Abstract
The expression of metastasis-associated protein 1 (MTA1) correlates well with tumor metastases; however, the associated molecular mechanism is not fully understood. Here, we explored the possibility of cross-talk between MTA1 and hypoxia-inducible factor-1α (HIF-1α), a key regulator of angiogenic factors. We observed that the expression of MTA1 was strongly induced under hypoxia in breast cancer cell lines such as MCF-7 and MDA-MB-231. When MTA1 was overexpressed, the transcriptional activity and stability of HIF-1α protein were enhanced. MTA1 and HIF-1α are physically associated in vivo and they were localized completely in the nucleus when coexpressed. MTA1 induced the deacetylation of HIF-1α by increasing the expression of histone deacetylase 1 (HDAC1). MTA1 counteracted to the action of acetyltransferase, ARD1, and it did not stabilize the HIF-1α mutant that lacks the acetylation site, K532R. These results indicate that acetylation is the major target of MTA1/HDAC1 function. Collectively, our data provide evidence of a positive cross-talk between HIF-1α and MTA1, which is mediated by HDAC1 recruitment, and indicate a close connection between MTA1-associated metastasis and HIF-1-induced tumor angiogenesis.
Keywords: HDAC1, HIF-1α, hypoxia, metastasis, MTA1
Introduction
Metastasis is a multistep event in which neoplastic cells detach from the primary tumor, migrate, disseminate, extravasate, and eventually proliferate at a discontinuous secondary site (Chambers et al, 2002; Fidler, 2003). To date, several human metastasis-associated genes have been shown to regulate the metastatic capacity of tumor cells (Yoshida et al, 2000; Debies and Welch, 2001). Among these genes, metastasis-associated protein 1 (MTA1) is closely associated with cancer metastasis. The rat mta1 gene was first identified using a differential cDNA library screening technique in a highly metastatic mammary adenocarcinoma cell line (Toh et al, 1994). The MTA1 gene encodes a protein of about 80 kDa, which includes functional domains such as a bromo-adjacent homology domain, an Egl-27 and MTA1 homology 2 domain, a conserved leucine zipper motif, a GATA-like zinc-finger motif, and an SWI3/ADA2/NCoR/TFI domain (Nawa et al, 2000). Elevated MTA1 levels in several tumor types, such as breast, colorectal, gastric, and esophageal carcinomas, appear to enhance metastasis, increase cell motility, and potentiate growth. Therefore, MTA1 may be an indicator of the potential aggressiveness of various tumors (Toh et al, 1999; Nicolson et al, 2003).
The MTA family is a group of structurally related proteins encoded by the same or different genes, including MTA1, MTA1s, MTA-ZG29p, MTA2, MTA3, and MTA3L (Kumar, 2003; Bowen et al, 2004). MTA proteins physically interact with histone deacetylase (HDAC) 1 and HDAC2 forming the nucleosome remodeling histone deacetylation (NuRD) complex, which plays an important role in histone deacetylation, alteration of chromatin structure, and transcriptional control (Kumar, 2003; Bowen et al, 2004). MTA family proteins form distinct complexes in which only one MTA molecule is present, which may be the basis for the functional specialization of different MTA proteins (Fujita et al, 2003; Yao and Yang, 2003). The tumor suppressor p53 is deacetylated by an HDAC1-containing MTA2 complex (Luo et al, 2000). As deacetylated p53 is ubiquitinated by MDM2 and degraded by proteasomes, the MTA2/HDAC1 complex is expected to modulate p53-mediated cell-growth arrest and apoptosis (Ito et al, 2002).
Hypoxia-inducible factor 1 (HIF-1) is the master transcriptional regulator that facilitates adaptation to low oxygen availability (Semenza, 2003). HIF-1 is composed of an HIF-1α subunit and an HIF-1β subunit, also known as the aryl hydrocarbon receptor nuclear translocator. Both HIF-1 subunits are members of the basic helix–loop–helix/PER–ARNT–SIM domain family of transcription factors. HIF-1α is constitutively transcribed and translated, but is rapidly degraded under normoxic conditions by the ubiquitin–proteasome pathway (Semenza, 2003; Mazure et al, 2004). Regulation of HIF-1α stability is mediated by a region that is referred to as the oxygen-dependent degradation (ODD) domain of HIF-1α, through various post-translational modifications (Mazure et al, 2004). HIF-1α is hydroxylated at proline residues 402 and 564 by the recently identified family of HIF prolyl hydroxylase domain (PHD) proteins, which require O2 (Bruick and McKnight, 2001; Epstein et al, 2001). Hydroxylated HIF-1α subsequently interacts with the tumor-suppressor von Hippel–Lindau protein (VHL), which targets it for proteasomal degradation (Semenza, 2003; Mazure et al, 2004). ARD1 may be another important enzyme that modifies HIF-1α by acetylating the Lys532 residue in the ODD domain of HIF-1α. Acetylation of HIF-1α can be coordinated with prolyl hydroxylation and ubiquitination, which leads to the proteasomal degradation of HIF-1α (Jeong et al, 2002).
The expression of HIF-1 is closely related to tumor growth and metastasis. When the expression of HIF-1α was analyzed by immunohistochemistry, it was overexpressed in colon, breast, gastric, lung, skin, ovarian, pancreatic, prostate, and renal carcinomas, relative to its expression in normal tissues. HIF-1α was overexpressed in only 29% of primary breast cancers, but in 69% of breast cancer metastases (Zhong et al, 1999). There is a statistically significant positive association between increased levels of HIF-1α protein and vascular endothelial growth factor (VEGF) expression in human colorectal carcinoma (Kuwai et al, 2003). HIF-1α expression correlated with high metastatic risk in a series of unselected patients with invasive breast cancer (Dales et al, 2005). As MTA1 and HIF-1α proteins are expressed in malignant metastatic tumor cells and are therefore expected to have important roles in tumor progression and metastasis during the development of cancer, we investigated the possibility of cross-talk between MTA1 and HIF-1α. Here, we show for the first time that MTA1 enhances the stability and transcriptional activity of HIF-1α by recruiting HDAC1 in human breast cancer cells. This may clarify the molecular mechanism by which MTA1 induces metastasis in various solid tumors.
Results
Expression of MTA1 is upregulated under hypoxia
First, we examined whether the expression of MTA1 is modulated under hypoxia. When the noninvasive and nonmetastatic human breast cancer cell line MCF-7 was exposed to an atmosphere of 0.1% O2, the expression of MTA1 protein was strongly enhanced after 0.5 h and up to 24 h. A similar pattern of expression was observed for HIF-1α protein (Figure 1A). However, the expression pattern of MTA1 mRNA differed from that of HIF-1α in that it was induced after 0.5 h and continued for 24 h, whereas the level of HIF-1α mRNA was consistent throughout the experimental period. A similar result was obtained in the human metastatic breast cancer cell line MDA-MB-231, except that the basal level of MTA1 expression was higher in MDA-MB-231 than in MCF-7 cells, which is consistent with a previous report (Toh et al, 1994; Matteucci et al, 2005). These results indicate that the expression of MTA1 is enhanced at the transcription level under hypoxia in breast cancer cells. The induction of MTA1 under hypoxia was also observed in other breast cancer cell lines, T47D and MDA-MB-435. The basal level as well as hypoxia-inducible level of MTA1 protein was largely different in these cell lines, but the level of MTA1 and HIF-1α was well correlated (Figure 1B).
Figure 1.
