Abstract
The Drosophila master sex-switch protein Sex-lethal (SXL) regulates the splicing and/or translation of three known targets to mediate somatic sexual differentiation. Genetic studies suggest that additional target(s) of SXL exist, particularly in the female germline. Surprisingly, our detailed molecular characterization of a new potential target of SXL, enhancer of rudimentary (e(r)), reveals that SXL regulates e(r) by a novel mechanism—polyadenylation switching—specifically in the female germline. SXL binds to multiple SXL-binding sites, which include the GU-rich poly(A) enhancer, and competes for the binding of CstF64 in vitro. The SXL-binding sites are able to confer sex-specific poly(A) switching onto an otherwise nonresponsive polyadenylation signal in vivo. The sex-specific poly(A) switching of e(r) provides a means for translational regulation in germ cells. We present a model for the SXL-dependent poly(A) site choice in the female germline.
Keywords: e(r), GU-rich element, RNA-binding proteins, polyadenylation, translation
Introduction
The choice of sexual identity is a fundamental biological process. Highly varied molecular mechanisms control sexual differentiation in well-studied metazoans (Ryner and Swain, 1995). In Drosophila melanogaster somatic cells, a hierarchy of alternative splicing events controls various aspects of sexual differentiation. The key sex determining genes (Sex-lethal (Sxl), transformer (tra), and double-sex (dsx)) are spliced differently in male (XY) and female (XX) flies (Schutt and Nothiger, 2000).
The master sex-switch protein SXL (Schutt and Nothiger, 2000) is an RNA-binding protein (Perez-Canadillas and Varani, 2001). SXL is absent in male flies and present in females. It blocks splice sites in three known pre-mRNAs by binding to adjacent uridine-rich sequences or Py-tracts, leading to exon skipping in Sxl, 3′ splice site switching in tra, and intron retention in msl2 (Black, 2003; Forch and Valcarcel, 2003). Moreover, SXL binds to uridine-rich sequence(s) in untranslated regions (UTRs) of the Sxl and msl2 mRNAs (Forch and Valcarcel, 2003), and represses translation. In female somatic cells, SXL allows synthesis of the TRA protein to mediate sexual differentiation and courtship behavior, and prevents synthesis of the MSL2 protein to allow proper dosage compensation (Schutt and Nothiger, 2000; Forch and Valcarcel, 2003).
In addition to its somatic functions, SXL also controls female germline development (Schutt and Nothiger, 2000). Absence of SXL in the female germline results in mitotic and meiotic defects, resulting in ovarian tumors or multicellular cysts of small undifferentiated cells (Schupbach, 1985; Salz et al, 1987; Steinmann-Zwicky et al, 1989) and in defects in chromosome pairing and meiotic recombination (Bopp et al, 1999). However, the mechanism of sex determination and differentiation in the germline is more complex and differs from the well-defined mechanism in somatic cells in several key aspects such as the nature of the sex-determining signal, the roles of Sxl, tra, and dsx, and dependence on an inductive signal from the soma (Mahowald and Wei, 1994; Steinmann-Zwicky, 1992; Schutt and Nothiger, 2000).
Several independent genetic studies suggest that additional targets of SXL exist, particularly in the female germline (Samuels et al, 1994; Kelley et al, 1995; Hager and Cline, 1997; Schutt and Nothiger, 2000; Fujii and Amrein, 2002; Vied et al, 2003). Whereas all known somatic targets of SXL are regulated at the level of splicing and/or translation, here we provide the first evidence that SXL regulates a new target, the enhancer of rudimentary (e(r)) mRNA, by polyadenylation switching in the female germline. Embryonic lethality of both sexes from RNA interference could explain why e(r) escaped previous genetic screens. Finally, we show functional significance of this regulation, and present a model for this sex-specific poly(A) switching.
