Abstract
Tissue regeneration requires the recruitment of adult stem cells and their differentiation into mature committed cells. In this study we describe what we believe to be a novel approach for tendon regeneration based on a specific signalling molecule, Smad8, which mediates the differentiation of mesenchymal stem cells (MSCs) into tendon-like cells. A biologically active Smad8 variant was transfected into an MSC line that coexpressed the osteogenic gene bone morphogenetic protein 2 (BMP2). The engineered cells demonstrated the morphological characteristics and gene expression profile of tendon cells both in vitro and in vivo. In addition, following implantation in an Achilles tendon partial defect, the engineered cells were capable of inducing tendon regeneration demonstrated by double quantum filtered MRI. The results indicate what we believe to be a novel mechanism in which Smad8 inhibits the osteogenic pathway in MSCs known to be induced by BMP2 while promoting tendon differentiation. These findings may have considerable importance for the therapeutic replacement of tendons or ligaments and for engineering other tissues in which BMP plays a pivotal developmental role.
Introduction
Tendon and ligament injuries are not common but present a major clinical challenge to orthopedic medicine. In the United States at least 200,000 patients undergo tendon or ligament repair each year (1). Until the present time, therapeutic options used to repair torn ligaments have consisted of autografts, allografts, and synthetic prostheses. To date, none of these alternatives has provided a successful long-term solution. Allogeneic implants may lead to an immune reaction (2, 3), whereas autografts may lead to a comorbid state at the donor site. The use of synthetic grafts, although very popular in the 1980s, frequently lead to degeneration and failure of the implant (4, 5). A few growth factors have been investigated for their ability to induce tendon or ligament formation; all belong to the bone morphogenetic protein (BMP) family, which includes growth differentiation factor 5 (GDF5), GDF6, and GDF7 (6). Indeed it has been demonstrated that GDF protein leads to ligament or tendon formation in vivo (6). Other works have shown the formation of bone and cartilage within GDF-induced tendon tissue in vivo (7). Some evidence supports the idea that regeneration of an injured tendon or ligament can be achieved using GDF proteins (8). Concomitantly, the use of adult mesenchymal stem cells (MSCs) has been studied as a platform for tendon healing. This unique stem cell population resides in the bone marrow and adipose tissue — and probably in other tissues — and has been extensively investigated during the last few decades. It has been shown that adult MSCs are able to differentiate into mesenchymal phenotypes including bone-forming osteoblasts, cartilage-forming chondrocytes, fat-storing adipocytes, and muscle- or tendon/ligament–forming cells (9–11). Nevertheless, it has also been shown that implantation of autologous adult MSCs (isolated from rabbit bone marrow) in a tendon defect did not provide any improvement in the tendon’s microstructure, although some of its biomechanical properties were improved by the presence of the implanted cells (12). Based on our current knowledge of ligament and tendon formation, we have chosen to combine the regenerative properties of MSCs with a potent inducer of tenocytic differentiation, an approach that has not to our knowledge been taken so far. Our research of the predominant signaling mediators of the BMP family, the Smads, has led us to identify such an inducer. The receptor-regulated Smads (R-Smads) Smad1, Smad5, and Smad8 are directly phosphorylated and activated by a BMP type I receptor (IA, IB, or activin-like receptor kinase 2 [ALK2], which is also the activin IA receptor; ActR-IA). Smad2 and Smad3 are phosphorylated and activated by activin or TGF-β type I receptors (ALK4/ActR-IB and ALK5/TβR-I, respectively) (13). Another type of Smad, the common mediator Smad, or Smad4, associates with activated R-Smads. The Smad complexes translocate into the nucleus and participate in the regulation of target genes. This occurs predominantly by the binding of other transcription factors and transcriptional regulation. The different Smad proteins are highly conserved in their MH1 and MH2 domains, whereas the connecting proline-rich linker region is divergent. The MH2 domain has effector functions, as determined by transcriptional 1-hybrid assays (transactivation activity); it creates mesoderm formation in Xenopus and osteoblast formation in the mesenchymal progenitor cell C3H10T1/2 (14–17). The MH1 domain interferes with the effector function of MH2 and exhibits a detectable DNA-binding capacity (15, 18). There is evidence that Smad1 and Smad5 æsignaling molecules are critically involved in osteogenic development (17, 19), but no clear evidence of the role of Smad8 in any differentiation pathway. In an attempt to elucidate the possible roles of Smads as mediators of mesenchymal differentiation, we have overexpressed Smad molecules in the well-studied murine MSC line C3H10T1/2. This cell line mimics human adult MSCs in their biological properties and readily differentiates into bone or cartilage tissue following the overexpression of either BMP2 or T-box factor, as previously reported (20, 21). Our in vitro and in vivo results have indicated a role of the Smad8 molecule in the tendon differentiation pathway. Therefore, we have attempted to induce tissue regeneration in an Achilles tendon partial-defect model by using C3H10T1/2 cells that were genetically engineered to overexpress this Smad molecule. Our aim was to achieve a new formation of tendon tissue without the undesired induction of bone or cartilage. Our investigations have led to the finding of what we believe to be a novel inducer of tenogenic differentiation and a possible new therapeutic platform for tendon regeneration based on genetically modified MSCs.
Results
Expression of BMP2 and the Smad8 linker plus MH2 domain leads to tenocytic cell differentiation in murine C3H10T1/2 cells.
Our goal was to understand the role of Smad8 during the differentiation of MSCs. As a model we used the well-established MSC line C3H10T1/2. Two routes of investigation were followed: the first aimed to determine whether Smad8 was able to induce differentiation on its own, and the second aimed to determine what was the influence of Smad8 expression on BMP2-induced differentiation. The parental C3H10T1/2 cells, or C3H10T1/2 constitutively expressing the BMP2 gene, were stably transfected with different Smad molecules as described in Methods. Endogenous Smad8 is expressed at extremely low levels in this cell line (22), and therefore we cloned Smad8 cDNA from a rat source (brain) by performing RT-PCR. We consider it unlikely that there are functional differences between rat and mouse Smad8 because only few amino acid exchanges have been detected (2 exchanges in the aminoterminal MH1 domain, 4 in the less conserved linker region, and 1 exchange in the carboxyterminal MH2 domain).
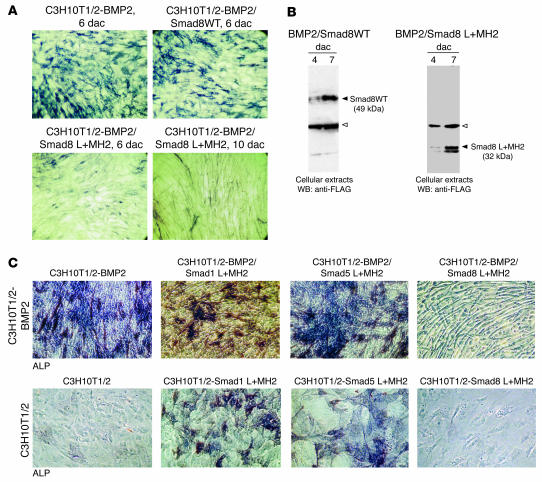
The biological outcome of Smad8 overexpression in recombinant BMP2-expressing MSCs (C3H10T1/2-BMP2 cells) was first determined by evaluating the differentiation potential of the transfected cells and comparing it with that of the parental cell line. When Smad1 or Smad5 molecules are stably transfected into C3H10T1/2 cells, stimulation of the BMP2-dependent osteogenic pathway is evident based on the number of alkaline phosphatase–positive (ALP-positive) osteoblast-like cells (17). We aimed to examine whether such an osteogenic effect is evident when the Smad8 molecule is transfected into the same cells. For the full-length Smad8 molecule, we observed no substantial difference between C3H10T1/2-BMP2 cells and the same type of cells expressing wild-type Smad8 (C3H10T1/2-BMP2/Smad8WT cells; Figure 1A, top panels), indicating that Smad8 either had not been activated by the BMP2 signalling pathway in C3H10T1/2 cells or was not biologically active. Since it has been demonstrated previously that R-Smad domains consisting of the carboxyterminal MH2 domain or the linker domain plus the MH2 domain (L+MH2) exert biological activity (14–17), we tested the Smad8 L+MH2 domain for biological activity in C3H10T1/2-BMP2 cells (Figure 1A, bottom panels). The most intriguing finding was that in vitro the differentiation pattern of BMP2/Smad8 L+MH2 progenitor cells displayed an elongated shape and did not exhibit the stellate-patterned phenotype of osteoblastic cells usually seen in C3H10T1/2 cells that express BMP2 (Figure 1A, top panels). The level of Smad8 expression was assessed in the genetically engineered C3H10T1/2-BMP2 cells by performing immunoblot analyses. The expression of full-length Smad8 and FLAG-tagged Smad8 L+MH2 proteins corresponded to expected sizes — 49 kDa and 32 kDa, respectively (Figure 1B). Interestingly, only Smad8 L+MH2 was able to elicit a morphological change in the C3H10T1/2-BMP2 cells, making them appear more like tenocytes (Figure 1, A and C). In the presence of BMP2 all C3H10T1/2 progenitors underwent mesenchymal differentiation and extensive matrix development resulting in multilayer growth. At day 10 after confluence, a relatively late stage of cultivation, residual ALP-positive cells were observed in cells embedded in the matrix and also on the top of the layer (Figure 1C). The presence of coexpressed L+MH2 domains from Smad1 and Smad5 had no influence on the morphological structure of the cells; however, the presence of Smad8 L+MH2 dramatically changed the morphology of the cells into an elongated cell phenotype (Figure 1C). C3H10T1/2 cells expressing Smad1 and Smad5 L+MH2 domains developed into osteoblast-like cells, while cells that expressed Smad8 L+MH2 did not possess comparable osteogenic capacity. Also, the development of a tenocyte-like morphology was not observed.
Figure 1. Tenogenic phenotype in C3H10T1/2-BMP2 cells due to forced expression of Smad8 L+MH2.
