Abstract
The Ter mutation causes primordial germ cell (PGC) loss on all mouse genetic backgrounds (Fig. 1a) with deficiency of PGCs1 starting at embryonic day 8. Ter is also a potent modifier of spontaneous testicular germ cell tumour (TGCT) susceptibility in the 129 family of inbred strains and increases TGCT incidence from a baseline rate of 5% in 129 to 94% in 129-Ter/Ter males2-4 (Figs. 1b & c). In 129, some of the remaining PGCs transform into undifferentiated pluripotent embryonal carcinoma (EC) cells2-6 and after birth they differentiate into various cells and tissues that compose TGCTs. Positional cloning of Ter revealed a point mutation that introduces a termination codon in the mouse ortholog (Dnd1) of the zebrafish dead-end (dnd) gene. PGC deficiency is corrected both with BACs that contain Dnd1 and with a Dnd1 encoding transgene. Dnd1 is expressed in fetal gonads during the critical period when TGCTs originate. DND1 has an RNA recognition motif (RRM) and is most similar to the apobec complementation factor (Acf), a component of the cytidine to uridine RNA editing complex. These results suggest that Ter may adversely affect essential aspects of RNA biology during PGC development. DND1 is the first protein with an RRM that is directly implicated as a heritable cause of spontaneous tumourigenesis. TGCT development of the 129-Ter strain models pediatric TGCT in humans. This work will have important implications for our understanding of the genetic control of TGCT pathogenesis and PGC biology.
Ter was mapped to mouse Chromosome 18 near Fgf1 by following inheritance of genetic markers and two phenotypes - tumour incidence and PGC deficiency/7,8. Testis weight was used as a surrogate quantitative phenotype for Ter in high resolution interspecific and intersubspecific backcrosses and intercrosses (Supplementary Fig. 1). 2753 meioses were examined from crosses between B6.129-Ter congenic strain and A/J, C57BL/6J, C3H/HeJ and MOLF/Ei strains to avoid recombination ‘hot’ and ‘cold’-spots that may exist in the genome of particular strains. Genotyping of F2 progeny with microsatellite markers narrowed the critical interval to 0.14 cM between 273 and 173RB (Fig. 2a). Four overlapping BACs spanned the Ter locus: RG-MBAC_173P21, RG-MBAC_282N17, RG-MBAC_273D11 and RG-MBAC_284F9 derived from the 129 strain.
Figure 2.
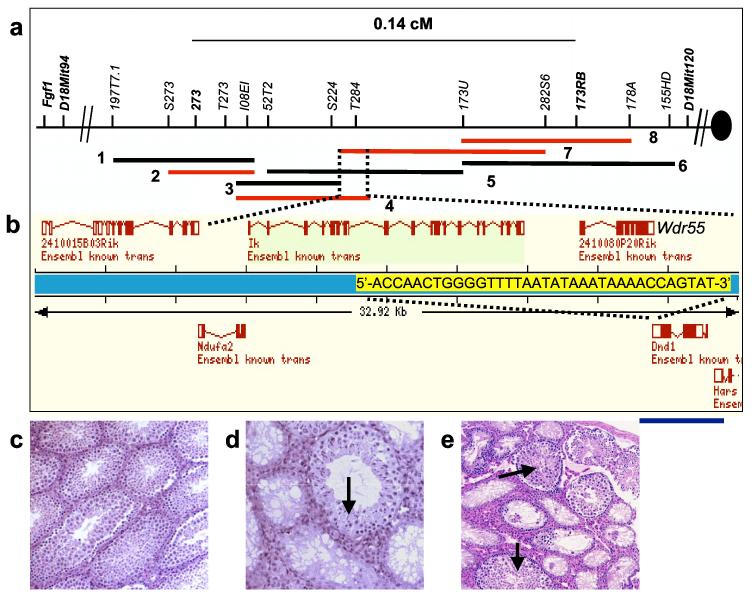
Positional cloning of Ter. (a) Physical map of the Ter locus. The 129-derived (RG-MBAC) BAC contig: 1 = 197J23, 2 = 173P21, 3 = 224K2, 4 = 284F9, 5 = 52N10, 6 = 368A6, 7 = 282N17, 8 = 273D11. (b) Ensembl gene map of the overlapping region of BACs 284F9 and 282N17. Overlapping bases of the last exons of Wdr55 and Dnd1 are marked in yellow. Blue line indicates the DNA fragment used in Tg(Dnd1)1Matn. (c) Testes histology of Ter/Ter with BAC 284F9 and sibling (d) also with transgene where rescue occurs in a subset of seminiferous tubules (arrow). (e) Histology of Ter/Ter testes with Tg(Dnd1)1Matn (arrow showing normal seminiferous tubule).
