than nonpest species to the nearest native relative (mean distance for pest = 4.09, SE = 0.28, n = 42; mean distance for nonpest = 3.37, SE = 0.12, n = 165, t-ratio = 2.47, df = 205; P = 0.014).
We also assessed an alternative explanation for the overall pattern: The geographic origins of invasive species differed from those of introduced, nonpest species, and this difference could be confounding our result. There were no significant differences between pest and nonpest introduced species in their areas of origin (χ2 = 7.26, 5 df, P = 0.20, Table 1); however, power to detect differences in origin between pest and nonpest species was relatively low (see Methods).
Table 1.
Areas of origin for species of introduced grasses in California
| Area of origin | Introduced nonpest | Introduced pest |
|---|---|---|
| Africa | 12 | 8 |
| Asia | 42 | 12 |
| Australia | 5 | 0 |
| Europe | 65 | 14 |
| Nonwestern North America | 19 | 3 |
| South America | 19 | 4 |
| Total | 163 | 42 |
χ2 = 8.50, 5 df; P = 0.13.
Discussion
We have demonstrated that introduced invasive environmental weed species with high ecological impacts are less closely phylogenetically related to members of the native community than are species that naturalize without having large effects on local native species’ diversity or richness. Similarly, we found that invasive grasses tend to be less closely phylogenetically related to their nearest native relative in an invaded community than noninvasive grasses, although the pattern was significant for only one measure of relatedness. These results support both Darwin’s naturalization hypothesis and the “escape from natural enemies” hypothesis and imply that, at least at the metacommunity scale of California, species that are more distantly related to the native community, are most likely to explode in population and become noxious weeds.
The two metrics of community relatedness, distance to nearest relative versus distance to the community as a whole, may reflect different kinds of ecological mechanisms underlying invasiveness. Distance to nearest taxon reflects interactions with a single species, likely the most phenotypically similar species, as envisioned by Darwin. We might expect that such similarity would play an important role at the establishment phase of invasions if limiting similarity is, indeed, the mechanism preventing establishment. A metric that captures overall community similarity is likely to be more reflective of dynamics and diverse interactions with multiple species. Multispecies interactions may be more important if multiple resources limit an invasive species, if natural enemies have broad (polyphagous) tastes that, nevertheless, have a phylogenetic signature (e.g., at the genus level), if a collection of related species has more impact than a single, closely related one, and/or if community-scale evolutionary naïveté to a particular interaction is an important mechanism (36). Our results suggest that the presence or absence of multiple, closely related species and not just a single most closely related species may more effectively limit the success of an invader once it has become established. Because our analyses considered only postestablishment population dynamics, they do not inform the importance of nearest taxon or community relatedness at the establishment phase (for which knowledge of failure to establish by introduced species would be key).
Phylogenetic relatedness of invaders to natives provides a method for identifying threats to native communities. As mentioned above, relatedness and evolutionary divergence of invaders may reflect novel ecological ways to use resources, novel “weapons” against cooccurring members, or ability to escape from enemies (12, 13, 17). We believe that the match between traits of invaders and those of natives, in addition to any single trait or characteristic of the invading species or the invaded community, may be key to understanding impacts of invaders. Because a large diversity of mechanisms may underlie invasion success (7), ecological novelty may be partially gauged by relatedness of the invader to natives and is a general metric that does not require knowledge of specific traits. In addition, use of ultrametric supertrees provides a phylogenetic context to communities and more effectively estimates the true divergence of invaders from native communities than simply comparisons at the genus level. Our results, coupled with recent findings in several other systems (9, 12, 33, 37), suggest that particular attention should be paid to newly introduced species for which there are no close relatives in the local biota.
Methods
We selected the grasses of California as our test group. The large number of grass species (489) and genera (128) in California and their dominance in wild grasslands of the state gave us reasonable power with which to test the importance of phylogenetic relatedness to invasiveness. Additionally, a recent supertree of the grasses (38) has elucidated many of the deeper phylogenetic relationships among grass genera. Although geopolitical boundaries are not always biologically meaningful, the California floristic province, as defined by Axelrod (39) and as used in The Jepson Manual (40), closely mirrors the vast majority of the area of the state of California, barring the southeastern Mojave desert; thus, the grass flora of California represents a biologically meaningful entity.