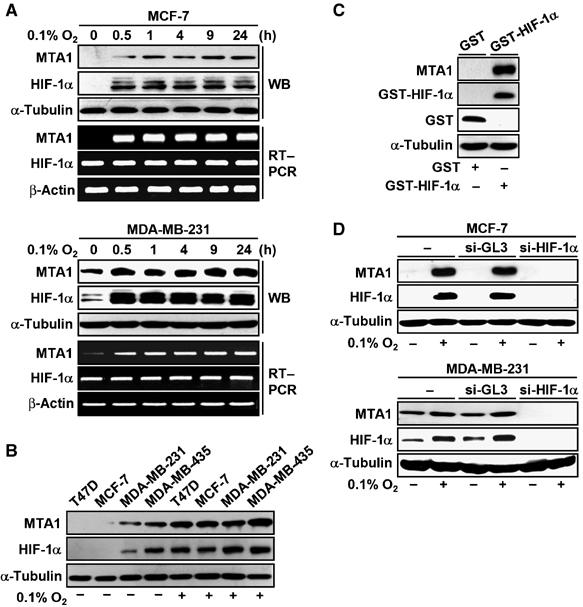
Expression of MTA1 is induced under hypoxia. (A) MCF-7 or MDA-MB-231 cells were incubated in hypoxia for the indicated time periods. The expression of HIF-1α and MTA1 was analyzed by Western blot (WB) analysis and RT–PCR. The expression of α-tubulin or β-actin was analyzed for control. (B) T47D, MCF-7, MDA-MB-231, and MDA-MB-435 cells were incubated under hypoxia or normoxia for 24 h as indicated. The expression of MTA1, HIF-1α, and α-tubulin was analyzed by WB analysis. (C) MCF-7 cells were transfected with 4 μg of each pEBG-HIF-1α or empty vector. The expression of MTA1, GST-HIF-1α, GST, and α-tubulin was analyzed by WB analysis. (D) MCF-7 or MDA-MB-231 cells were transfected twice at 24 h intervals with 200 nM of each control nonspecific siRNA, si-GL3, or human HIF-1α siRNA, si-HIF-1α. After 3 h of the last transfection, the cells were incubated under hypoxia or normoxia for 24 h. The expression of MTA1, HIF-1α, and α-tubulin was analyzed by WB analysis.
To examine whether HIF-1 was involved in the induction of MTA1, glutathione-S-transferase (GST)-fused HIF-1α chimera was introduced into MCF-7 cells under transfection condition that produced a significant level of the GST-fused HIF-1α under normoxia (Yoo et al, 2004a). MTA1 was strongly expressed in the presence of the GST-fused HIF-1α (Figure 1C) to compare with the GST alone. When the expression of HIF-1α was blocked by transfection of small interfering (si) RNA duplexes targeting HIF-1α, si-HIF-1α, the induction of MTA1 under hypoxia was eliminated in the breast cancer cells (Figure 1D). These results suggest that HIF-1α may lead the induction of MTA1 under hypoxia by a positive feedback mechanism.
MTA1 potentiates the transcriptional activity of HIF-1 by stabilizing HIF-1α protein
To explore the possibility of cross-talk between MTA1 and HIF-1α, we measured the transcriptional activity of HIF-1α using the HRE-tk-Luc reporter construct, which contains hypoxia response element (HRE) sequences of the 3′ enhancer in erythropoietin gene (Yoo et al, 2003, 2004b). Cotransfection of the MTA1 expression vector into MCF-7 cells activated the reporter activity in a dose-dependent manner. Similar results were obtained when a reporter containing the VEGF promoter was used (Figure 2A). The results indicate that MTA1 enhances the transcriptional activity of HIF-1, even in the absence of hypoxic stress. Next, we tested whether the enhanced transcriptional activity of HIF-1α in the presence of MTA1 was due to increase in the protein level of HIF-1α. We introduced MTA1 into MCF-7 and MDA-MB-231 cells, and measured the level of HIF-1α protein. The expression of HIF-1α, as well as that of VEGF, was increased upon expression of MTA1, which was comparable to their induction under hypoxia (Figure 2B). Although VEGF mRNA was greatly increased, HIF-1α mRNA levels were unchanged in the presence of MTA1 in both cell lines (Figure 2B). However, the induction of HIF-1α protein under hypoxia was not observed when the expression of MTA1 was suppressed by transfection of si-MTA1. The mRNA expression of MTA1, but not that of HIF-1α, was abolished in this condition (Figure 2C). Since these results suggest that MTA1 increases the stability of HIF-1α at the protein level, we measured HIF-1α protein in the presence of cycloheximide (CHX), which blocks de novo protein synthesis. As shown in Figure 2D, the integrity of HIF-1α in MD-MBA-231 cells was maintained either under hypoxia or in the presence of MTA1 for up to 60 min, while the basal-level expression of HIF-1α completely disappeared under normoxia in this period. Nuclear accumulation of HIF-1α was observed in the presence of MTA1 in immunofluorescence studies using red fluorescent protein-fused HIF-1α, Red-HIF-1α. Red-HIF-1α was barely detectable in the absence of CoCl2, whereas it accumulated only in the nucleus in the presence of CoCl2 in the experimental condition that we employed. Similarly, the level of Red-HIF-1α was enhanced and it accumulated in the nucleus when MTA1 was cotransfected (Figure 2E). The nuclear localization of MTA1 and HIF-1α was further confirmed in the cytoplasmic and nuclear fractions by Western blot analysis as shown in Figure 2F. These results demonstrate that MTA1 increases HIF-1α at the protein level, particularly in the nucleus, and suggest that MTA1 and HIF-1α may interact and colocalize in the nucleus.
Figure 2.
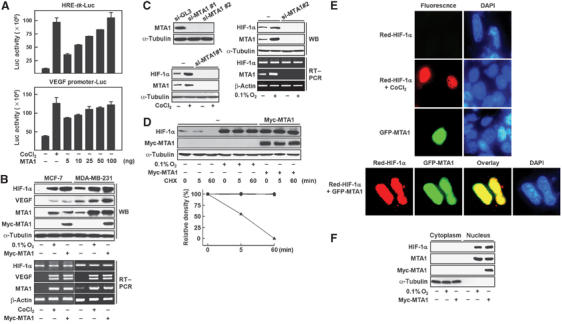
Transcriptional activity as well as protein stability of HIF-1α is enhanced in the presence of MTA1. (A) The HRE-tk-Luc (0.1 μg) or VEGF promoter (−2003∼23)-Luc (0.3 μg) reporter was cotransfected with the indicated amount of eucaryotic expression vector for MTA1 into MCF-7 cells. Transfected cells were incubated for 24 h in the presence or absence of 100 μM CoCl2. Luciferase activity was measured and normalized by β-gal activity. (B) MCF-7 or MDA-MB-231 cells were transfected with 3 μg of each pCMV-Myc-MTA1 or empty vector. Transfected cells were incubated under hypoxic stress, 0.1% O2 or 100 μM CoCl2, or normoxia for 24 h as indicated. The expression of HIF-1α, MTA1, Myc-MTA1, and VEGF was analyzed by Western blot (WB) analysis and RT–PCR. The expression of α-tubulin or β-actin was analyzed for control. (C) MDA-MB-231 cells were transfected twice at 24 h intervals with 200 nM of each control nonspecific siRNA (si-GL3) or human MTA1 siRNA (si-MTA1#1 or si-MTA1#2). After 3 h of the last transfection, the cells were incubated under hypoxic stress, 0.1% O2 or 100 μM CoCl2, or normoxia for 24 h as indicated. The expression of HIF-1α and MTA1 was analyzed by WB and RT–PCR. The expression of α-tubulin or β-actin was analyzed for control. (D) MDA-MB-231 cells were transfected with 3 μg of each pCMV-Myc-MTA1 (▪) or empty vector. The cells transfected with empty vector were incubated under hypoxia (•) or normoxia (▴) for 24 h as indicated. At the end of treatment, 10 μM cycloheximide (CHX) was added into the media for the indicated time periods. The expression of HIF-1α, Myc-MTA1, and α-tubulin was analyzed by WB analysis. The density of HIF-1α protein band was determined using an image analysis system. The values were normalized to that of α-tubulin and expressed as percent of the CHX-untreated control. (E) HEK293 cells that were plated the previous day on four-chamber slide glass were transfected with 0.5 μg of each Red-HIF-1α and/or GFP-MTA1. Transfected cells were treated with or without 100 μM CoCl2 for 24 h. Cells were fixed and visualized by immunofluorescence microscopy. DAPI was used to stain nuclei. (F) MCF-7 cells were transfected with 3 μg of each pCMV-Myc-MTA1 or empty vector. Transfected cells were incubated under hypoxia or normoxia for 24 h as indicated. Cytoplasmic and nuclear extracts were obtained and the expression of HIF-1α, MTA1, Myc-MTA1, and α-tubulin was analyzed by WB analysis.