We used BAC complementation tests to further reduce the size of the critical interval. Transgenic mice were generated with each of the four overlapping BACs. They were crossed to B6.129-Ter congenic mice to eventually generate Ter/Ter mice carrying individual transgenes. Mice with BACs 282N17 and 284F9 but not BACs 273D11 and 173P21 rescued the germ cell deficient phenotype (Supplementary Fig. 1), fully or partially restoring fertility in Ter/Ter mice as assessed histologically (Figs. 2c and d) and by breeding performance (not shown), thus demonstrating that the overlapping region between 282N17 and 284F9 harbours Ter.
Four genes are present within the overlapping region of BACs 282N17 and 284F9 (Fig. 2b). Comparison of the sequences of all the exons of these four genes from 129- Ter/Ter and 129-+/+ mice revealed a single base change (C to T) in the coding region of the Dead-end1 (Dnd1) gene (Figs. 3a and b), which is the zebrafish ortholog of dnd9. This substitution introduces a stop codon (R178X) in the coding regions of both isoforms of mouse Dnd1 encoded proteins (DND1) at amino acids 178 and 190, respectively (Fig. 3a; accession numbers BC034897 and AY321066). The mutation was found only in mice carrying Ter, but not in 129 substrains (129T2/SvEmsJ, 129S1/SvImJ, 129X1/SvJ) or other inbred strains (A/J, C57BL/6J, C3H/HeJ and MOLF/Ei). The base change introduces a new Dde1 restriction enzyme site within the Dnd1 sequence enabling mice with the Ter allele to be distinguished from wild-type (Supplementary Fig. 1).
Figure 3.
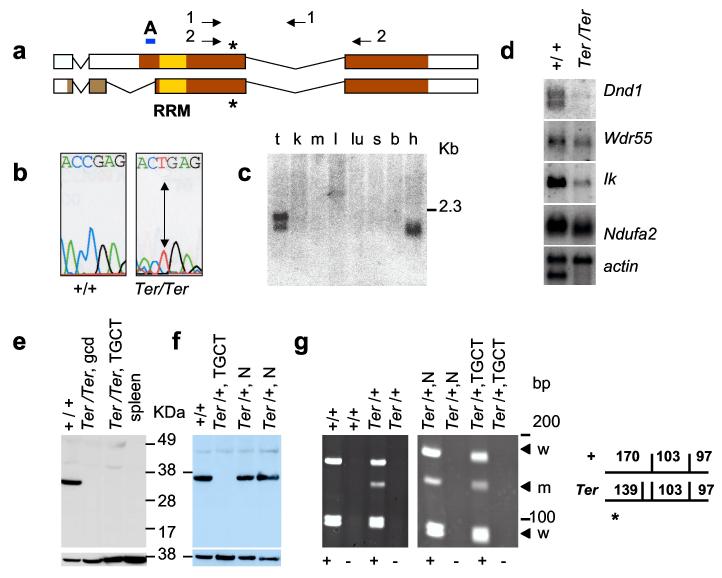
Expression of Dnd1 in normal tissues and TGCTs. (a) Comparison of DND1 isoforms with accession numbers BC034897 (top) and AY321066 (bottom). Colored regions indicate protein coding regions; yellow box marks RRM. Asterisk indicates amino acids 178 and 190, of the two isoforms, that is mutated (R178X) in Ter. Arrows indicate primers 4.13a (1) and A25 (2). A indicates the region used as peptide epitope to generate anti-DND1 antibody. (b) C to T mutation in Ter/Ter introduces stop codon. (c) Mouse tissue Northern blot for Dnd1. mRNAs are from testes (t), kidney (k), muscle (m), liver (l), lung (lu), spleen (s), brain (b) and heart (h). (d) Northern blot of mRNA from 129-+/+ testes and 129-Ter/Ter tumours. (e) Western blotting with anti-DND1 antibody of tissue lysates from normal (+/+) testes of 129-+/+, germ cell deficient (gcd) testes of B6.129-Ter/Ter congenic strain, bilateral TGCTs from 129-Ter/Ter mice, and spleen of 129-+/+. Lower panel shows control for loading using anti-β-actin. (f) Western blot with anti-DND1 of normal testes of 129-+/+ (+/+), tumour in the testes of 129-Ter/+ (Ter/+, TGCT), the normal contralateral testes (Ter/+, N), and normal testes of another 129-Ter/+ mouse (Ter/+, N). Lower panel shows the same blot re-hybridized with mouse anti-β-actin. (g) (left) RT-PCR of RNA from normal testes of 129-+/+ and 129-Ter/+ mice using primers A25 (2 in Fig. 3a) followed by Dde1 digestion. This produces fragments of 170, 103 and 97 bp from the + allele and 139, 103, 97 and 31 bp (not seen on gel) from the Ter allele. For controls, PCR reactions were performed on samples which included (+) or did not include Superscript II (-) during RT, as indicated below the panels. (right) Total RNA from normal and TGCT-bearing testes of the same 129-Ter/+ mouse was amplified by RT-PCR followed by Dde1 digestion and electrophoresis. The arrows indicate the wild-type (w) and Ter (m) allele.