The parental C3H10T1/2 and C3H10T1/2 constitutively expressing the BMP2 gene were stably transfected with different Smad molecules as described in Methods. (A) Histological analysis of ALP-positive cells at 6 and 10 days after confluence (dac). Note the elongated tenocyte-like appearance of BMP2/Smad8 L+MH2–expressing cells compared with BMP2/Smad8WT–expressing cells. (B) Recombinant expression of FLAG-tagged Smad8 variants in C3H10T1/2-BMP2 cells. Western immunoblotting of cellular extracts of C3H10T1/2-BMP2 cells. Open arrowhead indicates a nonspecific band; filled arrowhead indicates Smad8WT and Smad8 L+MH2 domains. WB, Western blot. (C) Smad8 L+MH2, but not Smad1 or Smad5 L+MH2, elicited a tenocyte-like morphology in C3H10T1/2-BMP2. Histological analysis (ALP) and phase-contrast microscopy. All cells represent day 10 after confluence. Top panels: In the presence of BMP2 all C3H10T1/2 cells underwent mesenchymal differentiation and extensive matrix development resulting in multilayer growth. At this relatively late stage of cultivation residual ALP-positive cells were observed embedded in matrix and on the top of the layer. The presence of coexpressed L+MH2 domains from Smad1 and Smad5 did not influence this morphology. The presence of Smad8 L+MH2 dramatically changed this morphology to an elongated cell phenotype. Bottom panels: Parental C3H10T1/2 cells expressing Smad1 and Smad5 L+MH2 developed an osteoblast-like appearance. Smad8 L+MH2–expressing cells did not possess a comparable osteogenic capacity. Also, development of an elongated cell phenotype was not observed. Magnification, ×40.
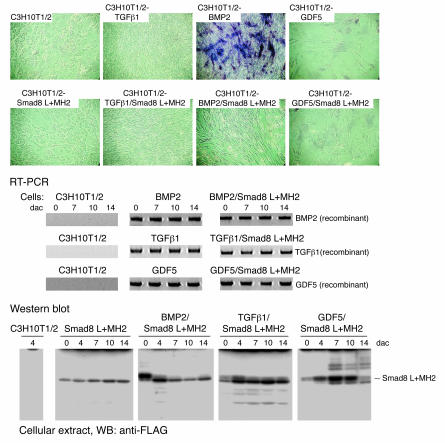
We also investigated the question of whether other ligands of the TGFβ/BMP family, such as TGFβ1 or GDF5, exert a similar change of morphology in the presence of the signalling mediator Smad8 L+MH2 (Figure 2). Only the combination of BMP2 and Smad8 L+MH2 led to the formation of the elongated tenocyte-like cell morphology at day 11 after confluence (Figure 2). Comparable expression of the ligands was shown by RT-PCR, and comparable Smad8 L+MH2 expression by Western analyses (Figure 2). Smad8 L+MH2 — either alone or in the presence of the ligands TGFβ1 or GDF5 — was not sufficient to induce the elongated tenocyte-like phenotype. Comparable to BMP2, TGFβ1 expression led to the formation of a cellular multilayer organization, but osteo- and/or chondrogenic differentiation in this multilayer was only observed in the presence of BMP2. In contrast, GDF5 induced monolayer growth only with relatively large cuboidal cells showing no tenocyte-like morphology (Figure 2). Marker gene expression assessing mesenchymal differentiation confirmed the histological analyses (data not shown). These results indicate that TGFβ1 and GDF5 are not efficient as BMP2 for the induction of tenogenic differentiation in the presence of Smad8 L+MH2.
Figure 2. Smad8 L+MH2 elicits a tenocyte-like morphology in C3H10T1/2-BMP2 cells, but not in C3H10T1/2-TGFβ1 or C3H10T1/2-GDF5 cells.
Histological analysis (ALP) and phase-contrast microscopy. All cells represent day 11 after confluence. As shown in Figure 1A, the expression of Smad8 L+MH2 led to an elongated tenocyte-like phenotype. This phenotype was not observed when Smad8 L+MH2 was expressed in parental C3H10T1/2 cells or in cells that stably express TGFβ1 or GDF5. TGFβ1 expression led to a dense multilayer growth, but neither osteo- and/or chondrogenic differentiation in vitro nor the formation of tenocyte-like cells in the presence of Smad8 L+MH2 was observed. Stable expression of GDF5 only allowed monolayer growth exhibiting large cellular phenotypes. Smad8 L+MH2 expression did not change this cellular phenotype. RT-PCR analyses show recombinant expression of TGFβ1 or GDF5 in the various cell lines. Western blot analyses show recombinant expression of Smad8 L+MH2 in the recombinant C3H10T1/2 lines.
Smad8 possesses the capacity for transactivation.
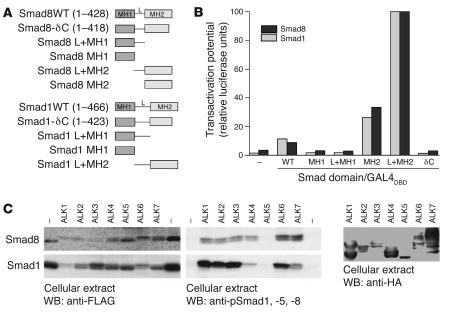
Since Smad8 has been described as a Smad molecule that mediates BMP signals (23–25), and because the forced expression of full-length Smad8 in C3H10T1/2-BMP2 cells was unable to stimulate BMP2-dependent osteogenesis, we questioned whether Smad8 possesses biological activity or interferes with BMP-dependent signalling. In the carboxyterminal end of its MH2 domain, Smad8 harbors an SSVS motif that has the potential to be phosphorylated for Smad activation (23, 24). In the mesenchymal progenitor cell line C2C12, however, various types of BMPs activate overexpressed Smad8 only marginally (25). In C3H10T1/2-BMP2 cells, full-length Smad8 did not substantially enhance osteogenic differentiation (Figure 1). To assess the general biological capacities of Smad8 and its domains in C3H10T1/2 cells, we compared the transactivation potential of Smad8 with that of the well-characterized Smad1 mediator. Full-length Smads as well as various Smad domains (Figure 3A) were fused in frame with the GAL4 DNA-binding domain. Using an upstream activation sequence–driven (UAS-driven) luciferase reporter, the hybrid Smad proteins were all recruited with the same efficiency to heterologous DNA (UAS) by means of the GAL4 DNA-binding domain, allowing us to investigate whether the Smad8 signaling mediator possesses a transactivation potential similar to that of Smad1 (Figure 3B). Similar to what has been demonstrated previously (16), hybrid proteins consisting of GAL4 fused to full-length Smad1 (Smad1WT) did not efficiently transactivate the GAL4 reporter or the full-length Smad8 (Smad8WT) (Figure 3B). This reflects the requirement for receptor- or ligand-induced conformational changes to disrupt the inhibitory interactions of the MH1 and MH2 domains necessary for Smads to become active.
Figure 3. Smad8 is activated by TGF-β/BMP type I receptors.
(A) Schematic representation of WT Smads and the Smad domains used for functional studies in HEK 293T and C3H10T1/2 cells. MH1 and MH2 are the major conserved Smad domains; “L” shows the linker region between them. The linker region is considerably smaller in Smad8 than in Smad1. (B) Results of reporter assays in HEK 293T cells, in which a GAL4 reporter with the GAL4 DNA-binding domain (GAL4DBD) fused to various forms of Smad proteins was used. Results are expressed as relative luciferase units normalized to β-gal activity and presented as percent of Smad L+MH2, which was arbitrarily set as 100%. Pooled data from at least 3 independent experiments are presented. Smad8 MH2 and L+MH2 domains exhibited a constitutive active transactivating potential comparable to that of Smad1. (C) Activation potential of Smad8 and Smad1 signalling mediators by constitutively active TGF-β/BMP receptors (ALK1–ALK7) in HEK 293T cells. FLAG-tagged Smad8 or Smad1 was transiently coexpressed with constitutively active HA-tagged type I receptors. Expression of all type I receptors was mediated by the identical vector (pcDNA3). Expression rate of the receptors was determined by Western blotting using anti-HA antibodies. Receptor bands indicate variations in the glycosylation of the ectodomain. Type I receptor–dependent phosphorylation of Smads was shown by anti-pSmad1, -5, and -8 antibodies, which also react with phosphorylated Smad8. Smad1 and Smad8 were phosphorylated by most type I receptors. In contrast to Smad1, Smad8 was also efficiently phosphorylated by ALK4 and ALK7 (TGFβ1 receptors).
The transactivating potential of individual Smad1 and Smad8 domains was assessed in the same experimental setup by comparing Smad1 and Smad8 MH1, L+MH1, MH2, and L+MH2 domains with Smad molecules lacking the carboxyterminal SSVS motif (δC). As expected, only the Smad1 and Smad8 MH2 and L+MH2 domains, which are not subjected to inhibition by the MH1 domain (Figure 3B), were constitutively active. Interestingly, the L+MH2 construct was much more efficient than the single MH2 construct because of the presence of the proline-rich linker domain. The increased biological activity of L+MH2 compared with MH2 alone has been observed in several previous studies (16, 17).
We also characterized the ability of constitutively active TGF-β/BMP type I receptors to induce phosphorylation in the carboxyterminal MH2 domain (Figure 3C). Immunoblotting with antibodies directed against the phosphorylated SSVS motif in Smad1, which recognizes phosphorylation of Smad8 as well, revealed that in human embryonic kidney (HEK) 293T cells Smad1 was efficiently phosphorylated by constitutively active ALK1, ALK2, and ALK3 (BMPR-IA) as well as by ALK6 (BMPR-IB). The TGF-β type I receptors ALK4, ALK5, and ALK7 are not or are only inefficiently able to do this, which is consistent with the idea that Smad1 is a signaling mediator for BMP signals. Other investigators have suggested the same for Smad8 (23, 24). This experiment (Figure 3C) was performed in HEK 293T cells, which allow a reproducible high level of transgene expression. In these cells ALK6 is the only BMP receptor (besides the TGFβ/activin receptor ALK7) that phosphorylates Smad8 to higher levels. In the mesenchymal progenitor cell line C3H10T1/2, on the other hand, ALK6 is not expressed at all and ALK3 seems to be the dominant BMP receptor (22).