Therefore, we examined whether HIF-1α and MTA1 are physically associated, which would facilitate the cross-talk of these proteins. As shown in Figure 3A, MTA1 and HIF-1α were reciprocally co-precipitated in the presence of CoCl2. To analyze the structural domain involved in this interaction, GST-fused HIF-1α deletion chimeras were tested (Figure 3B) (Yoo et al, 2004a). MCF-7 cells were transfected with the construct encoding the GST-fused full-length HIF-1α under transfection conditions that produced a significant level of the GST-fused HIF-1α in normoxic conditions. The immunoprecipitation results show that MTA1 interacted with each ODD, ID, and CTAD domain, but not with the N-terminus that contained amino acids 1–401, which is known to bind the aryl hydrocarbon receptor nuclear translocator (Figure 3C).
Figure 3.
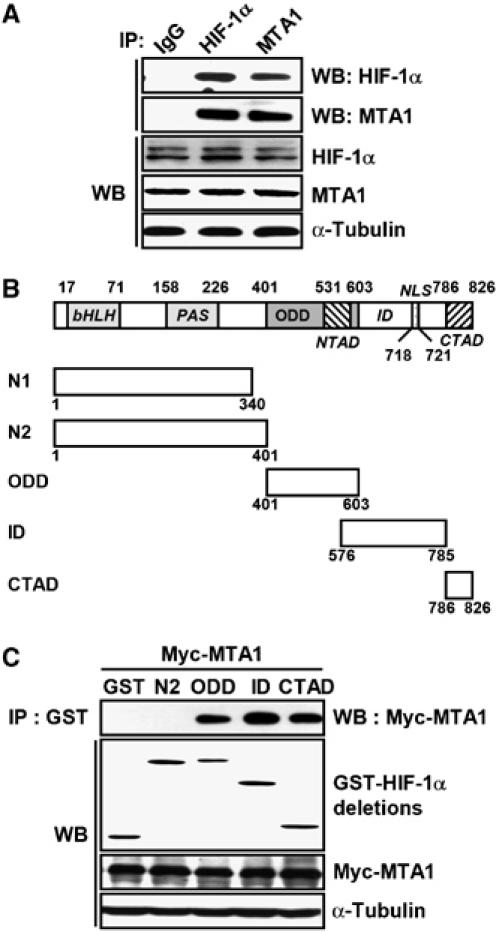
MTA1 interacts with HIF-1α in vivo. (A) MCF-7 cells were incubated in the presence of 100 μM CoCl2 for 24 h. Whole-cell lysates (500 μg) were immunoprecipitated (IP) with normal mouse IgG, anti-HIF-1α, anti-MTA antibodies, and then probed using anti-HIF-1α and anti-MTA1 antibodies. The expression of HIF-1α, MTA1, and α-tubulin was analyzed by Western blot (WB) analysis. (B) Schematic representation of full-length and deletion HIF-1α constructs containing the basic helix–loop-helix (bHLH)/PER–ARNT–SIM (PAS) (N1 and N2), ODD domain, inhibitory domain (ID), N-terminal transactivation domain (NTAD), and C-terminal transactivation domain (CTAD). Nuclear localization signal (NLS) was also indicated. (C) MCF-7 cells were transfected with 3 μg of each pEBG-HIF-1α deletion construct and pCMV-Myc-MTA1. Whole-cell lysates (500 μg) were IP with anti-GST antibody, and then probed using anti-Myc antibody. The expression of GST-HIF-1α deletions, Myc-MTA1, and α-tubulin was analyzed by WB analysis.
MTA enhances the association of HIF-1α with HDAC resulting in the deacetylation of HIF-1α
MTA1 is physically associated with HDAC1 in the NuRD complex and functions in chromatin remodeling (Toh et al, 2000; Yao and Yang, 2003). Therefore, we examined whether HDAC1 is involved in the MTA1-induced stabilization of HIF-1α. As shown in Figure 4A, the expression of HDAC1 was largely induced under hypoxia or in the presence of MTA1, which was similar to the induction of HIF-1α. Since there is a possibility that HDAC1 counteracts the acetylation of HIF-1α that leads to the degradation of the protein (Jeong et al, 2002), we examined whether trichostatin A (TSA), a potent specific inhibitor of HDAC, affected the stability of HIF-1α. TSA decreased the amount of HIF-1α that was induced by either hypoxia or MTA1 expression, indicating that HDAC1 activity is closely associated with the stability of HIF-1α (Figure 4A). Next, we examined the relative amounts of acetylated HIF-1α in the presence of MTA1 by immunoprecipitating acetylated HIF-1α. When cells were treated with MG132, an inhibitor of proteasomes, the level of acetylated HIF-1α increased significantly under normoxia (lane 2), but acetylated HIF-1α was not detected under hypoxia or in the presence of MTA1 (lanes 3 and 4). Similarly, when MG132 was treated with hypoxia or MTA1, the amount of acetylated HIF-1α was significantly decreased to compare with treatment of MG132 alone (lanes 8 and 9). However, TSA treatment significantly increased the amount of acetylated HIF-1α in the presence of MTA1 and MG132 (lane 6), suggesting that HDAC1 deacetylates HIF-1α and thereby blocks degradation of the protein (Figure 4B). As MTA1 protein is physically associated with HDAC1 (Toh et al, 2000; Yao and Yang, 2003), we examined the association between HIF-1α, MTA1, and HDAC1 under hypoxic conditions (Figure 4C). HDAC1 and HIF-1α were efficiently co-precipitated, indicating that these proteins are capable of forming a complex in vivo under hypoxic conditions. In summary, our data indicate that MTA1 inhibits the acetylation of HIF-1α by recruiting the HDAC1 protein, resulting in the stabilization of HIF-1α.
Figure 4.
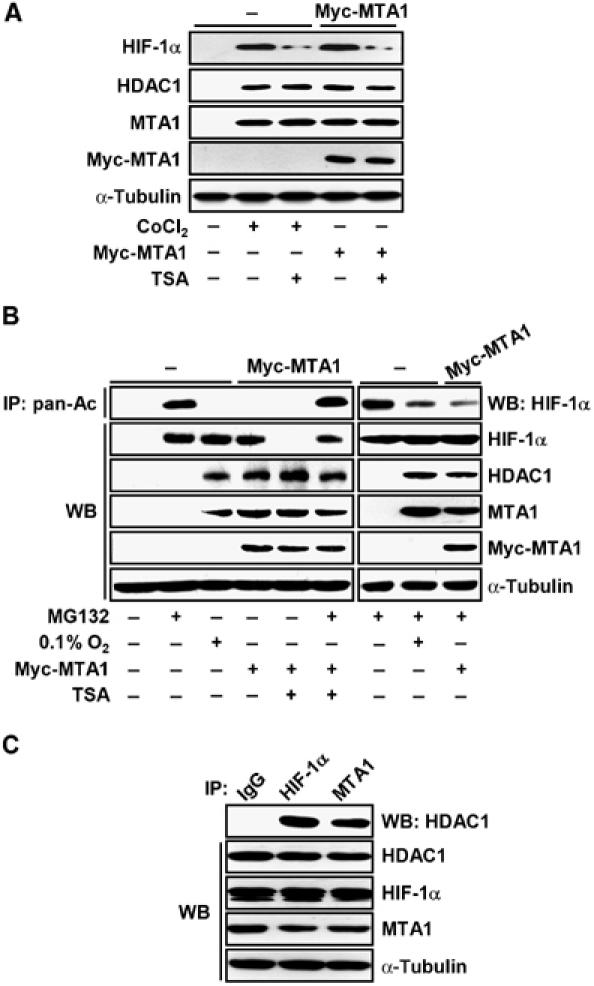
HDAC1 is involved in the MTA1-induced stabilization of HIF-1α. (A) MCF-7 cells were transfected with 3 μg of each pCMV-Myc-MTA1 or empty vector. Transfected cells were treated with or without 100 μM CoCl2 for 24 h as indicated. Cells were treated with or without 300 ng/ml TSA for 3 h as indicated before being harvested. The expression of HIF-1α, HDAC1, MTA1, Myc-MTA1, and α-tubulin was analyzed by Western blot (WB) analysis. (B) MCF-7 cells were transfected with 3 μg of each pCMV-Myc-MTA1 or empty vector. Transfected cells were incubated under hypoxia or normoxia for 24 h as indicated. Cells were treated with 10 μM MG132 or 300 ng/ml TSA for 1 h or 3 h, respectively, before cells were harvested. Whole-cell lysates (500 μg) were immunoprecipitated (IP) with anti-pan-Ac antibody, and then probed using anti-HIF-1α antibody. The expression of HIF-1α, HDAC1, MTA1, Myc-MTA1, and α-tubulin was analyzed by WB analysis. (C) MCF-7 cells were incubated in the presence of 100 μM CoCl2 for 24 h. Whole-cell lysates (500 μg) were IP with normal mouse IgG, anti-HIF-1α, anti-MTA1 antibodies, and then probed using anti-HDAC1 antibody. The expression of HIF-1α, HDAC1, Myc-MTA1, and α-tubulin was analyzed by WB analysis.