Results
Use of alternative poly(A) sites accounts for the sex-specific e(r) isoforms
As all known examples of SXL regulation involve its binding to uridine-rich sequences, we searched the entire Drosophila genome for potential high-affinity SXL-binding sites. The SXL consensus (UUUUUGUU(G/U)U(G/U)UUU(G/U)UU) used here for the search was described previously (Singh et al, 1995); a search using a shorter SXL-binding site (U8) (Sakashita and Sakamoto, 1994; Samuels et al, 1994) yielded a significantly larger and experimentally unmanageable number of hits in the genome and therefore the search was not pursued further. Seven of the candidates showed sex-specific mRNA isoforms (A Rahn and R Singh, unpublished results; details for the search algorithm will be published separately). Here, we report the detailed characterization of one of the seven candidates, e(r) (Wojcik et al, 1994). The GADFLY annotation database showed two e(r) transcripts (CT5770 and CT29800) resulting from an alternatively spliced exon and two alternative polyadenylation sites (Figure 1A). We found multiple potential SXL-binding sites in e(r): one adjacent to the 3′ splice site of exon 2 (Figure 1A(i)) and three downstream of the proximal polyadenylation site (Figure 1A(ii); 1, 2, and 3).
Figure 1.
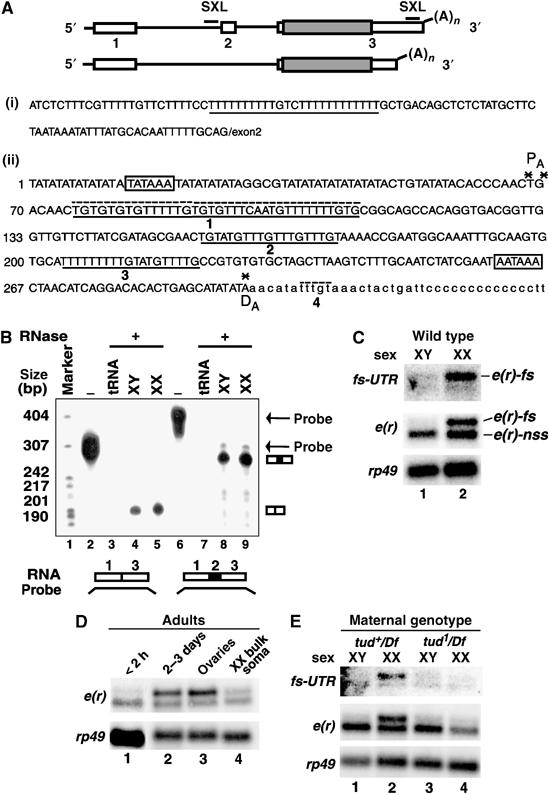
Female germline-specific poly(A) site switching accounts for the sex-specific isoforms of e(r). (A) (Top) Schematics of the two known transcripts of e(r), which differ with respect to inclusion/exclusion of exon 2 and use of alternative polyadenylation sites (Wojcik et al, 1994). Lines, introns; boxes, exons; shaded boxes, coding regions; (A)n, poly(A) tail; and SXL, potential SXL-binding sites. (i) Sequence of the 3′ end of intron 1. The potential SXL-binding site is underlined. (ii) Sequence of a portion of the 3′UTR region of e(r). Boxed nucleotides, alternative polyadenylation signals; asterisks, cleavage/polyadenylation sites; underlined nucleotides, potential SXL-binding sites (1, 2, and 3); dashed line above the sequence, potential GU-rich enhancer/CstF-64-binding sites (1 and 4). Lowercase residues downstream of the distal (DA) poly(A) site were sequenced in this study. (B) Exon 2 is alternately spliced, but not in a sex-specific manner. RNase protection was performed using labeled probes, shown at the bottom, corresponding to either exons 1 and 3 or 1, 2, and 3. Positions of the intact probes and relevant protected fragments are shown on the right. (C) The female-specific e(r)-fs transcript uses the downstream poly(A) site. A Northern blot with poly(A)+ RNA from male and female flies (lanes 1 and 2) was probed with either the e(r) cDNA or the fs-UTR probe corresponding to the sequence between the two poly(A) sites (PA and DA), represented by asterisks in (A(ii)). For this and subsequent figures, XY and XX indicate chromosomal sex. rp49 is a loading control. (D) The e(r)-fs transcript is primarily expressed in the ovaries of mature but not newly eclosed females. Northern blot of RNA isolated from newly eclosed (<2 h) adult XX flies (lane 1), 2- to 3-day-old adult XX flies (lane 2), isolated ovaries (lane 3), and the bulk somatic tissue (whole flies minus ovaries) of XX flies (lane 4). (E) (Top) Loss of the female germline results in the disappearance of the e(r)-fs transcript (lane 4). The poly(A)+ RNA was isolated from the progeny of tud mothers, and probed with the same probes as in panel C.