To confirm that Dnd1 is Ter, we used a 3.9 kb genomic DNA fragment derived from BAC 284F9 that encodes only the Dnd1 gene plus 860 bp and 510 bp flanking 5′ and 3′ ends, respectively, as a transgene (Tg(Dnd1)1Matn) to rescue the Ter/Ter phenotype. Ter/Ter males positive for Tg(Dnd1)1Matn, showed partial rescue of the germ cell deficient phenotype (Fig. 2e). Although many seminiferous tubules remained germ cell deficient, some contained immature and mature sperm, which is unprecedented in Ter/Ter males.
Double-stranded RNAs, which can result from expression of overlapping genes on opposite strands, can lead to loss of mRNA or translational silencing10,11. Dnd1 and Wdr55 (2410080P20Rik) are encoded on opposite strands of DNA and overlap by 35 bp in the 3′ ends of their last exon (Fig. 2b). To test whether the mutation impairs function of Wdr55 and indirectly leads to the Ter phenotype, we created mice with targeted deletion of Wdr55 (wdr55tm1Matn) (Supplementary Fig. 2). Mice homozygous for wdr55tm1Matn are embryonic lethal and die before E9.5. This is unlike Ter/Ter mice, which are viable but sterile. Moreover, double heterozygote males (+wdr55tm1Matn/Ter+) were not germ cell deficient and had gonad/body mass ratios ranging from 3.3 - 4.1, which is typical of wild-type but not Ter/Ter mice. Thus, Ter is not an allele of Wdr55. Therefore, functional complementation of Tg(Dnd1)1Matn transgenic mice together with normal testes in double heterozygotes demonstrates that the mutation in Dnd1 directly causes the Ter phenotype.
Expression of Dnd1Ter was characterized by Northern and Western blotting. Northern analysis revealed that transcripts of both Dnd1 isoforms are expressed in adult testes but one isoform in adult heart (Fig. 3c). Dnd1 transcripts were significantly decreased in tumours from 129-Ter/Ter males (Fig. 3d). Transcript levels of the other genes in the critical interval, Wdr55, Ik and Ndufa2, were also reduced to varying extents in tumour tissue suggesting that these genes also function in normal adult testes. The two DND1 protein isoforms differ at the first 33 and 45 amino acids of their N-terminus. A polyclonal antibody was generated against an 18 amino acid peptide of one isoform of DND1 (indicated by A, Fig. 3a). Western blotting of lysates from normal testes revealed a protein of 38 kDa, which is nearly the expected size (37.5 kDa) of the DND1 protein (Supplementary Fig. 1). This antibody also recognized a 62.5 kDa GST(glutathione Stransferase)-DND1 fusion protein. DND1 was not detected in protein lysates from germ cell deficient testes of three different B6.129-Ter/Ter or tumours of four different 129-Ter/Ter males (Fig. 3e). Thus, the various cell types of the tumours do not retain DND1 expression. However, a truncated DND1 protein was also not detected in Ter/Ter tumours. This rules out the possibility that tumour development is due to a dominantnegative acting oncogenic truncated DND1 protein. We note that the antibody we used detects one isoform of DND1 and cannot rule out the possibility that the other isoform is still expressed. However, that appears unlikely because the mutation introduces a stop codon in both isoforms of DND1.