MTA1/HDAC1 deacetylates the ODD domain of HIF-1α, which is counteracted by ARD1
We localized the region of HIF-1α protein that is modified by MTA1. As shown in Figure 5A, only the GST-fused ODD domain of HIF-1α, but no other domains, was present in acetylated form and it dramatically deacetylated in the presence of MTA1. Most GST-ODD domain was degraded within 5 min under normoxia, but the integrity of HIF-1α was maintained in the presence of either CoCl2 or MTA1 for up to 60 min when CHX was treated (Figure 5B). Previously, it has been reported that the ODD domain of HIF-1α is acetylated by ARD1 (Jeong et al, 2002). However, the observation is in conflict since overexpression or silencing of ARD1 was shown to have no impact on the stability and on the mRNA levels of the downstream target genes of HIF-1 (Bilton et al, 2005; Fisher et al, 2005). We tested whether the endogenous level of MTA1 affects the effect of ARD1 using several human cell lines. The basal level as well as CoCl2-inducible level of MTA1 protein was largely different, but was well correlated with the level of HIF-1α in these cell lines, suggesting that MTA1 may be associated with the expression level of HIF-1α (Figure 6A). The expression of ARD1 decreased the CoCl2-inducible level of HIF-1α protein in HEK293 cells, whereas HIF-1α protein was not reduced in the other cell lines such as HeLa and HepG2 (Figure 6B). Next, we tested whether a large amount of MTA1 induced stability of HIF-1α in the presence of ARD1 in HEK293 cells. As shown in Figure 6C, the stability of HIF-1α in HEK293 cells was restored by MTA1 expression in a dose-dependent manner, indicating that endogenous level of MTA1 in different cell lines may determine susceptibility for the ARD-1-induced degradation of HIF-1α. When the GST-fused HIF-1α was tested, the protein was present in acetylated form in the presence of ARD1 together with MG132 (lane 2), but the acetylation form disappeared when MTA1 was introduced (lane 4) (Figure 6D). Treatment with TSA decreased the recovered HIF-1α stability by MTA1, indicating that HDAC is involved in the action of MTA1 (Figure 6E). Since Lys532 of HIF-1α is the target site for acetylation by ARD1 protein (Jeong et al, 2002), we tested whether the stability of a mutant with a point mutation at this site, K532R, is affected by the expression of MTA1. The K532R mutant was stably expressed under normoxic condition (Figure 6F), which is consistent with a previous report (Jeong et al, 2002). Although wild-type HIF-1α was increased by MTA1 expression but decreased by TSA treatment, the expression of K532R was not altered under these conditions. Thus, Lys532 is probably the site that regulates the stability of HIF-1α through the deacetylation of HIF-1α that is induced by MTA1/HDAC1.
Figure 5.
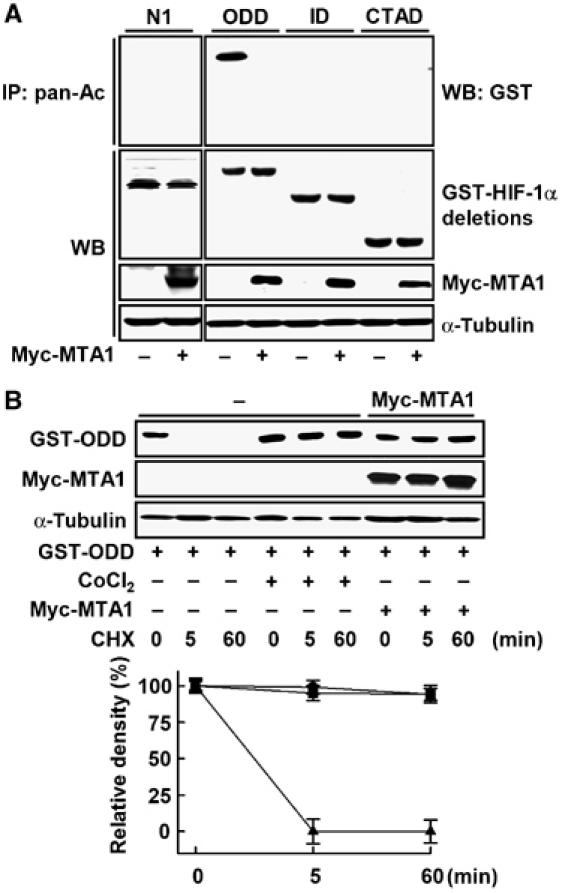
ODD domain of HIF-1α is deacetylated in the presence of MTA1. (A) MCF-7 cells were transfected with 3 μg each of the pEBG-HIF-1α deletion construct (Figure 3B), pCMV-Myc-MTA1, or empty vector. After 24 h of transfection, 500 μg of whole-cell lysates were immunoprecipitated (IP) with anti-pan-Ac antibody, and then probed using anti-GST antibody. The expression of GST-HIF-1α deletions, Myc-MTA1, and α-tubulin was analyzed by Western blot (WB) analysis. (B) MCF-7 cells were transfected with 3 μg each of pEBG-HIF-1α ODD (GST-ODD), pCMV-Myc-MTA1 (•), or empty vector. After 1 h of transfection, the cells transfected with empty vector were treated with (▪) or without (▴) 100 μM CoCl2 for 24 h. At the end of treatment, 10 μM cycloheximide (CHX) was added into the media for the indicated time periods. The expression of GST-ODD, Myc-MTA1, and α-tubulin was analyzed by WB analysis. The density of GST-ODD protein band was determined using an image analysis system. The values were normalized to that of α-tubulin and expressed as percent of the CHX-untreated control.
Figure 6.