To determine whether alternative splicing of exon 2 was the basis for the sex-specific isoform of e(r), we performed RNase protection analysis. We found that exon 2 was alternatively spliced (Figure 1B, lanes 4 and 5 versus 8 and 9), but was not spliced in a sex-specific manner (Figure 1B, lane 4 versus 5 and lane 8 versus 9). Next, we probed an RNA blot with the fs-UTR probe (Figure 1C), which corresponds to the sequence between the two polyadenylation sites of e(r) and includes potential SXL sites (Figure 1A(ii)). This probe hybridized to the longer isoform that was present in females (Figure 1C, lane 2, top panel), but not to the shorter isoform present in both sexes (lanes 1 and 2, middle panel). Hereafter, the shorter isoform will be referred to as e(r)-non-sex-specific (e(r)-nss) and the longer isoform will be referred to as e(r)-female-specific (e(r)-fs). Presence of non-sex-specific and female-specific isoforms of e(r) is reminiscent of the known SXL target, tra, although it is the sex-specific splicing regulation that contributes to the synthesis of the two isoforms of tra (Sosnowski et al, 1989). We conclude that use of two poly(A) sites, rather than alternative splicing, accounts for the sex-specific size difference of the e(r) transcripts.
e(r)-fs is restricted to the female germline
SXL controls sexual differentiation in both somatic and germ cells. To test whether e(r)-fs is synthesized in somatic cells or the germline, we dissected ovaries from the bulk somatic tissue of wild-type (wt; w1118) females, and analyzed e(r) expression by Northern analysis. Figure 1D shows that ovaries (lane 3), rather than the bulk somatic tissue (whole fly minus ovaries) (lane 4), were the primary source of the e(r)-fs transcript. However, the e(r)-fs transcript was barely detectable in newly eclosed (<2 h) females, which contain up to stage seven egg chambers (Figure 1D, lane 1). These data show that the e(r)-fs transcript is expressed specifically in the female germline during later stages of oogenesis.
As a complementary approach, we analyzed e(r) expression in the progeny of tudor (tud1/Df) flies, which lack a germline and thus are sterile (Boswell and Mahowald, 1985). Indeed, the e(r)-fs transcript was absent from the female progeny of tud1/Df flies (Figure 1E, lane 4 versus 2), demonstrating that e(r)-fs is a female germline-specific transcript.
SXL is necessary for the synthesis of e(r)-fs in the female germline
As the e(r) gene was identified as a potential target of SXL and showed a female germline-specific isoform, we tested whether synthesis of the e(r)-fs transcript in vivo was dependent on Sxl function, which is important both in somatic cells and in the germline (Schutt and Nothiger, 2000). To analyze the involvement of SXL specifically in the germline, we used two viable, female-sterile mutations of the sans fille (snf) gene (snf1621 and snf148), which disrupt Sxl function in the female germline but not in the soma (Salz, 1992; Nagengast et al, 2003). We found that female flies homozygous for either the snf1621 (Figure 2, lane 1) or snf148 (lane 3) alleles failed to synthesize the e(r)-fs transcript, indicating that loss of SXL expression by the snf mutations disrupted synthesis of e(r)-fs in the germline.
Figure 2.
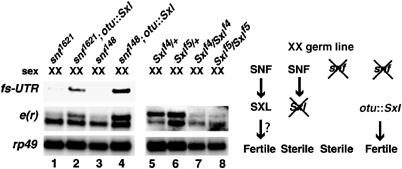
SXL function in germ cells is necessary for the sex-specific poly(A) switching of e(r). (Left panel) XX flies homozygous for the snf1621 or snf148 alleles, which disrupt SXL synthesis in the germline, do not express e(r)-fs (lanes 1 and 3). Expression of the SXL cDNA in snf mutant backgrounds under the control of the otu promoter restores the synthesis of the e(r)-fs transcript (lanes 2 and 4). Sxlf4 and Sxlf5 homozygotes show a loss of e(r)-fs expression (lanes 7 and 8). (Right panel) Schematics of SXL-related events and their disruption by specific mutations in the female germline. Disruption of SXL synthesis/function by mutations in Sxl or snf leads to female sterility, whereas expression of the Sxl cDNA under the control of the germline-specific otu promoter (otu∷Sxl) restores fertility in snf flies. Lanes 5–8 were not analyzed with the fs-UTR probe.