Although the California floristic province includes diverse habitats, many introduced species in California, because of their dispersal along roads and railroad rights-of way, are found in every county of the state (see distributions in the CalFlora on-line database). Thus, these introduced species interact with many species of natives. We recognize that, even within counties, plants may segregate according to habitat, but if propagule exchange occurs between these habitats, then we consider that interactions among species are probable at seed and seedling stages. For all these reasons, we feel that using the spatial scale of the state of California is appropriate (see also refs. 21, 22, and 33 for studies conducting analyses at the state level).
We assigned grass species to three categories: native (277 spp.), introduced nonpest (167 spp.), and introduced pest (46 spp.) species. We clarify that we consider as pests only noxious environmental weeds, that is, those that spread in rangelands and natural areas. We do not consider weeds solely limited to agricultural fields and cultivation that would therefore not experience the context of native community members. The Jepson Manual (40) and the CalFlora database (with its associated sources) were used to tabulate all native and nonnative grass species in California. Pest status was identified by using the California Exotic Pest Plant Council List of Noxious Weeds (see similar methods in ref. 33). The vast majority of species classified as pests in California are also considered pests by Exotic Plant Pest Councils in other states.
We were concerned that the area of origin of the invader might be confounded with phylogenetic relatedness. We therefore tabulated the origin of each invader to determine whether different origins were disproportionately represented in pest or nonpest grasses. Areas of origin were primarily identified by using The Jepson Manual (40), with some additional information from the Global Weed Compendium web site.
Supertree Construction.
We created a supertree of all of the grass species of California by using the grass supertree phylogeny of Salamin et al. (38) (semistrict consensus; Baum/Ragan; no reweighting; characters considered irreversible). Names of taxa were standardized with the U.S. Department of Agriculture Plants database (http://plants.usda.gov). Because not every genus of grass found in California is included in this supertree phylogeny, we had to drop 21 genera from our analysis (<10%, or 44 species, of the total number of species), including representatives of every class (native, introduced-nonpest and, invasive-pest) in this dropped group. The vast majority of these genera (16 of 21) had a single species in the California flora; Achnatherum had the most species with 17. Our final sample sizes were 237 native species, 165 introduced, nonpest species, and 42 introduced pest species. Species were attached to the supertree as polytomies at genus nodes by using the phylomatic tree retrieval tool (34) to form tree A. All subspecies were lumped into a single species to avoid pseudoreplication.
Like most supertrees, that of Salamin et al. (38) provides topology but not branch lengths. To characterize phylogenetic uniqueness, however, we needed some measure of divergence time among taxa. We therefore combined our larger branch-lengthless California supertree (A) with a smaller multigenus consensus tree from the Grass Phylogeny Working Group (GPWG), which was based on original sequence data (figure 4 in ref. 38); 7,128 bp), and which, therefore, did have branch-length estimates. We compared these trees to identify common nodes in the two trees (the most distal node on both trees that was basal to a set of common taxa exclusively descended from that node) and labeled these nodes in both trees with the same codes (using algorithm comnode in phylocom (www.phylodiversity.net/phylocom). We then used this labeled GPWG tree as an input tree in the program r8s (41) using nonparametric rate smoothing (NPRS) (root node set to 100 arbitrary time units) to obtain a chronogram for generic relationships and relative ages for the labeled internal nodes. Finally, we set the labeled interior nodes in our California grass supertree to the same ages and interpolated all other interior nodes to minimize variance in branch length (using the branch length adjuster (bladj) algorithm in phylocom (www.phylodiversity.net/phylocom). The resulting supertree (B) was ultrametric, having the topology of the Salamin et al. (38) large supertree, with branch lengths constrained by the GPWG tree (see similar methods in ref. 42). We believe the estimated branch lengths to be more accurate estimates of the true branch lengths than unitary branch lengths. The method we used for estimating the branch lengths (interpolating between age-estimated nodes) can be influenced by taxon sampling, but, because we had age estimates for so many nodes, the influence of taxon sampling will be slight.
To determine the degree to which variation in estimates of the best tree topology had an effect on our results, we also used a sample of 100 trees of the 985 most parsimonious trees that were the basis of the Salamin et al. (38) consensus tree to form a set of alternative trees (tree set C1–100; without branch lengths).
Comparison of Introduced-Pest with Introduced-Nonpest Species.