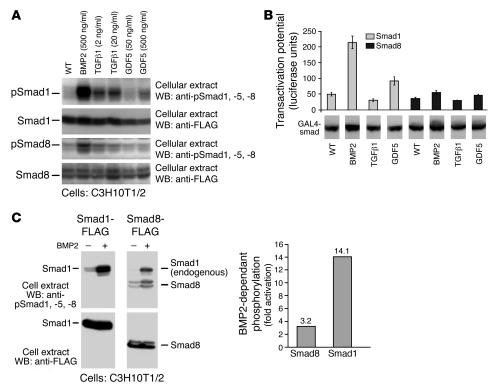
We therefore continued by investigating the levels of BMP2-, TGFβ1-, and GDF5-dependent activation of Smad1 and Smad8 in the C3H10T1/2 progenitors. The activation of Smad1WT or Smad8WT signalling mediators by BMP2, TGFβ1, and GDF5 was analyzed by blotting experiments using anti-pSmad1, -5, and -8 antibodies, which recognize phosphorylated forms of Smad1, Smad5, and Smad8, respectively (Figure 4A). When varying ligand concentrations were used, we observed that Smad1 was efficiently activated by BMP2, much less by TGFβ1, and only to a very reduced extent by GDF5. Furthermore, Smad8WT was only marginally activated by BMP2 and almost not at all by TGFβ1 and GDF5 (Figure 4A). In conclusion, the use of the biologically active form of Smad8 (Smad8 L+MH2) seems essential for tenogenic induction, since ligands of the TGFβ/BMP family only marginally activate this signalling mediator.
Figure 4. Smad8 exhibits a low activation potential by various ligands of the TGFβ/BMP family.
(A) FLAG-tagged Smad1 or Smad8 were transiently expressed in C3H10T1/2, and BMP2, TGFβ1, or GDF5 were added for 30 minutes at the indicated concentrations. After Western blotting, ligand-dependent phosphorylation of Smads was shown by antibodies specific for pSmad1, -5, and -8. The expression rate of the Smads was determined by Western blotting using anti-FLAG antibodies. (B) Evaluation of the Smad1 and Smad8 transactivation potential in HEK 293T cells, in which a GAL4 reporter with the GAL4 DNA-binding domain was fused to Smad1WT and Smad8WT proteins as described in Figure 3B. Pooled data from 3 independent experiments are presented. BMP2 led to efficient activation of the GAL4-Smad1 fusion protein, GDF5 was less effective, and TGFβ1 did not exhibit notable activation. The GAL4-Smad8 fusion protein was activated, albeit to a markedly lower extent, by BMP2, but not by TGFβ1 or GDF5. The expression level of the GAL4-Smad fusions was comparable as assessed by Western analyses of cellular extracts and blotting with anti-GAL4 antibodies. (C) Compared with Smad1, Smad8 exhibited a lower BMP2-dependent activation potential in C3H10T1/2. FLAG-tagged Smad1 or Smad8 were transiently expressed in C3H10T1/2, and BMP2 (200 ng/ml) was added for 30 minutes. Top: After Western blotting, BMP2-dependent phosphorylation of Smads is shown by anti-pSmad1, -5, and -8 antibodies. Bottom: Expression rates of the Smads were determined by Western blotting using anti-FLAG antibodies. In the graph at right, the level of BMP2-dependent Smad activation in C3H10T1/2 was evaluated by densitometric scanning.
This activation of Smad8 was further investigated in Figure 4B. We assessed the capacity of the ligands BMP2, TGFβ1, and GDF5 to activate Gal4-SmadWT fusion proteins. This level of activation was quantified by a Gal4 luciferase reporter in human HEK 293T cells as described above for Figure 3B. This test confirmed that BMP2 was the best ligand for the transactivation of Gal4-Smad1WT fusions. GDF5 exhibited a considerably lower capacity for Gal4-Smad1WT transactivation, while TGFβ1 did not show a considerable effect in this regard. In contrast, Gal4-Smad8WT fusions were activated only to a minor extent by BMP2, even less by GDF5, and not at all by TGFβ1, confirming the analyses in mesenchymal progenitor C3H10T1/2 by the anti-pSmad1, -5, and -8 antibodies in Figure 4A. The results of this experiment were obtained by comparable expression rates for all Gal4-Smad fusions tested as shown by Western blot analyses (Figure 4B).
The capacity of exogenously added BMP2 to activate recombinant Smad8WT compared with its capacity to activate recombinant Smad1WT is shown in Figure 4A and was evaluated in Figure 4C by densitometric scanning. Smad8WT was activated only 3-fold, compared with 14-fold in Smad1WT (Figure 4C). This higher level of Smad1 activation appears to be related to the C3H10T1/2 signaling system, which only seemed to activate Smad8 to a rather modest level. In conclusion, although Smad8 clearly possesses the capacity to act as an activator, full-length Smad8 seems remarkably inactive in this MSC line, and endogenous ligands/receptors that might mediate substantial Smad8 activation in these cells have not been identified.
C3H10T1/2-BMP2/Smad8 L+MH2 progenitor cells express tenogenic markers.
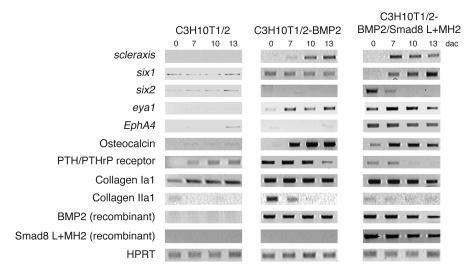
During in vitro cultivation, various modified progenitor cell lines underwent a developmental sequence from the undifferentiated phenotype to the BMP-dependent formation of mature osteoblasts and chondrocytes. Under the control of BMP2/Smad8 L+MH2, these cells developed into mature tenocyte-like cells. The time course of in vitro cultivation from confluence (termed day 0) to day 13 after confluence demonstrated most of this developmental sequence. Extensive matrix maturation was observed on days 4–10, during which the cells were able to grow in multilayers within a dense matrix. During the late stage of maturation (day 11 onward), mature cellular mesenchymal phenotypes were observed. In the presence of BMP2, biologically active Smad8 L+MH2 induced morphological characteristics in C3H10T1/2 progenitors that resembled those seen in tenocytes (Figure 1). As described earlier, the Smad8 L+MH2 domain exerted constitutive activity in C3H10T1/2-BMP2 cells (Figure 1). With the exception of scleraxis, however, there is no specific marker gene for tendons (26), although it should be noted that scleraxis transcripts are also detected in ligaments (27). Scleraxis expression seemed almost nonexistent in undifferentiated proliferating mesenchymal progenitors (Figure 5). During the matrix maturation period C3H10T1/2-BMP2/Smad8 L+MH2 cells seemed to enter tenogenic development relatively quickly, as indicated by the high level of scleraxis expression at day 7 after confluence compared with C3H10T1/2-BMP2 cells. Scleraxis was downregulated at the late stages of cultivation; an explanation for this could be that the in vitro cultivation conditions may not have been optimal for developed tenocytes. In contrast, in C3H10T1/2-BMP2 cells, scleraxis seemed to be upregulated when the cells underwent the late developmental steps toward becoming mature chondrocytes and osteoblasts. This could be explained by the fact that scleraxis may also be expressed in chondrocytes at a low level (26). A high rate of chondrocyte formation was observed at this time (days 10–13) in C3H10T1/2-BMP2 cells by using Alcian blue staining (data not shown). In conclusion, in C3H10T1/2-BMP2/Smad8 L+MH2 cells, scleraxis acted as a marker for tenogenic development because chondrogenesis and osteogenesis were not observed, whereas in C3H10T1/2-BMP2 cells, scleraxis may indicate the presence of maturing chondrocytes.
Figure 5. RT-PCR analyses of the expression levels of chondrogenic, osteogenic, and tenogenic markers in murine C3H10T1/2 cells.
Collagen Ia1, the PTH/PTHrP receptor, and osteocalcin are markers predominantly associated with osteogenesis, and collagen IIa1 is a marker associated with chondrogenic differentiation. Collagen Ia1, six1, six2, scleraxis, eya1, and EphA4 are tenogenic markers. Hypoxanthine guanine phosphoribosyl transferase (HPRT) was used to standardize e PCR conditions.
Among other genes, the tyrosine kinase receptor EphA4 has been reported to be upregulated in distal limb tendons (28, 29), along with the transcription factors six1 and six2 (30) and eya1 and eya2 (31). From our observations one may conclude that six2 together with EphA4 are expressed at the very early stages of tenogenic development. Interestingly, during the time at which six2 was downregulated (day 7 onward), six1 was upregulated. In addition, the receptor EphA4 showed a remarkably high specificity for the tenocytic BMP2/Smad8 L+MH2 progenitor cells. On the other hand, the osteogenic marker gene osteocalcin was downregulated only very moderately in C3H10T1/2-BMP2/Smad8 L+MH2 cells, although those cells did not possess any of the morphological characteristics of osteoblasts. Collagen I was not expressed differently in the tenogenic cells. This can be explained by the fact that collagen I is the major collagen secreted during osteogenesis and tenogenesis and is expressed at a high level in both cell types. In contrast, osteo- and chondrogenic markers, such as the PTH/PTHrP receptor and collagen II, were dramatically downregulated in the presence of Smad8 L+MH2. The PTH/PTHrP receptor is an important and specific early osteoblastic gene, and PTH sensitivity is one of the most important characteristics of osteoblasts. Moreover, Smad8 L+MH2 interfered with ALP activity, another hallmark of osteoblast development, and also severely interfered with the morphological appearance of stellate-patterned osteoblast-like cells (Figure 1C, bottom panels). In general, the expression of marker genes (Figure 5) supports the conclusion that constitutively active Smad8 L+MH2 suppresses cellular development into osteo- and chondrogenic lineages but initiates tenogenic differentiation. Based on these results we decided to investigate the in vivo differentiation of mesenchymal progenitor cells that express Smad8 L+MH2 and BMP2.
C3H10T1/2-BMP2/Smad8 L+MH2 progenitor cells form ectopic tendon tissue in vivo.