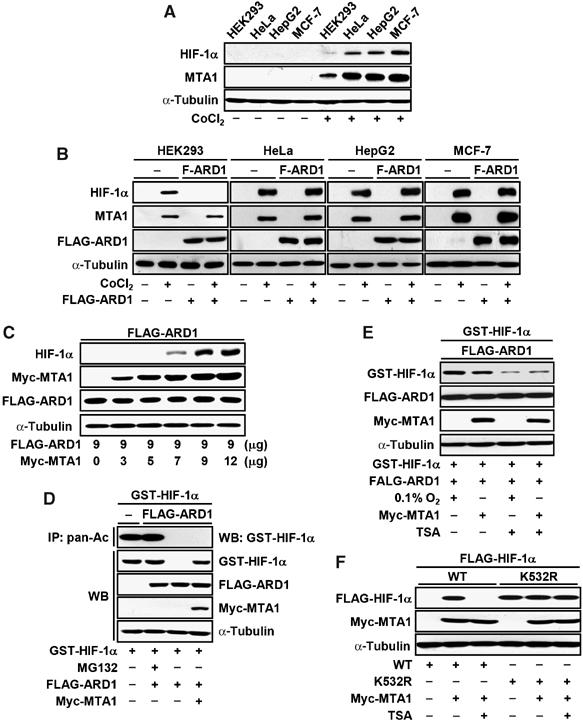
MTA1 counteracts to the ARD1-induced degradation of HIF-1α. (A) HEK293, HeLa, HepG2, and MCF-7 cells were treated with or without 100 μM CoCl2 for 24 h. The expression of HIF-1α, MTA1, and α-tubulin was analyzed by Western blot (WB) analysis. (B) HEK293, HeLa, HepG2, and MCF-7 cells were transfected with 3 μg of pCMV-Tag2C-FLAG-mARD1225 (F-ARD1) or empty vector. Transfected cells were incubated under hypoxia or normoxia for 24 h. The expression of HIF-1α, MTA1, FLAG-ARD1 (F-ARD1), and α-tubulin was analyzed by WB analysis. (C) HEK293 cells were transfected with 9 μg of pCMV-Tag2C-FLAG-mARD1225 (FLAG-ARD1) or the indicated amount of pCMV-Myc-MTA1. The expression of HIF-1α, Myc-MTA1, FLAG-ARD1, and α-tubulin was analyzed by WB analysis. (D) MCF-7 cells were transfected with 4 μg pEBG-HIF-1α (GST-HIF-1α), and 3 μg of each pCMV-Tag2C-FLAG-mARD1225 (FLAG-ARD1), pCMV-Myc-MTA1, or empty vector. Cells were treated with or without 10 μM MG132 for 1 h before being harvested. Whole-cell lysates (500 μg) were immunoprecipitated (IP) with anti-pan-Ac antibody, and then probed using anti-GST antibody. The expression of GST-HIF-1α, FLAG-ARD1, Myc-MTA1, and α-tubulin was analyzed by WB analysis. (E) MCF-7 cells were transfected with 4 μg pEBG-HIF-1α (GST-HIF-1α) with 3 μg of each pCMV-Tag2C-FLAG-mARD1225 or pCMV-Myc-MTA1 as indicated. After 1 h of transfection, cells were incubated under hypoxia or normoxia for 24 h as indicated. Cells were treated with or without 300 ng/ml TSA for 3 h before being harvested. The expression of GST-HIF-1α, FLAG-ARD1, Myc-MTA1, and α-tubulin was analyzed by WB analysis. (F) HEK293 cells were transfected with the indicated combinations of 1 μg of each p3XFLAGTM7.1-HIF-1α (WT), or p3XFLAGTM7.1-HIF-1α K532R (K532R), 3 μg of each Myc-MTA1 and empty vector. After 24 h of transfection, cells were incubated in the presence or absence of 300 ng/ml TSA for 3 h before being harvested. The expression of FLAG-HIF-1α, Myc-MTA1, and α-tubulin was analyzed by WB analysis.
Finally, we investigated whether MTA1 reduces the binding of HIF-1α to PHDs and VHL, because the acetylation of HIF-1α correlates with prolyl hydroxylation and ubiquitination (Jeong et al, 2002). Both VHL and PHDs bound strongly to HIF-1α in the presence of MG132, indicating that the binding of VHL to HIF-1α precedes the degradation of HIF-1α under normoxia, as described previously (Semenza, 2003; Mazure et al, 2004). However, the binding of HIF-1α to VHL or PHDs was largely decreased under hypoxia or in the presence of MTA1 (Figures 7A and B). Similar results were obtained when the ODD domain of HIF-1α was tested (Figure 7C). When the expression of MTA1 was suppressed by si-MTA1, these bindings and the amount of acetylated HIF-1α were significantly increased (Figure 7C), indicating that MTA1 plays a major role in the control of HIF-1α stability. Similarly, when MG132 was treated with hypoxia or MTA1, the binding of HIF-1α to VHL or PHD2 was significantly decreased to compare with MG132 alone. However, still a part of HIF-1α was acetylated and bound to VHL and PHD2, suggesting that a small portion of HIF-1α may undergo degradation process in the presence of hypoxia or MTA1. Taken together, these results suggest that hypoxia or MTA1 predominantly deacetylates HIF-1α and block the further processes of degradation such as hydroxylation and ubiquitination.
Figure 7.
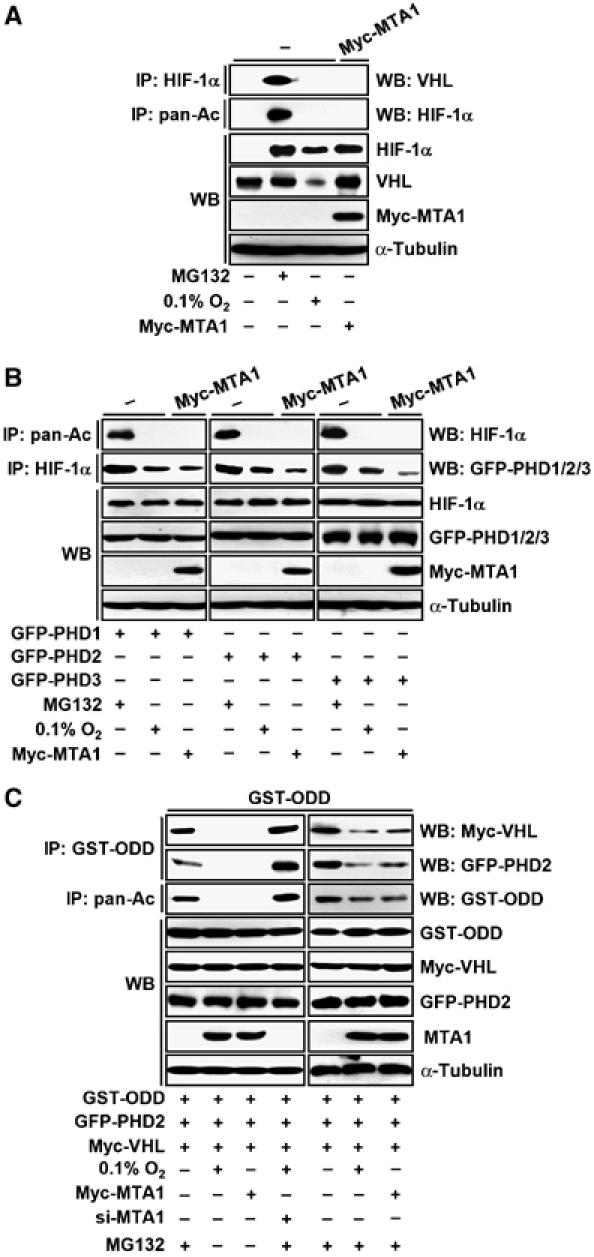
MTA1 interferes the association of HIF-1α and VHL and PHDs. (A) MCF-7 cells were transfected with 3 μg of each pCMV-Myc-MTA1 or empty vector. Transfected cells were incubated under hypoxia or normoxia for 24 h as indicated. Cells were treated with or without 10 μM MG132 for 1 h before being harvested. Whole-cell lysates (500 μg) were immunoprecipitated (IP) with anti-HIF-1α antibody, and then probed using anti-VHL antibody. The expression of HIF-1α, VHL, Myc-MTA1, and α-tubulin was analyzed by Western blot (WB) analysis. (B) MCF-7 cells were transfected with 3 μg of each pEGFP-N1-PHD1 (GFP-PHD1), pEGFP-N1-PHD2 (GFP-PHD2), or pEGFP-N1-PHD3 (GFP-PHD3), with or without 3 μg pCMV-Myc-MTA1 as indicated. Transfected cells were incubated under hypoxia or normoxia for 24 h. Cells were treated with or without 10 μM MG132 for 1 h before being harvested. Whole-cell lysates (500 μg) were IP with anti-pan-Ac and anti-HIF-1α antibodies, and then probed using anti-HIF-1α or anti-GFP antibodies. The expression of HIF-1α, GFP-PHDs, Myc-MTA1, and α-tubulin was analyzed by WB analysis. (C) MCF-7 cells were transfected with 3 μg of each pEBG-HIF-1α ODD (GST-ODD), pCMV-Myc-VHL, and pEGFP-N1-PHD2 (GFP-PHD2). For Myc-MTA1, transfection was repeated with 3 μg of each pCMV-Myc-MTA1 or empty vector after 24 h of the first transfection. After incubating for 24 h, the cells were exposed to hypoxia or normoxia for 24 h. For si-MTA1 transfection, 200 nM human MTA1 siRNA (si-MTA1) was transfected after 24 h of the first transfection, and repeated once after 21 h. After 3 h of the last transfection, the cells were incubated under hypoxia or normoxia for 24 h. Cells were treated with or without 10 μM MG132 for 1 h before being harvested. Whole-cell lysates (500 μg) were IP with anti-GST, or anti-pan-Ac antibodies, and then probed using anti-Myc, anti-GFP, or anti-GST antibodies. The expression of GST-ODD, Myc-VHL, GFP-PHD2, MTA1, and α-tubulin was analyzed by WB analysis.