129-Ter/+ mice have a modestly elevated TGCT incidence with 17% of the males having a unilateral tumour2. To test how Dnd1 is involved in tumour development in heterozygous males, we obtained testicular tumours and contralateral normal testes from two 129-Ter/+ mice and bisected each of these tissues to examine Dnd1 protein and RNA expression. DND1 was not detected in tumour protein lysates (Fig. 3f), although the contralateral normal testes of the 129-Ter/+ expressed DND1. To determine the mechanism of protein loss, we examined RNA from the 129-Ter/+ tumour samples. Because the tumour sizes were typically small, we used RT-PCR to amplify the fulllength Dnd1 cDNA and by sequencing detected both the normal wild-type and mutated transcripts from tumour tissues. We then developed an RT-PCR based assay (see Methods) to visually distinguish mRNA from the normal (+) and the Dnd1Ter allele (Fig. 3g, left). Using this method, the wild-type transcript was observed in tumours as well as in normal testes (Fig. 3g, right). As RT-PCR is a sensitive technique we were also able to amplify the Dnd1Ter transcript (Fig. 3g, labelled m) from the Ter/+ RNA. However, the Dnd1Ter transcript band is weaker compared to the wild-type allele. Taking into consideration both the Western (Fig. 3f) and RT-PCR data (Fig. 3g, left), it indicates that a second ‘hit’ has inactivated DND1 expression from the wild-type allele of 129-Ter/+ and thus resulted in tumour development. In the case of the two samples examined, this is likely due to a post-transcriptional mechanism such as translational silencing12,13.
The defect that underlies PGC deficiency and TGCT susceptibility in Ter males occurs during embryogenesis1. We therefore examined normal embryonic expression of Dnd1 and detected it at earlier stages than reported previously9. Whole-mount in situ hybridization using a Dnd1 specific RNA probe detected expression in the embryo and allantoic bud at E7.5, neuroectoderm at E8.5, and wide-spread expression in E9.5 embryo including the neural tube, head mesenchyme, first branchial arch and the hindgut, through which primordial germ cells are migrating (Supplementary Fig. 3). Dnd1 expression was also detected in the XY and XX genital ridges of E11.5 embryos (Figs. 4a & b). Dnd1 expression was up-regulated in the testis cords of the XY gonad between E12.5 and E14.5 (Figs. 4c & e), but down-regulated between E12.5 and E14.5 in XX gonads (Figs. 4d & f). Down-regulation of Dnd1 in the XX gonad occurred progressively from anterior to posterior, similar to the wave of meiotic entry of XX germ cells14,15. The contrasting patterns of Dnd1 expression in XX versus XY gonads may account for the differential susceptibility to teratocarcinogenesis in female versus male 129-Ter/Ter mice.
Figure 4.

Expression of Dnd1 in embryonic gonads. (a) Whole-mount in situ hybridization of E11.5 XY and (b) E11.5 XX genital ridge (g) and mesonephros (m), in which Dnd1 expression is localized to the gonad. (c) Whole-mount in situ hybridization of an E12.5 XY gonad (t), in which Dnd1 is localized to testis cords (tc), whereas Dnd1 expression in (d) E12.5 XX gonad (o) has a broad distribution throughout the gonad. (e) Whole-mount in situ hybridization of an E14.5 XY gonad, in which Dnd1 is localized to testis cords, in contrast to (f) E14.5 XX gonads in which Dnd1 expression is reduced. (g) and (h) are section in situ hybridization of an E12.5 XY and E13.5 XY gonad respectively. Dnd1 is specific to developing testicular cords in the E12.5 and E13.5 XY gonads. Scale bars represent 500 μm.