To determine how novel an introduced species is relative to natives, we calculated the relative phylogenetic distance between each nonnative grass species and all native grass species to get a mean distance for each nonnative species to members of the native community. Using our rate-adjusted California grass supertree B, we calculated the mean phylogenetic distance from each invasive species (n = 42) to all native species (n = 235) and from each noninvasive introduced species (n = 165) to all native species, using the function icomdist in phylocom (ref. 18 and www.phylodiversity.net/phylocom). These two sets of numbers were compared by using a t test (i.e., invasive aliens vs. noninvasive aliens). This process was repeated for all supertrees from most parsimonious input trees (tree set C, branch lengths of 1.0). We also made these comparisons on tree A, where all branches were assumed equal, although we consider this a less rigorous analytical approach. If there is a difference in distance between these two classes of introduced species, then it suggests that the match between native species and introduced species is important in predicting invasiveness. Finally, we repeated these same analyses, but instead of calculating the mean distance of each introduced species to the whole native community, we calculated the mean phylogenetic distance to the nearest native relative in the native community. The first set of analyses provides a community-wide perspective, whereas an analysis of nearest taxon might more closely address whether limiting similarity to a single taxon, as proposed by Darwin and others, is important in determining invasiveness. Finally, we conducted identical analyses at the site level, using species lists from five University of California reserves located in diverse California habitats. Despite small numbers of grass species (40 < N < 53), in three of five reserves for which we found significant differences, pest exotics were significantly less related to natives than nonpest exotics at the nearest taxon, whole-community level, or both (data not presented).
Comparison of Areas of Origin.
Areas of origin of nonnative species were identified by using The Jepson Manual, with some additional information from the Global Weed Compendium web site. For two species, area of origin was unknown. A χ2 analysis was used to compare the proportion of pest and nonpest species from each continental area. Species with any African range were categorized as African in origin, even if they had additional ranges in Europe (n = 5 spp.) and plants with a South American and nonwestern U.S. distribution were categorized with South America (n = 6 spp). These wide-ranging species were also classified the other way and analyses rerun, with the same qualitative result (0.13 < P < 0.24; 5 df for all tests). To determine the robustness of the results for Table 1, we determined that a power level of 0.80 at α = 0.05 was reached when total sample size was 310 species; this number represents a 50% increase in sample size over the current study, and we estimate that the power we currently have is only ≈60%.
Acknowledgments
We thank M. Rejmanek and M. Sanderson for insightful conversations on methods; J. DiTomaso for help with information on the invasive status of some species; and M. Rejmanek, M. Sanderson, D. Simberloff, J. Stachowicz, J. Rudgers, P. C. Wainwright, and anonymous reviewers who commented on and improved the manuscript. This work was supported by Biological Invasions, Integrative Graduate Education and Research Traineeship Program, National Science Foundation Grants DGE 0114432 (to S.Y.S.) and DEB-0212873 (to C.O.W.).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mack R. N., Lonsdale W. M. Bioscience. 2001;51:95–102. [Google Scholar]
- 2.Sagoff M. J. Agric. Environ. Ethics. 2005;18:215–236. [Google Scholar]
- 3.Byers J. E., Noonburg E. G. Ecology. 2003;84:1428–1433. [Google Scholar]
- 4.Kolar C. S., Lodge D. M. Science. 2002;298:1233–1236. doi: 10.1126/science.1075753. [DOI] [PubMed] [Google Scholar]
- 5.Booth M. S., Caldwell M. M., Stark J. M. J. Ecol. 2003;91:36–48. [Google Scholar]