Discussion
Metastasis is a multistep process that allows tumor cells to overcome barriers to local invasion, intravasation, survival in circulation, arrest in capillaries, extravasation, and finally outgrowth to produce macrometastases at distant organs (Chambers et al, 2002; Fidler, 2003). Metastasis requires the concerted actions of multiple genes, such as MST1, NM23, stromelysin-3, KAI-1, MKK4, and MTA1 (Yoshida et al, 2000; Debies and Welch, 2001). Although MTA1 is overexpressed in a variety of human metastatic cancer cell lines and cancerous tissues, the role of this protein in particular steps of the metastatic process has not yet been clarified. In this study, we demonstrate that MTA1 enhances the stability and transcriptional activity of HIF-1α, the major transcriptional regulator of hypoxic stress.
Hypoxic conditions in rapidly growing tumors allow malignant cells to form hypoxic vascular solid tumors, which are aggressive and metastatic (Semenza, 2003; Vaupel, 2004). Under hypoxic conditions, HIF-1α expression is upregulated at the protein level, because ubiquitin–proteasome-mediated degradation is decreased (Semenza, 2003). Epigenetic modifications of HIF-1α, such as hydroxylation and acetylation, play important roles in the regulation of HIF-1α protein stability (Jeong et al, 2002; Mazure et al, 2004). However, the role of ARD1, an acetyltransferase, in the regulation of HIF-1α is in conflict since both overexpression and silencing of ARD1 were shown to have no impact on the stability and on the mRNA levels of the downstream target genes of HIF-1 (Bilton et al, 2005; Fisher et al, 2005). In this report, we found that MTA1 expression level was closely associated with the ARD1 function. In the cell line that expresses a low level of MTA1 such as HEK293, the ARD1-induced HIF-1α degradation was significant, whereas ARD1 did not function in the other cell lines such as HeLa and HepG2 that express a high level of HIF-1α (Figure 6B). Indeed, ARD1 decreased protein level as well as acetylation status of the GST-fused full-length HIF-1α under normoxia in our study (Figure 6D), supporting the role of ARD1 in the regulation of HIF-1α stability. The fact that MTA1 competes for the ARD1-induced HIF-1α degradation (Figure 6C) indicates that the acetylation of HIF-1α may be the major target of MTA1 function. During the preparation of this manuscript, Kim et al (2006) reported several ARD1 variants that have different effects on stability and acetylation of HIF-1α. Consistent with our result, mARD1225, which was used in this investigation, strongly induced degradation of HIF-1α, whereas mARD1235 and hARD1235 had much weaker effect on the stability of HIF-1α. Tissue/cell distribution and specific roles of these ARD1 variants in the regulation of hypoxia as well as tumor progression in connection with MTA1 and HDAC1 need to be investigated.
Microarrray analyses revealed that HDAC1 regulates both activation and repression of target genes (Glaser et al, 2003; de Ruijter et al, 2005). However, it has not been clear whether HDAC1 can play a role in activating transcription and if so what the underlying mechanisms are. In our study, we found that the MTA/HDAC complex could induce protein stability by deacetylating of important transcription factors, in addition to its established role in repression of transcription by chromatin remodeling. Our finding may provide a potential clue for the HDAC1-induced transcriptional activation. The dual function of the MTA/HDAC complexes may result from heterogeneity in subunit composition of these complexes. Identification and characterization of the components of the MTA1 complex that is associated with each functional subset should provide valuable insight into the control of intracellular signaling in response to hypoxia and tumor metastasis.
The NuRD complex is a multisubunit protein complex containing both HDAC and chromatin remodeling ATPase activities (Bowen et al, 2004). Recently, it was demonstrated that the NuRD complex contains MTA1, which physically interacts with HDAC1 and HDAC2 (Xue et al, 1998; Yao and Yang, 2003). As the hypoacetylation of core histones is associated with transcriptionally inactive genes, complexes containing HDAC activity are thought to participate in transcriptional repression (Bowen et al, 2004). Indeed, the expression of the 92-kDa type IV collagenase, MMP-9, was repressed by the MTA1-dependent recruitment of HDAC2 to the distal promoter region of MMP-9 (Yan et al, 2003). HDAC complexes are associated with the MTA1-mediated repression of the transcriptional function of the estrogen receptor (ER), which is induced by heregulin (Mazumdar et al, 2001). MTA1 represses the MTA1-interacting protein coactivator (MICoA)-mediated stimulation of the estrogen receptor elements, resulting in the functional inactivation of the ER pathway (Mishra et al, 2003). MTA1 represses the transactivation function of the ER by interacting with cellular factors such as MAT1, a cyclin-dependent kinase-activating kinase complex ring finger factor, and NRIF3, a nuclear receptor coregulator (Talukder et al, 2003, 2004). However, in this report, we show that MTA1 enhanced the transcriptional activity of HIF-1α, indicating that the MTA1/HDAC1 complex may play a role as a coactivator (Figure 2A). The observation that the CTAD domain, which provides the transactivation function of HIF-1α, participates in the interaction with MTA1, may support this notion (Figure 3C), although the details of this mechanism remain to be determined. Similarly, Kato et al (2004) reported that HDAC7 induces the transcriptional function of HIF-1 by recruiting the coactivator p300 onto the CTAD domain of HIF-1α.
The previous observations that the expression level of the MTA1 gene was elevated in rapidly growing mammary adenocarcinomas (Nicolson et al, 2003) and that 30% of transgenic mice expressing MTA1 developed focal hyperplastic nodules in the breast, and about 7% exhibited mammary tumors within 18 months (Bagheri-Yarmand et al, 2004), strongly suggest a role for MTA1 in mammary gland tumorigenesis. Recently, MTA3 was identified as an estrogen receptor-regulated gene and its expression is downregulated in ER-negative breast tumors (Mishra et al, 2004). Further, in the absence of ER or MTA3, the expression of the transcriptional repressor Snail, a master regulator of epithelial to mesenchymal transitions, was aberrant (Fujita et al, 2003; Mishra et al, 2004). These observations may provide a link between the expression of the MTA family and estrogen action in mammary tumor progression since ERα is the major ER in the mammary epithelium, and a major percentage of breast cancers are ERα-positive and estrogen-dependent. It has been well established that ER expression is associated with low invasiveness and low motility. Therefore, ER expression has become an important prognostic marker for human breast cancer (Platet et al, 1998). Here, our study demonstrates that MTA1 expression is induced under hypoxia which results in stabilization of HIF-1α (Figures 1 and 2). Since ERα is degraded under hypoxia via a proteasome-dependent pathway (Stoner et al, 2002) and the downregulation of ERα under hypoxia involves the physical interaction of ERα and HIF-1α (Cho et al, 2005), we postulate that the MTA family decreases the stability of ERα, disrupts the ERα-mediated epithelial differentiation, and subsequently induces metastatic phenotypes. This notion is supported by the previous observations that HIF-1α is overexpressed in breast cancer metastases (Zhong et al, 1999), and HIF-1α-positive tumors express significantly lower levels of ERα (Kurebayashi et al, 2001). This link between MTA1 and HIF-1α may provide a mechanism for the metastatic progression of ERα-negative mammary tumors, which has important clinical implications for both prognosis and anti-estrogen therapy against mammary gland cancer.
An increasing number of structurally diverse HDAC inhibitors are emerging as an exciting new class of potential anticancer agents (Drummond et al, 2005). Importantly, here we demonstrate a close connection between MTA1/HDAC1-associated metastasis and HIF-1-induced tumor angiogenesis and suggest a potential use of HDAC inhibitors to treat progressive and metastatic human cancers.
Materials and methods
Cell culture and hypoxic treatment
Human breast adenocarcinoma cell lines, MCF-7, T47D, MDA-MB-231, and MDA-MB-435, human embryonal kidney cell line, HEK293, human cervical carcinoma cell line, HeLa, and human hepatocellular carcinoma cell line, HepG2, were obtained from American type culture collection. Cells were maintained in Dubelco's modified Eagle's medium containing 10% fetal bovine serum (FBS) at 37°C in a 5% CO2/95% air incubator. Cells were exposed to hypoxia (0.1% O2) by incubating cells at 37°C in 5% CO2/10% H2/85% N2 anaerobic incubator (Forma Scientific). Hypoxia was also induced chemically by treating cells with 100 μM CoCl2 (Sigma Chemical). When exposed to hypoxic condition, cells were incubated in media containing 1% FBS.