To visualize PGCs in 129-Ter/Ter embryos, we introduced GFP (green fluorescent protein) transgene driven by the PGC-specific Oct-4 promoter16, GOF-1/PE/EGFP, in the 129-Ter strain. At E12.5, the number of GFP-labelled PGCs was significantly reduced in the urogenital ridges of 129-Ter/Ter compared to 129-+/+ embryos (Fig. 1d to f). However, the few surviving PGCs in Ter/Ter embryos had successfully migrated to the genital ridges1 and tumours eventually occur in the testes of Ter/Ter mice. Therefore, Dnd1 is not essential for PGC migration in the mouse. Loss of PGCs in 129-Ter/Ter embryos precedes development of EC cells and TGCTs. Inactivation of Dnd1 expression in PGCs during embryogenesis in 129-Ter/Ter mice is therefore implicated as the causal event that drives PGCs to exit the germ-line and transform to EC cells and TGCTs.
Figure 1.

Gonadal phenotypes of Ter/Ter males. (a) Testis of 4-week old Ter/Ter male showing lack of germ cells (arrow points to Sertoli cell only phenotype) in the seminiferous tubules. (b,c) Histological sections through testicular germ cell tumours (TGCTs) from 4-week old 129-Ter/Ter male mice. Tissue types observed includes cartilage (arrow in b) and neuroepithelia (* in c). Arrow in (c) indicates germ cell deficient seminiferous tubules adjacent to neuroepithelial cells of the tumour. (d) GFP-tagged PGCs. Genital ridges of E12.5 embryos showing GFP expression in PGCs of 129-+/+;Oct4-GFP male, (e) 129-Ter/Ter;Oct4-GFP male, and (f) 129-Ter/Ter;Oct4-GFP female. The arrow indicates one of the genital ridges of the dissected embryo.
The mouse DND1 protein shows sequence identity to proteins with RNA recognition motifs (RRMs), with highest similarity to mouse apobec-1 complementation factor (Acf, XP_129159; 34% overall amino acid identity and 48% identity in the RRM) (Supplementary Fig. 4). Acf encodes the RNA-binding subunit of the RNA editing enzyme complex (editosome) that converts specific cytidines to uridines in the apolipoprotein B transcript and other RNAs17. Several RNA binding proteins are anomalously expressed in certain cancers18-23 but it is unclear whether these are causes or consequences of tumourigenesis. DND1 may be an RNA or DNA-binding subunit of other apobec-like proteins24 involved in nucleic-acid editing and the Ter mutation raises the possibility that TGCT susceptibility is a consequence of aberrant nucleic-acid editing.
Various aspects of RNA biology play central roles in the development of the PGC lineage25-27. We show that functional inactivation of mouse Dnd1 gene results in severe germ cell deficiency and increased TGCT susceptibility. Our findings implicate a causal role of RNA biology in TGCT susceptibility as DND1 has an RRM motif28 and is most similar to Acf, which is involved in RNA editing. Extrapolating from the findings in Ter mice, although germ cell tumours present clinically in infants and young adults, it is apparent that genetic and environmental influences during embryogenesis increase the susceptibility of PGCs to tumourigenesis. 129-Ter mice are a valuable model system to study the initiating events of tumourigenesis because the cell of origin of the tumours and the time of onset is defined and now one of the defects, the Ter mutation in Dnd1, that predisposes to PGC transformation has been identified.
Methods
Nomenclature. The ortholog of Ter in zebrafish is the dead-end (dnd) gene9. Based on orthology with dnd, we call the mouse gene Dead-end, with gene and allele symbol Dnd1 and Dnd1Ter, respectively. Mouse Dnd1 accession numbers are BC034897 and AY321066. The gene 2410080P20Rik (NM_026464) on the opposite strand and partially overlapping with Dnd1 was named Wdr55. Accession numbers for Ik and Ndufa2 are NM_011879 and NM_010885, respectively.
Generation of genetic and physical maps. Polymorphic microsatellites were subcloned from the BACs using colony hybridization and mapped to Chr 18 (primers to amplify the polymorphic markers are listed in Supplementary Fig. 4). Accession numbers of the BACs are: AC027276 for RG-MBAC_173P21; AC027278 for RG-MBAC_284F9; AC087795 for RP23-181H5 (which is the C57BL/6J equivalent of RG-MBAC_282N17); AC087772 for RP23-326L17 (which is the C57BL/6J equivalent of RG-MBAC_2849F). BAC RG-MBAC_273D11 was not sequenced.