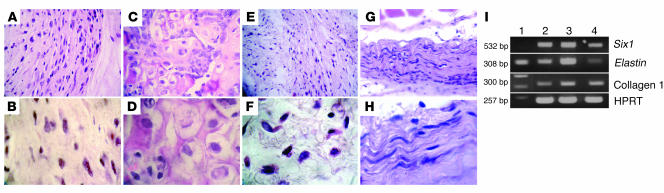
Parental C3H10T1/2 progenitors, progenitors that coexpressed Smad8 L+MH2 and BMP2, and C3H10T1/2 cells that expressed either Smad8 L+MH2 or BMP2 were implanted s.c. in C3H/HeN mice as described in Methods. Thirty days after implantation, histological analysis of implanted C3H10T1/2-BMP2/Smad8 L+MH2 cells revealed the formation of a mass of dense tissue that consisted of spindle-shaped tenocytes (Figure 6, G and H). In comparison, in transplants of parental C3H10T1/2 and C3H10T1/2-Smad8 L+MH2 progenitor cells, only a mass of nonspecific connective tissue was formed (Figure 6, A, B, E, and F) C3H10T1/2 cells that overexpressed only the BMP2 gene differentiated into bone and cartilage tissue when implanted in vivo (Figure 6, C and D), similar to what has been previously described (20). RT-PCR performed on implants derived from C3H10T1/2-BMP2/Smad8 L+MH2 cells detected the expression of genes usually found in tendon/ligament tissues, like Six1, Elastin, and Collagen I (Figure 6I), while the other implants showed only collagen I expression (data not shown).
Figure 6. C3H10T1/2-BMP2/Smad8 L+MH2 cells induce ectopic tendon formation after s.
. implantation. The forced expression of Smad8 L+MH2 in C3H10T1/2-BMP2 cells led to differentiation into tenocytes at murine ectopic sites 30 days after s.c. cell transplantation. (A–H) H&E-stained sections of ectopically implanted MSCs. (A and B) C3H10T1/2 progenitors. (C and D) C3H10T1/2-BMP2 cells. (E and F) C3H10T1/2-Smad8 L+MH2 cells. (G and H) C3H10T1/2-BMP2/Smad8 L+MH2 s.c. implants. Magnification, ×40 (top panels), ×100 (bottom panels). C3H10T1/2-BMP2/Smad8 L+MH2 implants demonstrated a completely different morphology than the other implants, highly resembling neotendon tissue. C3H10T1/2 progenitors and C3H10T1/2-Smad8 L+MH2 showed nonspecific mesenchymal tissue morphology, while the C3H10T1/2-BMP2 implants contained cartilage and bone foci. (I) RT-PCR analysis of C3H10T1/2-BMP2/Smad8 L+MH2 cell implants 30 days after implantation. Lane 1, DNA ladder; lanes 2 and 3, engineered cell implants; lane 4, mouse ligament tissue. All 3 genes are associated with tendon/ligament tissues. Results obtained from the other implants showed only Collagen I expression and no expression of Six1 and Elastin genes (not shn).
C3H10T1/2-BMP2/Smad8 L+MH2 progenitor cells induce Achilles tendon repair.
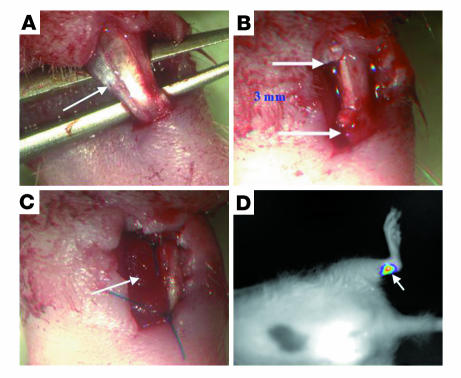
C3H10T1/2 progenitors that coexpressed Smad8 L+MH2 and BMP2 or C3H10T1/2 cells that expressed either Smad8 L+MH2 or BMP2 were implanted within a 3-mm partial defect of Achilles tendon in 12 nude rats as described in Methods (Figure 7, A–C). The survival of implanted cells at the implantation site was monitored using a noninvasive cooled charge-coupled device (CCCD) tracking system. Because the cells had been infected with a retroviral luciferase vector before implantation, a positive luciferase signal indicated the presence of engineered cells in the surgically treated tendon (Figure 7D). The rats were killed 4, 5, or 7 weeks after implantation, and their Achilles tendons were analyzed. The engineered C3H10T1/2-BMP2/Smad8 L+MH2 cells, which for these investigations were infected with adeno–β-gal before implantation, were identified in the defect site by applying an immunohistochemical stain (Figure 8, G–I; arrowheads). A histological analysis demonstrated filling of the defect with tenocyte-like cells that were closely oriented in the same axis as the rat tendon cells (Figure 8, G–I). In comparison, the defect implanted with C3H10T1/2-Smad8 L+MH2 cells was filled with nonspecific connective tissue (Figure 8, D–F). The implantation of C3H10T1/2 BMP2 cells led to the formation of cartilage and bone foci in the defect region (Figure 8, A–C). Isolation of the defect site implanted with C3H10T1/2-BMP2/Smad8 L+MH2 from cryosections of surgically treated tendon, which was achieved using laser capture microdissection (LCM) technique, enabled us to perform RT-PCR analysis on RNA extracted exclusively from the implantation site. Our results detected the expression of murine collagen I, a major gene expressed in tendon, within this site (Figure 9). Moreover, a double quantum filtered (DQF) MRI analysis was used to evaluate tendon tissue regeneration at the defect site. The motion of water molecules in tendon is anisotropic due to their interaction with the oriented collagen fibers. This anisotropic motion results in a residual 1H dipolar interaction for H2O and residual 2H quadrupolar interaction for D2O. The magnitudes of the 1H dipolar splitting (vD) as well as the 2H quadrupolar splitting (vQ) are related to the density of the collagen fibers and their ordering. The 1H dipolar interaction as well as the 2H quadrupolar interaction are best measured by DQF techniques, which detect the signals only from nuclei experiencing anisotropic motion. Both the line shape and the intensity of the DQF signals depend on the creation time τ. It has been shown that the creation time can be used as a contrast mechanism to differentiate between different ordered tissues like tendon. The sensitivity of the DQF MRI to the ordering of the tissue has been previously used to monitor the healing process of rabbit Achilles tendon (32–34). The intact tendon has a maximal signal at τmax of 0.4 ms, while the ruptured tendon gives a much lower signal, with the maximum at much longer creation times. The DQF analysis performed in this study demonstrated the presence of a higher level of oriented collagen fibers within the defect site implanted with C3H10T1/2-BMP2/Smad8 L+MH2–engineered cells (Figure 10, E and F) even more than that found in the nonoperated contralateral site (Figure 10, A and B). Moreover, the DQF signal in the tendons implanted with C3H10T1/2-Smad8 L+MH2 (Figure 10, I and J) or C3H10T1/2-BMP2 cells (Figure 10, G and H) was rather similar to those implanted only with collagen sponge (Figure 10, A and B) and was much lower than that detected in those defects implanted with C3H10T1/2-BMP2/Smad8 L+MH2 cells. These results demonstrated that MSC progenitors that overexpressed BMP2/Smad8 L+MH2 were able to survive and induce tendon tissue regeneration within the defect site.
Figure 7. Achilles tendon regeneration model.
An Achilles tendon partial-thickness defect was created in athymic rats. Adult athymic rats (4 months old) were anesthetized as described in Methods. The Achilles tendon (indicated by arrow) was separated from the plantaris and soleus tendons (A), and a 3-mm partial-resection defect was created in its lateral substance (B, arrows indicate the span of the defect). Cells (3 × 106) were seeded onto a collagen sponge (arrow), which was then placed within the tendon defect and sutured to the tendon (C). The skin was closed in a routine manner using 2-0 Mersilk. (D) Noninvasive monitoring of cell survival in the implantation site. C3H10T1/2-BMP2/Smad8 L+MH2 progenitor cells expressing the luciferase gene were implanted in an Achilles tendon defect in the rat. CCCD analysis demonstrated a positive luciferase signal detected in the implantation site (arrow), indicating the survival of implanted cls.
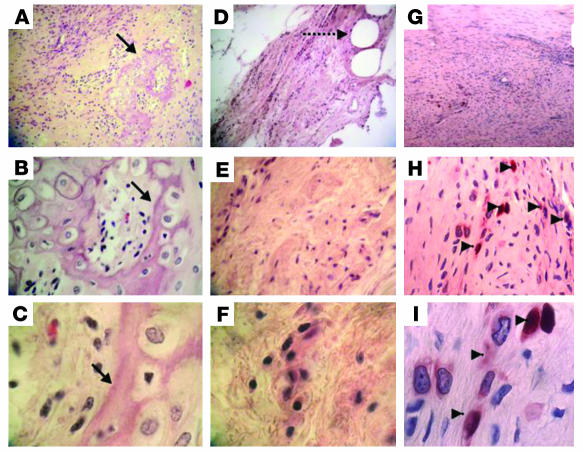
Figure 8. Neotendon tissue formation in a partial-defect model in the Achilles tendon of a rat.
C3H10T1/2-BMP2 (A–C), C3H10T1/2-Smad8 L+MH2 (D–F), and C3H10T1/2-BMP2/Smad8 L+MH2 cells infected with adeno-LacZ (G–I) were implanted in an Achilles tendon defect in nude rats (3 × 106 cells per implant). (A–F) Histological (H&E) and (G–I) immunohistochemical (anti-LacZ) staining was performed 4 weeks after implantation. (A–C) Tendon implanted with C3H10T1/2-BMP2 cells demonstrated cartilage and bone foci formation within the tendon tissue (arrows). (D–F) Tendon implanted with C3H10T1/2-Smad8 L+MH2 cells showed mesenchyme-like tissue formation at the site of implantation (dashed arrow shows suture used to hold collagen scaffold in the defect site). (G–I) Tendon implanted with C3H10T1/2-BMP2/Smad8 L+MH2 cells infected with adeno-LacZ prior to implantation showed LacZ-positive cells in the tenogenic implant (arrowheads). Cells seemed to be arranged parallel to the tendon long axis. Magnification, ×10 (top panels), ×40 (middle panels), ×100 (bottom panels).
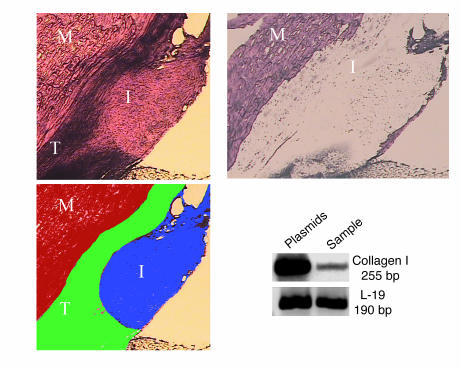
Figure 9. Collagen I mRNA is expressed in the site of C3H10T1/2-BMP2/Smad8 L+MH2 progenitor cell implantation in the Achilles tendon.