Western blotting, immunoprecipitation, and immunofluorescence
Cells were lysed in a lysis buffer containing 150 mM NaCl, 50 mM Tris, pH 7.4, 5 mM EDTA, 1% NP-40, and protease inhibitors for 30 min on ice, and whole-cell lysates were obtained by subsequent centrifugation. The protein concentration was quantified by bicinchoninic acid assay (Pierce). In total, 50 μg of protein from whole-cell lysates were subjected to 8–15% sodium dodecylsulfate–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Blocking was performed in 5% (w/v) non-fat-dried milk in phosphate-buffered saline containing 0.1% Tween-20. The membrane was then incubated with specific antibodies against HIF-1α (sc-13515, sc-10790, or sc-8711), MTA1 (sc-9445, sc-9446, or sc-17773), VEGF, VHL, Myc, green fluorescence protein (GFP), HDAC1 (Santa Cruz Biotech), GST (Amersham Pharmacia), or α-tubulin (Oncogene). Subcellular fractionation was performed as previously described (Yeo et al, 2005). Immunoprecipitation was performed as previously described using anti-HIF-1α, anti-MTA1, anti-GST, anti-Myc, and normal rabbit IgG, or normal mouse IgG (Santa Cruz Biotech) (Yoo et al, 2004b). To detect the acetylated HIF-1α, 500 μg whole-cell lysates were incubated with 1 μg anti-pan-Ac antibody (Santa Cruz Biotech), and then precipitated by adding 40 μl protein-A agarose slurry, and probed with anti-HIF-1α antibody. The expression of α-tubulin was monitored as a control. Immunofluorescence study was carried out basically as described previously using Red-HIF-1α or GFP-MTA1 (Yoo et al, 2004b). One representative of at least three independent experiments with similar results is shown.
Plasmids and transient transfection
The HRE-tk-Luc, containing four copies of the erythropoietin hypoxia-responsive element, that is, 5′-GATCGCCCTACGTGCTGTCTCA-3′ and the VEGF promoter (−2068 to +50)-Luc reporter constructs have been described previously (Yoo et al, 2003, 2004a). The eucaryotic expression vectors for the p3XFLAGTM7.1-HIF-1α, the pEBG-HIF-1α (GST-HIF-1α), the truncated pEBG-HIF-1α constructs, pEGFP-C7-MTA1 (GFP-MTA1), pEGFP-N1-PHD1 (GFP-PHD1), pEGFP-N1-PHD2 (GFP-PHD2), pEGFP-N1-PHD3 (GFP-PHD3), and pCMV-Tag2C-FLAG-mARD1225 (FLAG-ARD1) were described previously (Jeong et al, 2002; Metzen et al, 2003; Yao and Yang, 2003; Yoo et al, 2003, 2004a, 2004b; Kim et al, 2006). The far-red fluorescent protein-tagged-HIF-1α (Red-HIF-1α) were constructed by inserting PCR-amplified fragments into pHcRed1-C1 (Clontech). The FLAG-tagged HIF-1α mutant (K532R) expression plasmid was constructed with p3XFLAGTM7.1-HIF-1α as template using a PCR-based mutagenesis kit (Clontech). Specific oligonucleotides
(forward, 5′-TAGTGATATGGTCAATGAATTCAGGTTGGAAT TGGTAGAAAAACTTT-3′;
reverse, 5′-AAAGTTTTTCTACCAATTCCAACCTGAATTCA TTGACCATATCACTA-3′)
were used to replace a lysine residue at amino acid 532 (AAG) with arginine (AGG). The full-length MTA1 constructs were constructed by inserting corresponding PCR-amplified fragments into the EcoRI/XhoI sites of pCMV-Myc (Clontech). All of the new constructs were verified by DNA sequencing.
For transient expression of protein, MCF-7, T47D, MDA-MB-231, MDA-MB-435, and HeLa cells were seeded in six-well culture plate (4 × 105 cells/well), or 100-cm2 dish (1.6 × 106 cells/dish), HEK293 cells were seeded in six-well culture plate (8 × 105 cells/well) or 100-cm2 dish (3 × 106 cells/dish), or HepG2 cells were seeded in six-well culture plate (8 × 105 cells/well), and incubated overnight. The cells were transfected with expression vectors or corresponding amount of empty vectors using Polyfect® (Qiagen) or WelFect-EXTMPLUS (WelGENE Inc., Korea). For reporter gene analysis, MCF-7 cells were seeded in 12-well culture plate (1.5 × 105 cells/well) and transfected with reporter plasmid (0.1–0.3 μg), β-galactosidase (β-gal) expression vector (0.2 μg) with various amount of MTA1 expression vector using Polyfect® (Qiagen). At the end of treatment, luciferase activity was determined using an Analytical luminescence luminometer. Luciferase activity was normalized for transfection efficiency using the corresponding β-gal activity. Data shown are the mean±s.d. of three independent determinations.
Transfection of siRNA duplexes
The siRNA duplexes targeting HIF-1α
(si-HIF-1α, 5′-CCUAUAUCCC AAUGGAUGAUGTT-3′ and
5′-TTGGAUAUAGGGUUACCUACUAC-3′),
MTA1 (si-MTA1#1, 5′-CCCUGUCAGUCUGCUAUAATT-3′ and
5′-UUAUAGCAGACUGACAGGGTT-3′; si-MTA1#2, 5′-AAGACCCUGCUGGCAGAUAAATT-3′ and
5′-UUUAUCUGCCAGCAGGGUCU UTT-3′)
(Mishra et al, 2004), and control nonspecific siRNA, si-GL3, were synthesized and purified by Shamchully Pharm Co. (Korea). The transfection of siRNAs was performed twice at 24 h intervals with OligofectamineTM reagent (Invitrogen) according to the manufacturer's protocol.
Reverse transcriptase-polymerase chain reaction
Total RNA was prepared using RNeasy kit (Qiagen Inc.). PCR reaction was performed as described previously with specific primers for
MTA1 (forward: 5′-GAAATATGGTGGCTTGAAAAT-3′,
reverse: 5′-ACTTTGGTGGGGTAGGACTTC-3′),
HIF-1α (forward: 5′-CAAGTGCATCATTAAGACTG-3′,
reverse: 5′-GGCTGCTCCAGGTCCTGGCG-3′),
VEGF (forward: 5′-TGGAGTTTGCTAAACGTCTG-3′,
reverse: 5′-GTAGCTTATCTACTTTTGTC-3′), and
β-actin (5′-CGTGGGCCGCCCTAGGCACCA-3′,
reverse: 5′-TTGGCTTAGGGTTCAGGGGGG-3′)
(Yoo et al, 2003). Genes were analyzed under the same conditions used to exponentially amplify the PCR products. The expression of β-actin was monitored as a control. One representative of at least three independent experiments with similar results is shown.
Acknowledgments
We are grateful to Drs Kyu-Won Kim, Eric Metzen, and Wen-Ming Yang for the ARD1, the GFP-PHD1, 2 and 3, and the GFP-MTA1 plasmid, respectively. This work was supported by a grant from the Ministry of Science and Technology (M1-0311-00-0089).