Creation of BAC and Tg(Dnd1)1Am transgenics. BACs were purified, linearized and microinjected into FVB fertilized one-cell embryos29. PCR analysis to test for presence of the BAC transgenes was carried out using the primers BAC-51-F 5′-cttaactatgcggcatcagagc-3′ and BAC-201-R 5′-gccagctggcgtaatagcgaag-3′ which amplified a 173 bp fragment from the vector-insert junction. Mice carrying the 129-derived chromosomal segment spanning D18Mit232, D18Mit94 and D18Mit17 (Ter/+) and positive for BAC transgenes were intercrossed. Genotyping for the 129-derived chromosomal segment identified Ter/Ter mice bearing individual BAC transgenes. To generate Tg(Dnd1)1Matn, BAC 284F9 was digested with BglII and Nhe1 and subcloned into pBlueScript II KS+. The 3.9 kb fragment encoding Dnd1 was identified by hybridization with 32P-labelled PCR product of primers 4.13a-F and 4.13a-R (Supplementary Methods). The 3.9 kb fragment was purified, linearized, and microinjected into FVB fertilized one-cell embryos. Primers designed against the vectorinsert boundary, A16.1-F (5′-gtcatccttgcacgctgcgcacg-3′) and A16.1-R (5′gcttgatatcgaattcctg-3′), were used for PCR genotyping for presence of Tg(Dnd1)1Matn. Founder mice were crossed to the B6.129-Ter congenic strain. Mice carrying the 129-derived chromosomal segment spanning D18Mit232, D18Mit94 and D18Mit17 (Ter/+) and positive for the Tg(Dnd1)1Matn transgene were intercrossed to generate Ter/Ter mice bearing Tg(Dnd1)1Matn transgene. Testes weight measurements were taken for male progeny and stained sections were examined histologically.
Gonad/body (g/b) weight ratios and histology. Adult mice (4 weeks of age or older) were sacrificed and the body weight and testes weight recorded. The g/b ratio was calculated as gonad weight/body weight × 1000. Testes or tumours were preserved in 10% phosphate-buffered formalin for at least 48 h. Sections (5 μm) were stained with hematoxylin and eosin.
In situ hybridization. Embryos and gonads were fixed in 4% PFA for 12 hours at 4°C, washed in 0.1% Tween 20/PBS, dehydrated in 100% methanol, and stored at 20°C until used. Full length cDNA of Dnd1 (image clone ID: 3470697) was used as probe. In situ hybridizations on mouse embryos were performed as described previously30.
Generation of anti-DND1 antibody and GST-DND1. Rabbit polyclonal antibody was generated against the DND1 peptide spanning amino acids 16 to 33. GST(glutathione Stransferase)-DND1 fusion protein was made by cloning Dnd1 cDNA in-frame into pGEX-2TK (Amersham Biosciences). GST-DND1 fusion protein was induced by IPTG (isopropyl-β-D-thiogalactoside) and affinity purified using Glutathione Sepharose 4B.
Assays to distinguish the Dnd alleles. To distinguish the mRNA transcripts from the wild-type and Ter alleles of Dnd1 total RNA from 129-+/+ and 129-Ter/+ testes and tumours were treated with DNase1 before converting to cDNA using Superscript II Rnase H Reverse Transcriptase (Invitrogen). PCR was performed using the primers A25-F 5′-cgagctgacggtggacgggctgcc-3′ and A25-R 5′-ctgcctagcttcatccgttgac-3′ (primers 2, Fig. 3a), which were designed to span exons 2 and 3 of Dnd1 to yield a 396 base pair product. The RT-PCR products were digested with Dde1 before electrophoresis on a 7% acrylamide gel. Dde1 digestion of RT-PCR product produces fragments of sizes: 170 bp, 103 bp and 97 bp from the + allele and 139 bp, 103 bp, 97 bp and 31 bp from the Ter allele. The 31 bp fragment was not observed on the gel (Fig. 3g). Controls to exclude amplification of contaminating DNA was performed on parallel RNA samples by leaving out Superscript II during reverse transcription reaction (indicated in the lanes marked -). The primers Dnd13-F (5′-caggagccagaaaggtatcaata-3′) and Dnd13-R (5′cttaacccatttagggtacctgt-3′) were used to amplify a 1059bp Dnd1 transcript by RT-PCR for sequencing.