Upper left panel depicts the implantation site in the host Achilles tendon as viewed under a light microscope. Lower left panel shows a schematic representation of the different histological components in the field of view. Upper right panel depicts the histological section after the implantation region was removed from the Achilles tendon using LCM. Blots show the gel electrophoresis results of RT-PCR performed on RNA extracted from the microdissected sample. Ribosomal protein L-19 served as a control. M, muscle; T, host tendon; I, implantation area. Magnification, ×4.
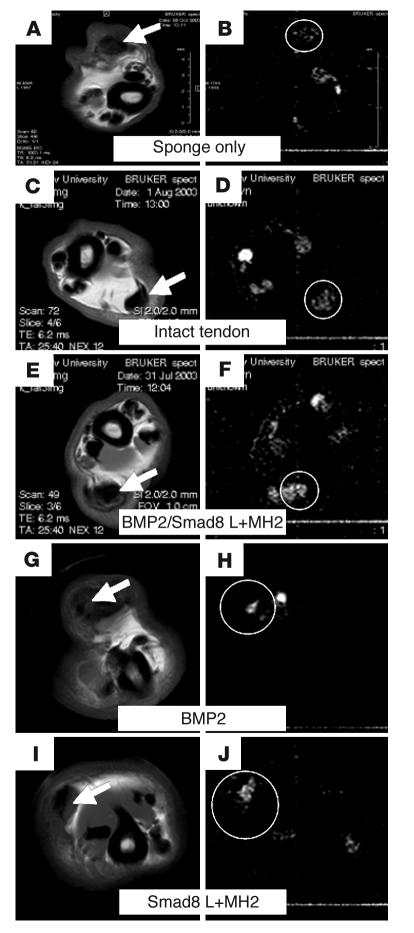
Figure 10. Tendon-defect repair demonstrated by micro-MRI.
(A, C, E, G, and I) MSME axial sections of Achilles tendon of a rat. (B, D, F, H, and J) DQF images of the same axial sections. Tendon was either implanted with collagen sponge without cells (A and B), left intact (C and D), or implanted with C3H10T1/2-BMP2/Smad8 L+MH2 cells (E and F), C3H10T1/2-BMP2 cells (G and H), or C3H10T1/2-Smad8 L+MH2 cells (I and J). DQF images showed ordered collagen fiber formation in the defect site implanted with C3H10T1/2-BMP2/Smad8 L+MH2 cells at a higher level than that found in the contralateral tendon or in the other experimental groups. Arrows indicate the Achilles tendon at the site of implantation. Circles highlight the DQF signal at the site of implantation or, in the case of no treatment, at the tendon.
Discussion
Our results indicate that a particular Smad signalling cascade is involved in the differentiation of MSCs to tenocytes. It has been reported that members of the BMP family of signalling mediators (GDF5, GDF6, and GDF7) implanted at ectopic sites are able to induce tendon-like structures and expression of a tenocytic marker gene (6). It has also been shown that BMP signals from the mesenchyme appear to repress areas of tendon formation, assuring that only specific domains are responsible for the fate of the tendon (27). Experiments using GDF5- and GDF6-deficient mice have shown that these factors are associated with joint formation (28, 35) and that the disruption of tendon formation observed in GDF5-deficient mice is probably secondary to the joint defect (29, 30). It cannot be excluded that another BMP family member may be responsible for tendon induction, especially since BMP4 is expressed in substantial levels in tendons and since ectopic application of recombinant human follistatin, an antagonist of certain TGF-β superfamily proteins including activin and BMP4, results in the loss of tendon, implicating crucial signaling by TGF-β superfamily members for tendon development (31). Our study is the first to our knowledge to indicate that a particular Smad signaling molecule downstream of the TGF-β/BMP family of secreted factors might play a decisive role in tenogenesis. In addition, this is the first report to our knowledge that attempts to combine the BMP signaling pathway with the repair properties of MSCs for tendon repair in vivo.
Constitutively active TGF-β/BMP type I receptors may activate Smad8 (Figure 3C), and full-length Smad8 and its domains exhibit a transactivating potential similar to the related R-Smad1 (Figure 3B). Interestingly, a tendon differentiation pattern was observed only when forced expression of the active biological form of Smad8 (Smad8 L+MH2) was induced in BMP2-expressing MSCs. Such a pattern was not evident when the full-length cDNA of Smad8 was expressed in the same cells. Based on our results, it is our hypothesis that Smad8 plays a pivotal role in the signalling cascade that leads to tenocytic differentiation of MSCs; however, this cascade is not directly activated by BMP2 but by another factor, which is still unknown.
In general, the BMP-dependent weak activation of Smad8 has been reported in several biological systems, and we also observed it here. In C3H10T1/2 progenitors, overexpressed Smad8 was much less activated by BMP2 than by Smad1: 3-fold as opposed to 14-fold (Figure 4A). In this study the level of Smad8WT activation was not sufficient to obtain a similar physiological response compared with Smad8 L+MH2, which resulted in tenocyte-like cell formation (Figure 1). Nevertheless, there is evidence that in C2C12 cells BMP4 induces ALP activity equally well in the presence of recombinantly expressed Smad8, Smad1, and Smad5. In the same study Smad5 was phosphorylated most efficiently by GDF5, another BMP family member, whereas Smad1 and Smad8 were only weakly phosphorylated or not phosphorylated at all (25). Interestingly, BMP6 induces ALP activity in C2C12 cells through the activation of Smad1 and Smad5, but not Smad8 (32). In the latter study it was also documented that Smad8 is neither phosphorylated nor translocated into the nucleus as a result of BMP6 stimulation. Smad8 seems to be different also in regard to protein stability. It has recently been demonstrated that overexpression of Smurf1 negatively regulates mouse embryonic lung branching morphogenesis by specifically reducing the protein levels of Smad1 and Smad5 but not Smad8 (33). Indeed, Smad8 may require interactions between a unique BMP superfamily member and ALK for efficient activation in normal cell physiology, and these interactions need to be clarified by further investigations. Alternatively, one might envisage a different mode for efficient Smad8 activation. Recently it was shown that a special component of the extracellular matrix is able to activate Smad signalling without the BMP pathway (36). If it is confirmed, the components of that special extracellular matrix may activate BMP signalling pathways without the BMP receptor system; our results that Smad8 is not efficiently activated by the TGFβ/BMP pathway could be explained by such a system.
When ectopically implanted, C3H10T1/2-BMP2/Smad8 L+MH2 cells seem to give rise only to tenocytes, although one would expect there to be at least some formation of bone and cartilage due to the recombinant BMP2 secreted into the muscle tissue. This should lead to the migration of host mesenchymal progenitors and the concomitant development of bone and cartilage. The complete absence of bone and cartilage particles could be explained if these implanted C3H10T1/2-BMP2/Smad8 L+MH2 cells synthesize and secrete an inhibitor for BMP2. Indeed, preliminary microarray analysis data document that BMP2/Smad8 L+MH2 cells synthesize considerable amounts of follistatin, a potential BMP2 inhibitor (our unpublished observations). Thus host mesenchymal progenitors probably do not markedly migrate and react to BMP2 secreted from ectopic BMP2/Smad8 L+MH2 cells. Nevertheless, in an autocrine manner, the secreted BMP2 seems able to activate endogenous signalling pathways in BMP2/Smad8 L+MH2 cells. These signals are necessary for the cells to undergo efficient tenogenic development. The endogenous BMP2-dependent signalling capacity may be lower, allowing the constitutively active Smad8 L+MH2 domain to exert its tenogenic capacity. We assume that the high abundance of constitutively active Smad8 L+MH2 molecules within the cells overtook any other signalling cascade that usually leads to osteogenesis (via Smad1 or Smad5) or to chondrogenesis. Since we have seen that the overexpression of Smad8 L+MH2 in C3H10T1/2 cells alone was not sufficient to induce differentiation into tenocytes, we can assume that upregulation of other genes downstream to BMP2 is necessary for Smad8 to exert its induction of tenocytic differentiation. We have shown that genetically modified MSCs that overexpress an active form of the Smad8 molecule formed new tendon tissue in ectopic regions and induced tendon regeneration in injury sites in which the cells have been implanted. Overexpression of the Smad8 L+MH2 molecule in BMP2-expressing progenitor cells generated an elongated morphology that did not resemble the osteoblastic stellate pattern usually seen in MSCs that express BMP2 (Figure 1). In addition, RT-PCR analysis of the engineered cells revealed upregulation of the tendon-specific markers six1, six2, eya1, EphA4, and scleraxis (Figure 5). Interestingly, the six and eya genes, together with the dachshund (dach) gene, form a network of genetically interacting proteins. It has been shown that six1 is required for the development of the murine kidney, muscle, and inner ear and exhibits synergistic genetic interactions with eya factors. The phosphatase function of eya switches the function of six1–dach from repression to activation, causing transcriptional activation through recruitment of coactivators. This provides a molecular mechanism for activation of specific gene targets in mammalian organogenesis, which could also play a role in tendon and ligament formation (37).
When C3H10T1/2-Smad8 L+MH2/BMP2 progenitors were implanted s.c. or into the Achilles tendon defect, no bone or cartilage formation was detected, as would have been anticipated based on previous reports on MSCs that express BMP2 (20). Instead, our histological analysis demonstrated the formation of dense connective tissue that was characteristic of ligament or tendon tissue (Figure 6). Regeneration was achieved in the tendon-defect model as observed by DQF MRI (Figure 10). The results demonstrated extensive formation of collagen fibers compared with tendons implanted with C3H10T1/2-Smad8 L+MH2 or C3H10T1/2-BMP2. The DQF MRI signal was even more prominent than the one detected in the intact tendon.
Recently, there have been several efforts to use different BMP family members to heal ligaments or tendons; however, the use of such factors has often been accompanied by bone and/or cartilage formation and a concomitant mineralization that reduces the tensile strength and stability of the tendons (8, 34). No bone or cartilage formation was seen in the BMP2/Smad8 L+MH2 progenitors; therefore we infer that the approach taken in the present study could be advantageous for tendon regeneration.