- 6.Stohlgren T. J., Binkley D., Chong G. W., Kalkhan M. A., Schell L. D., Bull K. A., Otsuki Y., Newman G., Bashkin M., Son Y. Ecol. Monogr. 1999;69:25–46. [Google Scholar]
- 7.Levine J. M., Vila M., D’Antonio C. M., Dukes J. S., Grigulis K., Lavorel S. Proc. R. Soc. London B; 2003. pp. 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Ruijven J., De Deyn G. B., Berendse F. Ecol. Lett. 2003;6:910–918. [Google Scholar]
- 9.Ricciardi A., Atkinson S. K. Ecol. Lett. 2004;7:781–784. [Google Scholar]
- 10.Vivanco J. M., Bais H. P., Stermitz F. R., Thelen G. C., Callaway R. M. Ecol. Lett. 2004;7:285–292. [Google Scholar]
- 11.Callaway R. M., Thelen G. C., Rodriguez A., Holben W. E. Nature. 2004;427:731–733. doi: 10.1038/nature02322. [DOI] [PubMed] [Google Scholar]
- 12.Fargione J., Brown C. S., Tilman D. Proc. Natl. Acad. Sci. USA. 2004;101:414. [Google Scholar]
- 13.Mack R. N. Int. J. Plant Sci. 2003;164:S185–S196. [Google Scholar]
- 14.Darwin C. London: John Murray; 1859. p. 490. [Google Scholar]
- 15.Strong D. R., Lawton J. H., Southwood T. R. E. Insects on Plants: Community Patterns and Mechanisms. Cambridge, MA: Harvard Univ. Press; 1984. [Google Scholar]
- 16.Harvey P. H., Pagel M. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 17.Cavender-Bares J., Ackerly D. D., Baum D. A., Bazzaz F. A. Am. Nat. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- 18.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. In: Annual Review of Ecology and Systematics. Futuyma D. J., editor. Vol. 33. Palo Alto, CA: Annual Reviews; 2002. pp. 475–505. [Google Scholar]
- 19.Webb C. O., Gilbert G. S., Donoghue M. J. Ecology. 2006 doi: 10.1890/0012-9658(2006)87[123:psmssa]2.0.co;2. in press. [DOI] [PubMed] [Google Scholar]
- 20.Rejmanek M., Richardson D. M. Ecology. 1996;77:1655–1661. [Google Scholar]
- 21.Rejmanek M. In: Invasive Species and Biodiversity Management. Sandlund O. T., Schei P. J., Vilken A., editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 79–102. [Google Scholar]
- 22.Daehler C. C. Am. Nat. 2001;158:324–330. doi: 10.1086/321316. [DOI] [PubMed] [Google Scholar]
- 23.Cassey P., Blackburn T. M., Duncan R. P., Lockwood J. L. J. Anim. Ecol. 2005;74:250–258. [Google Scholar]
- 24.Duncan R. P., Williams P. A. Nature. 2002;417:608–609. doi: 10.1038/417608a. [DOI] [PubMed] [Google Scholar]
- 25.Webb C. O. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 26.Crooks J. A. Oikos. 2002;97:153–166. [Google Scholar]
- 27.Mack R. N. Ann. Mo. Bot. Gard. 2003;90:77–90. [Google Scholar]
- 28.Callaway R. M., Thelen G. C., Barth S., Ramsey P. W., Gannon J. E. Ecology. 2004;85:1062–1071. [Google Scholar]
- 29.Ridenour W. L., Callaway R. M. Plant Ecol. 2003;169:161–170. [Google Scholar]
- 30.Anagnostakis S. L. Mycologia. 1987;79:23–37. [Google Scholar]
- 31.Caffrey C., Smith S. C. R., Weston T. J. Condor. 2005;107:128–132. [Google Scholar]
- 32.Patterson K. B., Runge T. Am. J. Med. Sci. 2002;323:216–222. doi: 10.1097/00000441-200204000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Lockwood J. L., Simberloff D., McKinney M. L., Von Holle B. Biol. Invasions. 2001;3:1–8. [Google Scholar]
- 34.Maddison W. P., Slatkin M. Evolution. 1991;45:1184–1197. doi: 10.1111/j.1558-5646.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- 35.Maddison D. R., Maddison W. P. macclade 4.0: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2000. [DOI] [PubMed] [Google Scholar]
- 36.Callaway R. M., Ridenour W. M., Laboski T., Weir T., Vivanco J. M. J. Ecol. 2005;93:576–583. [Google Scholar]
- 37.Ricciardi A. Can. J. Fish. Aquat. Sci. 2001;58:2513–2525. [Google Scholar]
- 38.Salamin N., Hodkinson T. R., Savolainen V. Syst. Biol. 2002;51:136–150. doi: 10.1080/106351502753475916. [DOI] [PubMed] [Google Scholar]
- 39.Axelrod D. I. In: Terrestrial Vegetation of California. Barbour M., Major J., editors. New York: Wiley; 1977. pp. 139–194. [Google Scholar]
- 40.Hickman J. C. The Jepson Manual: Higher Plants of California. Berkeley, CA: Univ. of California Press; 1993. [Google Scholar]
- 41.Sanderson M. J. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 42.Moles A. T., Ackerly D. D., Webb C. O., Tweddle J. C., Dickie J. B., Pitman A. J., Westoby M. Proc. Natl. Acad. Sci. USA. 2005;102:10540–10544. doi: 10.1073/pnas.0501473102. [DOI] [PMC free article] [PubMed] [Google Scholar]