Supplementary Material
Acknowledgements
We thank Hans Scholer for the GOF-1/DPE/EGFP construct, A. Kong and W. Cosme-Blanco for technical help, G. Lozano for critical reading of the manuscript, and members of the Nadeau lab for suggestions. Services of the Trans-NIH Mouse Initiative were used for sequencing BACs encoding the Ter locus. This project was supported by NCI grant CA093754 to A.M, CA75056 to J.H.N. and with funds from the NCI, NIH, under Contract N01-C0-12400. D.C. and B.C. are funded by a grant from NIH-HL-063054. Veterinary resources, DNA sequencing and Genetically Engineered Mouse Facility were supported by the Cancer Center Support (Core) Grant, CA16672. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
Footnotes
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Sakurai T, Iguchi T, Moriwaki K, Noguchi M. The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Develop. Growth Differ. 1995;37:293–302. doi: 10.1046/j.1440-169X.1995.t01-2-00007.x. [DOI] [PubMed] [Google Scholar]
- 2.Noguchi T, Noguchi M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 1985;75:385–392. [PubMed] [Google Scholar]
- 3.Stevens LC. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst. 1973;50:235–242. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Stevens LC, Mackensen JA. Genetic and environmental influences on teratocarcinogenesis in mice. J. Natl. Cancer Inst. 1961;27:443–453. [Google Scholar]
- 5.Stevens LC. Origin of testicular teratomas from primordial germ cells in mice. J. Natl. Cancer Inst. 1967;38:549–552. [PubMed] [Google Scholar]
- 6.Donovan PJ, de Miguel MP. Turning germ cells into stem cells. Curr. Opin. Genet. Dev. 2003;13:463–471. doi: 10.1016/j.gde.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Asada Y, Varnum DS, Frankel WN, Nadeau JH. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat. Genet. 1994;6:363–368. doi: 10.1038/ng0494-363. [DOI] [PubMed] [Google Scholar]
- 8.Sakurai T, Katoh H, Moriwaki K, Noguchi T, Noguchi M. The ter primordial germ cell deficiency mutation maps near Grl-1 on mouse chromosome 18. Mamm. Genome. 1994;5:333–336. doi: 10.1007/BF00356550. [DOI] [PubMed] [Google Scholar]
- 9.Weidinger G, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 10.Mello CC, Conte D. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 11.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 12.Shao J, Sheng H, Inoue H, Morrow JD, DuBois RN. Regulation of constitutive cyclooxygenase-2 expression in colon carcinoma cells. J. Biol. Chem. 2000;275:33951–33956. doi: 10.1074/jbc.M002324200. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein. CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 14.Yao HH, DiNapoli L, Capel B. Meiotic germ cells antagonize mesonephric cell migration and testis cord formation in mouse gonads. Development. 2003;130:5895–5902. doi: 10.1242/dev.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol. 2003;262:303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- 16.Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. Embo J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 2000;20:1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Z, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 19.Barbouti A, et al. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23) Cancer Res. 2003;63:1202–1206. [PubMed] [Google Scholar]
- 20.Drabkin HA, et al. DEF-3 (g16/NY-LU-12), an RNA binding protein from the 3p21.3 homozygous deletion region in SCLC. Oncogene. 1999;18:2589–2597. doi: 10.1038/sj.onc.1202601. [DOI] [PubMed] [Google Scholar]
- 21.Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20:6544–6550. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- 22.Jinawath N, Furukawa Y, Nakamura Y. Identification of NOL8, a nucleolar protein containing an RNA recognition motif (RRM), which is overexpressed in diffuse-type gastric cancer. Cancer Sci. 2004;95:430–435. doi: 10.1111/j.1349-7006.2004.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuei D-J, et al. RBMY, a male germ cell-specific RNA-binding protein, activated in human liver cancers and transforms rodent fibroblasts. Oncogene. 2004;23:5815–5822. doi: 10.1038/sj.onc.1207773. [DOI] [PubMed] [Google Scholar]
- 24.Wedekind JE, Dance GSC, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 25.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNA polII-dependent transcription in primordial germ cells. Curr. Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Moore FL, et al. Human pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. U S A. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 28.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 29.Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 30.Henrique D, et al. A digoxigenin labeled RNA probe for Sox9 was detected using an alkaline phosphatase-conjugated anti-digoxigenin antibody. Nature. 1995;375:787–790. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