In the present study we used a stem cell–based approach, which proved to be highly effective in repairing the tendon defect because the engineered progenitors were able to engraft into the injury site, as demonstrated using a noninvasive CCCD system (Figure 7). This was confirmed by an in situ immunohistochemical study performed on the surgically treated tendon samples (Figure 8). After their engraftment, the cells were involved in tendon tissue regeneration, which was evident for at least 7 weeks after implantation. The immunohistochemical analysis was able to identify only a small number of cells within the regenerated tendon. This could be attributed to the fact that an adenoviral vector was used to label the cells prior to implantation. Adenoviral infection is known to last only a few weeks, and therefore it is possible that some of the cells have lost the LacZ transgene by the time of the tissue harvest. It is also possible that only a small number of implanted C3H10T1/2-BMP2/Smad8 L+MH2 cells survived within the defect after all, meaning that the tendon regeneration detected by the DQF MRI system could be due to the activity of host cells as well. Such a phenomenon was also evident when C3H10T1/2-BMP2 cells were used for bone fracture repair (20, 38). Although robust bone formation was achieved, only a few of the implanted cells were identified at the implantation site. It is our hypothesis that MSCs have a major initial role in the induction and orchestration of a tissue formation process. This was clearly shown when fracture repair using C3H10T1/2-BMP2 cells was compared with non-MSCs (CHO cells), which overexpressed BMP2 as well. The results showed that the bone repair using the mesenchymal progenitors led to better organized and localized bone formation than the one achieved using non-MSCs (38). However, the use of induction elements such as Smad8 and BMP2 is essential for tissue regeneration because the transplantation of MSCs alone may not have promoted the complete regeneration of damaged tendon tissue (12).
In conclusion, our study demonstrates that genetically modified MSCs that overexpress the active form of the Smad8 molecule and BMP2 could serve as a regenerative platform for the repair of torn ligaments or tendons, as demonstrated in this study in the Achilles tendon model. Our results suggest that the Smad8 molecule inhibits the osteogenic pathway induced by BMP2 and promotes the tendon/ligament differentiation route. Since BMP and Smad molecules play important roles in other nonskeletal tissues such as nerve and liver (39, 40), it is highly conceivable that a novel platform in which interacting Smad signalling pathways are combined may have implications for the development of new therapeutic applications for nonskeletal tissues as well.
Methods
DNA constructs.
Smad8 full-length cDNA was cloned from the brain of a 5-day-old rat using the primers Smad8-FLAG-fw and Smad8-rev. We also constructed Smad1 and Smad8 variants consisting of the linker and the MH2 or MH1 domain using the primer pairs to perform a PCR from full-length Smad clones. Unique Bam HI and Sal I sites in the forward and reverse primer sequences allowed directional integration in the expression vector pMT7T3. The expression of cDNAs in this vector is controlled by a long-terminal repeat of the myeloproliferative virus (41). The integrity of the constructs was confirmed by sequencing. The FLAG tags were added to full-length Smads and their variants at the amino terminus to detect recombinant proteins in a Western blot analysis.
Cell culture.
Murine C3H10T1/2 MSCs were routinely cultured in tissue-culture flasks containing DMEM supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, and antibiotics (50 U/ml penicillin and 50 mg/ml streptomycin). To assess cell differentiation in vitro, the cells were plated at a density of 5–7.5 × 103 cells/cm2. After the cells had reached confluency (arbitrarily termed day 0), 50 μg/ml ascorbic acid and 10 mM β-glycerophosphate were added as specified by Owen et al. (42).
DNA transfection.
The cells were transfected using FUGENE6 according to the manufacturer’s protocol (Roche Diagnostics Corp.). C3H10T1/2-BMP2 cells were obtained by transfection with pSV2pac followed by selection with puromycin (5 μg/ml). The features of these C3H10T1/2-BMP2 cells have been described previously (20, 38, 41, 43, 44). The stable expression of Smads and their variants in C3H10T1/2-BMP2 cells was enabled using cotransfection with pAG60, which conferred resistance to G418 (750 μg/ml). Individual clones were picked, propagated, and tested for recombinant expression by RT-PCR. Approximately 10 individual cellular clones were picked and tested for expression of recombinant Smads by RT-PCR. Clones that demonstrated a high level of transgene expression were propagated and frozen. Selected cell clones were expanded in the presence of puromycin or puromycin/G418, and the selected pressure was maintained during subsequent manipulations.
Preparation of RNA and RT-PCR.
Total cellular RNAs were prepared using Trizol reagent according to the manufacturer’s protocol (Invitrogen Corp.). Five micrograms of total RNA were reverse transcribed, and cDNA aliquots were subjected to PCR. RT-PCR was normalized by the transcriptional levels of hypoxanthine guanine phosphoribosyl transferase (HPRT). The HPRT-specific 5′ and 3′ primers were GCTGGTGAAAAGGACCTCT and AAGTAGATGGCCACAGGACT, respectively. The following 5′ and 3′ primers (listed respectively) were used to evaluate osteo-/chondrogenic or tenogenic differentiation: collagen 1a1, GCCCTGCCTGCTTCGTG and CGTAAGTTGGAATGGTTTTT; collagen 2a1, CCTGTCTGCTTCTTGTAAAAC and AGCATCTGTAGGGGTCTTCT; osteocalcin, GCAGACCTAGCAGACACCAT and GAGCTGCTGTGACATCCATAC; Smad8-FLAG, TTCCGGATCCGCCACCATGGACTACAAGGATGACGATGACAAGCACCCCAGCACCCCCATCAGCTCC and TTCCGTCGACTGGAAATGCAGCTGAGCACATGAC; vector-borne recombinant human BMP2, ACGAGGTCCTGAGCGAGTTC and GATCGAAGCTCAATTAACCCTCAC; PTHR, GTTGCCATCATATACTGTTTCTGC and GGCTTCTTGGTCCATCTGTCC; Six1, AGTTCTCGCCTCACAATCAC and AGGCCTGGAAGAGAATAGTTT; Six2, GGCTCAAGGCGCACTACATC and ACTGCCATTGAGCGAGGAAG; scleraxis, AGCCATCACCCGCCTGTCCATCTC and CGCTGCGCACACTCATC; and EphA4, GCCGCCGGGTACACGACACT and AAGCCCCATGGTTTCAGCAATCTC; and Elastin, TTGGCCCTGGTGGTGTTACT and ATATTTAGCAGCTTTGGATG.
Immunoblot analysis.
Cell pellets of transfected HEK 293T or C3H10T1/2 cells (shock-frozen in liquid nitrogen) were lysed in 1% Nonidet P-40, 150 mM NaCl, 20 mM Tris (pH 7.5), 2 mM EDTA, 50 mM NaF, 1 mM Na2P2O7, and 1 mM phenylmethylsulfonyl fluoride supplemented with protease inhibitors (Protease Inhibitor Mixture Tablets; Roche Molecular Biochemicals). Vectors encoding the constitutively active HA-tagged TGFβ/BMP type I receptors ALK1–ALK6 in pCDNA3 were obtained from P. ten Dijke (Leiden University Medical Center, Leiden, The Netherlands) and C. Heldin (Uppsala University, Uppsala, Sweden) (45), and 1 vector encoding ALK7 was obtained from C. Ibanez (Karolinska Institute, Stockholm, Sweden) (46). Immunoblotting with appropriate antibodies was used to analyze cell extracts. FLAG antibodies were purchased from Sigma-Aldrich and HA antibodies (SC-805) from Santa Cruz Biotechnology Inc.; pSmad1, -5, and -8 antibodies (#9511), which recognize phosphorylated forms of Smad1, Smad5, and Smad8, were obtained from Cell Signaling Technology. Human recombinant BMP2 from E. coli was isolated in the lab and used for induction studies. To evaluate activation levels of Smads, exposed films were scanned with the Epson scanner 1680 Pro and quantified by analysis on a PC using the public domain NIH Image program ImageJ (version 1.33h; http://rsb.info.nih.gov/nih-image/). For analysis of recombinant Smad8 L+MH2 protein expression of stably transfected C3H10T1/2 cell lines, protein content of the cell lysates was determined by the Bradford method, and 25 μg total protein was applied per lane. That equal protein amounts were transferred to the PVDF membranes was confirmed by Ponceau S stain.
Reporter assays.
Reporter assays were performed using the GAL4 DNA-binding domain fused to various forms of Smad proteins as described previously (16). Briefly, in total 1,000 ng DNA was transfected in each well of a 24-well plate seeded with 100,000 HEK 293T cells the day before transfection. The DNA mix included a plasmid expressing β-gal under the strong constitutive RSV promoter for normalization purposes and a reporter plasmid encoding firefly luciferase under the control of 5 GAL4 binding sites upstream of a minimal TATA box (pG5 luc; Promega) and combinations of effector plasmids. When appropriate, transfection reactions were supplemented with an empty vector. Cells were harvested 24 hours after transfection and lysed with a detergent lysis solution (100 mM K-phosphate, pH 7.8; 0.2 % Triton X-100; 0.5 mM DTT). β-Gal activity was measured using the Luminescent β-Galactosidase Kit II from Clontech Laboratories Inc., and luciferase activity was measured using the luciferase assay system from Promega. All results were expressed as luciferase activity normalized with β-gal values (relative luciferase units). Expression of Gal4-SmadWT fusion proteins was assessed using a GAL4 monoclonal antibody (SC-510; Santa Cruz Biotechnology Inc.).
Histological analysis.
C3H10T1/2 MSC clones were cultured in vitro up to 14 days after confluency and were subjected to ALP histochemical staining. In vitro synthesis of ALP was visualized by staining the cells with Fast BCIP/NBT (Sigma-Aldrich).
Tissue samples obtained from the in vivo experiments were fixed in 4% formalin overnight, passed through a graded series of ethanol, and embedded in paraffin. Sections 5 μm thick were cut from each paraffin block using a motorized microtome (Leica Microsystems). H&E staining was performed as described previously (37).
Cell labeling.
To identify genetically engineered MSCs within the site of implantation in vivo, the cells were infected either with an adenovector encoding for the marker gene β-gal or with a retrovirus encoding for the luciferase gene. When the cells had reached 80% confluency, they were infected in vitro with a recombinant adenovirus encoding for β-gal at an MOI of 100. The efficiency of the infection was estimated after infection by using X-gal staining (data not shown) (37). A retrovirus encoding the luciferase gene was obtained by transfecting HEK 293 cells with PCL-ampho and PLNC-Luc plasmids. The HEK 293 cells were transfected upon reaching 80% confluency, the supernatant was collected from the MSC culture, and luciferase expression was estimated by performing a luciferase assay (Promega Inc.; data not shown).