References
- Bagheri-Yarmand R, Talukder AH, Wang RA, Vadlamudi RK, Kumar R (2004) Metastasis-associated protein 1 deregulation causes inappropriate mammary gland development and tumorigenesis. Development 131: 3469–3479 [DOI] [PubMed] [Google Scholar]
- Bilton R, Mazure N, Trottier E, Hattab M, Dery MA, Richard DE, Pouyssegur J, Brahimi-Horn MC (2005) Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1α and is not induced by hypoxia or HIF. J Biol Chem 280: 31132–31140 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC (2002) Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2: 563–572 [DOI] [PubMed] [Google Scholar]
- Cho J, Kim D, Lee S, Lee Y (2005) Cobalt chloride-induced estrogen receptor α down-regulation involves hypoxia-inducible factor-1α in MCF-7 human breast cancer cells. Mol Endocrinol 19: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Dales JP, Garcia S, Meunier-Carpentier S, Andrac-Meyer L, Haddad O, Lavaut MN, Allasia C, Bonnier P, Charpin C (2005) Overexpression of hypoxia-inducible factor HIF-1α predicts early relapse in breast cancer: retrospective study in a series of 745 patients. Int J Cancer 116: 734–739 [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, Meinsma RJ, Bosma P, Kemp S, Caron HN, van Kuilenburg AB (2005) Gene expression profiling in response to the histone deacetylase inhibitor BL1521 in neuroblastoma. Exp Cell Res 309: 451–467 [DOI] [PubMed] [Google Scholar]
- Debies MT, Welch DR (2001) Genetic basis of human breast cancer metastasis. J Mammary Gland Biol Neoplasia 6: 441–451 [DOI] [PubMed] [Google Scholar]
- Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC (2005) Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol 45: 495–528 [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Fidler IJ (2003) The pathogenesis of cancer metastasis: the ‘seed and soil' hypothesis revisited. Nat Rev Cancer 3: 453–458 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Etages SD, Hayes L, Crimin K, Li B (2005) Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J Biol Chem 280: 17749–17757 [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA (2003) MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113: 207–219 [DOI] [PubMed] [Google Scholar]
- Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK (2003) Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2: 151–163 [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21: 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW (2002) Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell 111: 709–720 [DOI] [PubMed] [Google Scholar]
- Kato H, Tamamizu-Kato S, Shibasaki F (2004) Histone deacetylase 7 associates with hypoxia-inducible factor 1α and increases transcriptional activity. J Biol Chem 279: 41966–41974 [DOI] [PubMed] [Google Scholar]
- Kim SH, Park JA, Kim JH, Lee JW, Seo JH, Jung BK, Chun KH, Jeong JW, Bae MK, Kim KW (2006) Characterization of ARD1 variants in mammalian cells. Biochem Biophys Res Commun 340: 422–427 [DOI] [PubMed] [Google Scholar]
- Kumar R (2003) Another tie that binds the MTA family to breast cancer. Cell 113: 142–143 [DOI] [PubMed] [Google Scholar]
- Kurebayashi J, Otsuki T, Moriya T, Sonoo H (2001) Hypoxia reduces hormone responsiveness of human breast cancer cells. Jpn J Cancer 92: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwai T, Kitadai Y, Tanaka S, Onogawa S, Matsutani N, Kaio E, Ito M, Chayama K (2003) Expression of hypoxia-inducible factor-1α is associated with tumor vascularization in human colorectal carcinoma. Int J Cancer 105: 176–181 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381 [DOI] [PubMed] [Google Scholar]
- Matteucci E, Locati M, Desiderio MA (2005) Hepatocyte growth factor enhances CXCR4 expression favoring breast cancer cell invasiveness. Exp Cell Res 310: 176–185 [DOI] [PubMed] [Google Scholar]
- Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R (2001) Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol 3: 30–37 [DOI] [PubMed] [Google Scholar]
- Mazure NM, Brahimi-Horn MC, Berta MA, Benizri E, Bilton RL, Dayan F, Ginouves A, Berra E, Pouyssegur J (2004) HIF-1: master and commander of the hypoxic world. A pharmacological approach to its regulation by siRNAs. Biochem Pharmacol 68: 971–980 [DOI] [PubMed] [Google Scholar]
- Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Burgel T, Jelkmann W, Acker H, Fandrey J (2003) Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J Cell Sci 116: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Mazumdar A, Vadlamudi RK, Li F, Wang RA, Yu W, Jordan VC, Santen RJ, Kumar R (2003) MICoA, a novel metastasis-associated protein 1 (MTA1) interacting protein coactivator, regulates estrogen receptor-α transactivation functions. J Biol Chem 278: 19209–19219 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Talukder AH, Gururaj AE, Yang Z, Singh RR, Mahoney MG, Franci C, Vadlamudi RK, Kumar R (2004) Upstream determinants of estrogen receptor-α regulation of metastatic tumor antigen 3 pathway. J Biol Chem 279: 32709–32715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawa A, Nishimori K, Lin P, Maki Y, Moue K, Sawada H, Toh Y, Fumitaka K, Nicolson GL (2000) Tumor metastasis-associated human MTA1 gene: its deduced protein sequence, localization, and association with breast cancer cell proliferation using antisense phosphorothioate oligonucleotides. J Cell Biochem 79: 202–212 [PubMed] [Google Scholar]
- Nicolson GL, Nawa A, Toh Y, Taniguchi S, Nishimori K, Moustafa A (2003) Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clin Exp Metast 20: 19–24 [DOI] [PubMed] [Google Scholar]
- Platet N, Prevostel C, Derocq D, Joubert D, Rochefort H, Garcia M (1998) Breast cancer cell invasiveness: correlation with protein kinase C activity and differential regulation by phorbol ester in estrogen receptor-positive and -negative cells. Int J Cancer 75: 750–756 [DOI] [PubMed] [Google Scholar]
- Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3: 721–732 [DOI] [PubMed] [Google Scholar]
- Stoner M, Saville B, Wormke M, Dean D, Burghardt R, Safe S (2002) Hypoxia induces proteasome-dependent degradation of estrogen receptor α in ZR-75 breast cancer cells. Mol Endocrinol 16: 2231–2242 [DOI] [PubMed] [Google Scholar]
- Talukder AH, Gururaj A, Mishra SK, Vadlamudi RK, Kumar R (2004) Metastasis-associated protein 1 interacts with NRIF3, an estrogen-inducible nuclear receptor coregulator. Mol Cell Biol 24: 6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder AH, Mishra SK, Mandal M, Balasenthil S, Mehta S, Sahin AA, Barnes CJ, Kumar R (2003) MTA1 interacts with MAT1, a cyclin-dependent kinase-activating kinase complex ring finger factor, and regulates estrogen receptor transactivation functions. J Biol Chem 278: 11676–11685 [DOI] [PubMed] [Google Scholar]
- Toh Y, Kuninaka S, Endo K, Oshiro T, Ikeda Y, Nakashima H, Baba H, Kohnoe S, Okamura T, Nicolson GL, Sugimachi K (2000) Molecular analysis of a candidate metastasis-associated gene, MTA1: possible interaction with histone deacetylase 1. J Exp Clin Cancer Res 19: 105–111 [PubMed] [Google Scholar]
- Toh Y, Kuwano H, Mori M, Nicolson GL, Sugimachi K (1999) Overexpression of metastasis-associated MTA1 mRNA in invasive oesophageal carcinomas. Br J Cancer 79: 1723–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh Y, Pencil SD, Nicolson GL (1994) A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem 269: 22958–22963 [PubMed] [Google Scholar]
- Vaupel P (2004) The role of hypoxia-induced factors in tumor progression. Oncologist 9: 10–17 [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861 [DOI] [PubMed] [Google Scholar]
- Yan C, Wang H, Toh Y, Boyd DD (2003) Repression of 92-kDa type IV collagenase expression by MTA1 is mediated through direct interactions with the promoter via a mechanism, which is both dependent on and independent of histone deacetylation. J Biol Chem 278: 2309–2316 [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM (2003) The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J Biol Chem 278: 42560–42568 [DOI] [PubMed] [Google Scholar]
- Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO (2005) Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol 19: 950–963 [DOI] [PubMed] [Google Scholar]
- Yoo YG, Cho S, Park S, Lee MO (2004a) The carboxy-terminus of the hepatitis B virus X protein is necessary and sufficient for the activation of hypoxia-inducible factor-1α. FEBS Lett 577: 121–126 [DOI] [PubMed] [Google Scholar]
- Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu DY, Seong JK, Lee MO (2003) Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1α through activation of mitogen-activated protein kinase pathway. J Biol Chem 278: 39076–39084 [DOI] [PubMed] [Google Scholar]
- Yoo YG, Yeo MG, Kim DK, Park H, Lee MO (2004b) Novel function of orphan nuclear receptor Nur77 in stabilizing hypoxia-inducible factor-1α. J Biol Chem 279: 53365–53373 [DOI] [PubMed] [Google Scholar]
- Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW (2000) Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst 92: 1717–1730 [DOI] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW (1999) Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res 59: 5830–5835 [PubMed] [Google Scholar]