Ectopic implantation model.
All animal experiments were conducted according to the guidelines and approval of the Hadassah Medical Centre and the Hebrew University ethics committee. Aliquots containing 5 × 106 cells were prepared in a suspension (300 μl) of 1 × PBS. The cells were injected s.c. into the sacral region of female C3H/HeN mice (4–8 weeks old). Before implantation, the mice were anesthetized with a ketamine-xylazine mixture (30 μl/mouse) and given cefazolin antibiotic (Cefamezin, 5 mg/mouse; Teva Pharmaceuticals); both drugs were administered intraperitoneally. Skin over the implantation site was swabbed with chlorhexidine gluconate (0.5%) before cell implantation. At 10, 20, or 30 days after implantation, the mice were killed and the implants were excised.
Achilles tendon regeneration model.
A partial-thickness defect in the Achilles tendon was created in athymic rats. Twelve adult athymic rats (each 4 months old) were anesthetized with a ketamine-xylazine mixture (75 mg/kg ketamine and 10 mg/kg xylazine, injected intraperitoneally). Each rat received an intraperitoneal injection of Rimadyl (5 mg/kg) to reduce any postoperative pain or inflammatory response. At the surgery site, the skin was shaved and swabbed with chlorhexidine gluconate (0.5%). The Achilles tendon was separated from the plantaris and soleus tendons, and a 3-mm-long partial-resection defect was created in its lateral region. Genetically engineered MSCs (3 × 106 cells) were seeded onto a collagen sponge (Duragen; Integra Life Sciences), which was then placed into the tendon defect and attached to the tendon using a 6-0 polypropylene monofilament nonabsorbable suture (Figure 7). The skin was closed in a routine manner using 2-0 Mersilk. The tension on the tendons returned to approximately normal. The rats were allowed to move immediately after surgery while in their cages. Four weeks after implantation, the rats were killed by administration of CO2. The Achilles tendon was excised and processed for histological and immunohistochemical analyses.
LCM and real-time RT-PCR.
Samples of Achilles tendon implanted with genetically modified MSCs were embedded in OCT and snap-frozen in liquid N2. Ten-micron sections were obtained from each frozen block using a cryostat. The frozen sections were briefly air-dried on uncoated glass slides and washed with 70% ethanol. The sections were kept on dry ice or at –80°C until they were subjected to LCM. Just before the procedure, the sections were fixed in 70% ethanol for 10 seconds and stained with H&E by immersion using the following protocol: 10 seconds deionized H2O, 30 seconds hematoxylin, 10 seconds deionized H2O, 10 seconds 70% ethanol, 1 minute eosin (alcoholic), 10 seconds 95% ethanol, 10 seconds 100% ethanol, and 30 seconds xylene (47). LCM was performed using an Arcturus PixCell II apparatus. The settings of the laser used in this study were as follows: spot diameter, 15 μm; pulse duration, 50 ms; power, 50 mW. The microdissected area included the implantation site within the host tendon tissue. Every section was dropped onto a separate cap, and 2,500 to 3,000 pulses were used for each cap. RNA was extracted from the isolated cells using Trizol reagent as described recently by Zhan et al. (48). The entire amount of RNA was reverse transcribed to cDNA as described by Moutsatsos et al. (20). Two microliters of cDNA was subjected to real-time PCR using the Roche Light Cycler (Roche Diagnostics Corp.) according to the manufacturer’s protocol. PCR was performed in 20 μl final volume using the Light Cycler Fast Start Master SYBR Green I kit (Roche Diagnostics Corp.) with specific primers for the housekeeping gene RPL19 and murine collagen I.
DQF MRI of a partial defect in a rat tendon.
Proton (1H) DQF MRI provides new contrast enhancement based on the nonzero average of the dipolar interaction caused by the anisotropic motion of water molecules; this is determined by the interaction between the water molecules and the ordered collagen fibers. DQF MRI provides high-contrast signals, differentiating the tissues of tendon, bone, skin, and muscle. Tendons, which produce a weak signal in standard MRI sequences, are highlighted in the 1H DQF image (45, 46).
A partial defect was created in the Achilles tendon of nude rats as described above, and a collagen I sponge loaded with engineered MSCs was implanted within each defect. At 1 or 5 weeks after implantation, the rats were killed. Both the surgically treated leg and the intact control leg were subjected to a micro-MRI analysis, and multi-slice-multi-echo (MSME) images were obtained. The same samples were further subjected to DQF MRI, in which only the tendons were analyzed (46, 47).
Both MSME and DQF images were acquired at the level of the induced defect and at the corresponding level in the contralateral tendon. The following parameters were used: for sagittal MSME images, data matrix 256 × 128, FOV 2 × 2 cm, TR/TE 1 s/6.2 ms, slice thickness 1 mm, averages 4 (black signal indicates short T2 and bright signal indicates long T2); for axial MSME images, data matrix 256 × 128, FOV 1 × 1 cm, TR/TE 1 s/6.2 ms, slice thickness 2 mm, averages 12 (black signal indicates short T2 and bright signal indicates long T2); for sagittal DQF images, data matrix 128 × 128, FOV 2 × 2 cm, slice thickness 1 mm, creation times 0.4, 0.6, and 0.8 ms, averages 144 (bright signal indicates short T2); and for axial DQF images, data matrix 128 × 128, FOV 1 × 1 cm, slice thickness 2 mm, creation time 0.4 ms.
In vivo molecular imaging.
To evaluate the survival of implanted cells in the tendon defect, we used a noninvasive tracking system. Since the cells had been infected with a retroviral luciferase vector before implantation, we were able to detect the luciferase signal within the site of implantation using a CCCD tracking system (Roper Chemiluminescence Imaging System described by Bar et al.; ref. 49) consisting of a CCCD camera (model LN/CCD-1300EB; Roper Scientific Inc.) equipped with an ST-133 controller and a 50-mm Nikon lens (Nikon Inc.). In this system, a pseudocolor image represents light intensity (blue signifies least intense and red most intense). The integrated light is the result of 2-minute exposure and acquisition. This CCCD camera converts photons to electrons with an efficiency of approximately 90% in the wavelength emission range of luciferin and converts approximately 1.8 electrons per count. Thus every count represents 2 photons. The measurement is a total integrated signal of a constant exposure interval. Whenever the exposure conditions (including time, F-stop, position of stage, binding ratio, and time after injection with luciferin) are kept identical, the measurements are comparable. The measurement can be verified by using a progressively larger region of interest (ROI) and by subtracting backgrounds that are the same. An increase in the ROI has no appreciable effect on the total intensity that is measured. Before light detection, the rats were anesthetized with a ketamine-xylazine mixture, which was injected intraperitoneally at 1 μl/g body weight. Ten minutes before monitoring the light emission, the animals were given intraperitoneal injections of beetle luciferin (Promega) in PBS (126 mg/kg body weight). It usually takes 10 minutes for luciferase activity to reach its peak (49). After the image had been superimposed in real time over the grayscale image of the rat, the animal was placed in a dark box, which was supplemented with a controlled light so that pictures of the background image could be obtained, and the animal was exposed to the CCCD system. The composite image was transferred to a personal computer by using a plug-in module.
In situ detection β-gal expression (immunohistochemical analysis).
Surgically treated tendons were fixed in 4% formalin for 24 hours, embedded in paraffin, and cut into 5-mm-thick sections. The sections were then transferred to glass slides (Superfrost; Menzel-Gläser). The slides were heated at 65°C for 45 minutes; this was followed by deparaffinization, which was accomplished by soaking the slides in xylene 2 times for 5 minutes. The tissue sections were subsequently hydrated in a series of baths containing descending grades of ethanol and rinsed in PBS. Endogenous peroxidase activity was removed by treatment with 0.1% H2O2 for 30 minutes. The sections were incubated with blocking solution (CAS block; Zymed) for 30 minutes at room temperature. Primary β-gal antibody (polyclonal rabbit anti-mouse, affinity-purified Cortex CR2029RAP; Cortex Biochem), diluted 1:100 in PBS, was applied to the slides for 1 hour at room temperature. After incubation with the primary antibody the slides were rinsed in PBS, and a secondary goat anti–rabbit IgG antibody (biotin-conjugated; Zymed, 626140), diluted 1:100 in PBS, was applied to the slides at room temperature for 30 minutes. After the slides had been washed with PBS (2 times for 5 minutes), they were incubated with horseradish peroxidase conjugated to streptavidin and given an application of 3-amino-9-ethyl-carbazole dye for visualization with light microscopy. The slides were stained with hematoxylin, washed, and mounted.
Acknowledgments
We would like to thank P. ten Dijke, C. Heldin, and C. Ibanez for vectors encoding constitutively active TGF-β/BMP type I receptors. This study was funded in part by GENOSTEM (EU-FR6-integrated project).
Footnotes
Nonstandard abbreviations used: ALK, activin-like receptor kinase; ALP, alkaline phosphatase; BMP, bone morphogenetic protein; CCCD, cooled charge-coupled device; DQF, double quantum filtered; GDF, growth differentiation factor; HEK, human embryonic kidney; LCM, laser capture microdissection; L+MH2, R-Smad linker domain plus the MH2 domain; MSC, mesenchymal stem cell; MSME, multi-slice-multi-echo; R-Smad, receptor-regulated Smad.
Conflict of interest: The authors have declared that no conflict of interest exist
Andrea Hoffmann and Gadi Pelled contributed equally to this work.
See the related Commentary beginning on page 863.
References
- 1.Pennisi E. Tending tender tendons. Science. 2002;2955:1011. doi: 10.1126/science.295.5557.1011. [DOI] [PubMed] [Google Scholar]
- 2.Jackson D.W., Simon T.M., Kurzweil P.R., Rosen M.A. Survival of cells after intra-articular transplantation of fresh allografts of the patellar and anterior cruciate ligaments. DNA-probe analysis in a goat model. J. Bone Joint Surg. Am. 1992;74:112–118. [PubMed] [Google Scholar]
- 3.Sabiston P., Frank C., Lam T., Shrive N. Allograft ligament transplantation. A morphological and biochemical evaluation of a medial collateral ligament complex in a rabbit model. Am. J. Sports Med. 1990;18:160–168. doi: 10.1177/036354659001800209. [DOI] [PubMed] [Google Scholar]
- 4.Olson E.J., et al. The biochemical and histological effects of artificial ligament wear particles: in vitro and in vivo studies. Am. J. Sports Med. 1988;16:558–570. doi: 10.1177/036354658801600602. [DOI] [PubMed] [Google Scholar]
- 5.Woods G.A., Indelicato P.A., Prevot T.J. The Gore-Tex anterior cruciate ligament prosthesis. Two versus three year results. Am. J. Sports Med. 1991;19:48–55. doi: 10.1177/036354659101900108. [DOI] [PubMed] [Google Scholar]
- 6.Wolfman N.M., et al. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-β gene family. J. Clin. Invest. 1997;100:321–330. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helm G.A., et al. A light and electron microscopic study of ectopic tendon and ligament formation induced by bone morphogenetic protein-13 adenoviral gene therapy. J. Neurosurg. 2001;95:298–307. doi: 10.3171/jns.2001.95.2.0298. [DOI] [PubMed] [Google Scholar]
- 8.Aspenberg P., Forslund C. Enhanced tendon healing with GDF 5 and 6. Acta Orthop. Scand. 1999;70:51–54. doi: 10.3109/17453679909000958. [DOI] [PubMed] [Google Scholar]
- 9.Caplan A.I. Mesenchymal stem cells. . J. Orthop. Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 10.Bruder S.P., Fink D.J., Caplan A.I. Mesenchymal stem cells in bone development, bone repair, and skeletal regeneration therapy. J.Cell. Biochem. 1994;56:283–294. doi: 10.1002/jcb.240560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis J.E., et al. A quadripotential mesenchymal progenitor cell isolated from the marrow of an adult mouse. J. Bone Miner. Res. 1999;14:700–709. doi: 10.1359/jbmr.1999.14.5.700. [DOI] [PubMed] [Google Scholar]
- 12.Awad H.A., et al. Autologous mesenchymal stem cell-mediated repair of tendon. Tissue Eng. 1999;5:267–277. doi: 10.1089/ten.1999.5.267. [DOI] [PubMed] [Google Scholar]
- 13.Heldin C.H., Miyazono K., Ten Dijke P. TGF-β signalling from cell membrane to nucleus through Smad proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 14.Liu F., et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 15.Baker J.C., Harland R.M. A novel mesoderm inducer, Madr2 functions in the activin signal transduction pathway. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 16.Meersseman G., et al. The C-terminal domain of Mad-like signal transducers is sufficient for biological activity in theXenopus embryo and transcriptional activation. . Mech. Dev. 1997;61:127–140. doi: 10.1016/s0925-4773(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 17.Ju W., et al. The bone morphogenetic protein 2 signaling mediator Smad1 participates predominantly in osteogenic and not in chondrogenic differentiation in mesenchymal progenitors C3H10T1/2. J. Bone Miner. Res. 2000;15:1889–1899. doi: 10.1359/jbmr.2000.15.10.1889. [DOI] [PubMed] [Google Scholar]
- 18.Akimaru H., et al. Drosophila CBP is a co-activator ofcubitus interruptus inhedgehog signalling. . Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura R., et al. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 1998;273:1872–1879. doi: 10.1074/jbc.273.4.1872. [DOI] [PubMed] [Google Scholar]
- 20.Moutsatsos I.K., et al. Exogenously regulated stem cell-mediated gene therapy for bone regeneration. Mol. Ther. 2001;3:449–461. doi: 10.1006/mthe.2001.0291. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann A., et al. The T-box transcription factor Brachyury mediates cartilage development in mesenchymal stem cell line C3H10T1/2. J. Cell Sci. 2002;115:769–781. doi: 10.1242/jcs.115.4.769. [DOI] [PubMed] [Google Scholar]
- 22.Kaps C., et al. Distinct roles of BMP receptors type IA and IB in osteo-/chondrogenic differentiation in mesenchymal progenitors (C3H10T1/2). Biofactors. . 2004;20:71–84. doi: 10.1002/biof.5520200202. [DOI] [PubMed] [Google Scholar]
- 23.Kawai S., et al. Mouse smad8 phosphorylation downstream of BMP receptors ALK-2, ALK-3, and ALK-6 induces its association with Smad4 and transcriptional activity. Biochem. Biophys. Res. Commun. 2000;271:682–687. doi: 10.1006/bbrc.2000.2704. [DOI] [PubMed] [Google Scholar]
- 24.Valcourt U., Gouttenoire J., Moustakas A., Herbage D., Mallein-Gerin F. Functions of transforming growth factor-beta family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J. Biol. Chem. 2002;277:33545–33558. doi: 10.1074/jbc.M202086200. [DOI] [PubMed] [Google Scholar]
- 25.Aoki H., et al. Synergistic effects of different bone morphogenetic protein type I receptors on alkaline phosphatase induction. J. Cell Sci. 2001;114:1483–1489. doi: 10.1242/jcs.114.8.1483. [DOI] [PubMed] [Google Scholar]
- 26.Cserjesi P., et al. Scleraxis: A basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121:1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- 27.Schweitzer R., et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 28.Settle S.H., Jr., et al. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 29.Mikic B., Schalet B.J., Clark R.T., Gaschen V., Hunziker E.B. GDF-5 deficiency in mice alters the ultrastructure, mechanical properties and composition of the Achilles tendon. J. Orthop. Res. 2001;19:365–371. doi: 10.1016/S0736-0266(00)90018-4. [DOI] [PubMed] [Google Scholar]
- 30.Clark R.T., et al. GDF-5 deficiency in mice leads to disruption of tail tendon form and function. Connect. Tissue Res. 2001;42:175–186. doi: 10.3109/03008200109005648. [DOI] [PubMed] [Google Scholar]
- 31.D’Souza D., Patel K. Involvement of long- and short-range signalling during early tendon development. Anat. Embryol. (Berl.). 1999;200:367–375. doi: 10.1007/s004290050286. [DOI] [PubMed] [Google Scholar]
- 32.Ebisawa T., et al. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. 1999;112:3519–3527. doi: 10.1242/jcs.112.20.3519. [DOI] [PubMed] [Google Scholar]
- 33.Shi W., et al. Overexpression of Smurf1 negatively regulates mouse embryonic lung branching morphogenesis by specifically reducing Smad1 and Smad5 proteins. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L293–L300. doi: 10.1152/ajplung.00228.2003. [DOI] [PubMed] [Google Scholar]
- 34.Aspenberg P., Forslund C. Bone morphogenetic proteins and tendon repair. Scand. J. Med. Sci. Sports. 2000;10:372–375. doi: 10.1034/j.1600-0838.2000.010006372.x. [DOI] [PubMed] [Google Scholar]
- 35.Storm E.E., et al. Limb alterations inbrachypodism mice due to mutations in a new member of the TGFβ-superfamily. . Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 36.Jadlowiec J.A., Zhang X., Li J., Campbell P.G., Sfeir C. Extracellular matrix–mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of bone morphogenetic protein. J. Biol. Chem. . 2005;281:5341–5347. doi: 10.1074/jbc.M506158200. [DOI] [PubMed] [Google Scholar]
- 37.Turgeman G., et al. Engineered human mesenchymal stem cells: a novel platform for skeletal cell mediated gene therapy. J. Gene Med. 2001;3:240–251. doi: 10.1002/1521-2254(200105/06)3:3<240::AID-JGM181>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 38.Gazit D., et al. Engineered pluripotent mesenchymal cells integrate and differentiate in regenerating bone: a novel cell-mediated gene therapy. J. Gene Med. 1999;1:121–133. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<121::AID-JGM26>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 39.Helm G.A., Alden T.D., Sheehan J.P., Kallmes D. Bone morphogenetic proteins and bone morphogenetic protein gene therapy in neurological surgery: a review. Neurosurgery. 2000;46:1213–1222. doi: 10.1097/00006123-200005000-00038. [DOI] [PubMed] [Google Scholar]
- 40.Duncan S.A., Watt A.J. BMPs on the road to hepatogenesis. Genes Dev. 2001;15:1879–1884. doi: 10.1101/gad.920601. [DOI] [PubMed] [Google Scholar]
- 41.Ahrens M., et al. Expression of human bone morphogenetic proteins -2 or -4 in murine mesenchymal progenitor C3H10Tν cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol. 1993;12:871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- 42.Owen T.A., et al. Progressive development of the rat osteoblast phenotype in vitro, reciprocal relationships in the expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 43.Hollnagel A., Ahrens M., Gross G. Parathyroid hormone enhances early and suppresses late stages of osteogenic and chondrogenic development in a BMP-dependent mesenchymal differentiation system (C3H10T1/2). J. Bone Miner. Res. 1997;12:1993–2004. doi: 10.1359/jbmr.1997.12.12.1993. [DOI] [PubMed] [Google Scholar]
- 44.Bächner D., et al. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2). Dev. Dyn. 1998;213:398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 45.Fechete R., Demco D.E., Blumich B., Eliav U., Navon G. Anisotropy of collagen fiber orientation in sheep tendon by 1H double-quantum-filtered NMR signals. J. Magn. Reson. 2003;162:166–175. doi: 10.1016/s1090-7807(02)00200-8. [DOI] [PubMed] [Google Scholar]
- 46.Ikoma K., et al. Evaluation of collagen fiber maturation and ordering in regenerating tendons employing H-1 double quantum filtered NMR spectroscopy. J. Orthop. Res. 2003;21:149–156. doi: 10.1016/S0736-0266(02)00092-X. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen T.X., et al. Laser capture microdissection-based in vivo genomic profiling of wound keratinocytes identifies similarities and differences to squamous cell carcinoma. Oncogene. 2003;22:3964–3976. doi: 10.1038/sj.onc.1206614. [DOI] [PubMed] [Google Scholar]
- 48.Zhan G., Shaheen F., Mackiewicz M., Fenik P., Veasey S.C. Single cell laser dissection with molecular beacon polymerase chain reaction identifies 2A as the predominant serotonin receptor subtype in hypoglossal motoneurons. Neuroscience. 2002;113:145–154. doi: 10.1016/s0306-4522(02)00137-9. [DOI] [PubMed] [Google Scholar]
- 49.Bar I., et al. Molecular imaging of the skeleton: quantitative real-time bioluminescence monitoring gene expression in bone repair and development. J. Bone Miner. Res. 2003;18:570–57. doi: 10.1359/jbmr.2003.18.3.570. [DOI] [PubMed] [Google Scholar]