In addition, we examined e(r) expression in Sxlf4 and Sxlf5 homozygous females. We found that the female-sterile Sxlf4 and Sxlf5 alleles, which prevent proper nuclear localization of SXL in the germline but permit normal somatic function (Bopp et al, 1993), also resulted in a loss of the e(r)-fs transcript (Figure 2, lanes 7 and 8). As a complementary approach, we expressed SXL in the snf mutant backgrounds using a germline-specific ovarian tumor (otu) promoter (Hager and Cline, 1997), which is known to restore female fertility in these lines (Nagengast et al, 2003). We found that the germline-specific expression of SXL in snf flies indeed restored female fertility (Nagengast et al, 2003; data not shown) and the expression of the e(r)-fs transcript (Figure 2, lanes 2 and 4). As controls, ovaries isolated from snf1621 flies contained only the e(r)-nss transcript (data not shown), indicating that tumorous ovaries transcribe e(r), process it to produce e(r)-nss, but fail to switch it to e(r)-fs. Furthermore, our analysis with the suppressor, Su(Sxlf4)46 (Bopp et al, 1999), of the female-sterile Sxlf4 mutation showed that it rescued the poly(A) switching of e(r) (Supplementary Figure S1, lane 3 versus 2). Based on what we know about the suppressor (see details in Supplementary data), these results are consistent with our model that SXL regulates e(r), and support the idea that the suppressor acts by influencing an event(s) upstream of SXL, to restore its activity and/or localization. Therefore, our combined results show that SXL function in the female germline is required for the generation of the e(r)-fs transcript, and that the Sxl cDNA alone is sufficient to restore the expression of the e(r)-fs isoform in the snf mutant germlines, which lack SXL function and express only the e(r)-nss transcript.
SXL-binding site is important for poly(A) site choice in vivo
We asked what sequence features were important for sex-specific poly(A) site choice: specifically, whether the potential SXL-binding site(s) was important for the default use of the proximal poly(A) site in males, for poly(A) switching in females, or both; whether the order (or relative positions) of the two poly(A) sites was important for the observed sex-specific pattern; and whether the female-specific poly(A) switching required repression of the proximal site or activation of the distal site. To address these issues, we developed an in vivo reporter system in which various combinations of the wt or mutant 3′UTRs of e(r) were joined downstream of the coding region of a heterologous (EGFP) sequence (to distinguish it from the endogenous e(r) transcript) and placed under the control of the e(r) promoter for proper expression. These constructs were introduced into flies using standard P-element-mediated transformation, and their polyadenylation patterns in male and female flies were analyzed by Northern analysis using an EGFP probe.
First, whereas nontransgenic lines gave no signal (Figure 3, lanes 1 and 2), transgenic lines with the wt e(r) 3′UTR (wt construct) showed a shorter non-sex-specific transcript (Figure 3, lanes 3 and 4) and a longer female-specific transcript (Figure 3, lane 4). Thus, our reporter faithfully recapitulates the sex-specific poly(A) site choice observed for the endogenous e(r) transcript. Second, mutation of the first potential SXL-binding site (Figure 1A(ii)), which also corresponds to a GU-rich enhancer element for the proximal poly(A) site (Zhao et al, 1999), activated the distal (DA) site in males (Figure 3, lane 7); males lack SXL and do not normally use the distal site (DA) for the wt 3′UTR. The poly(A) pattern was identical in males and females (Figure 3, lane 8). Another mutation (mutant2; see Figure 4A) of this GU-rich element only partially activated the DA site in males, and showed little, if any, further switching in females (Figure 3, lanes 9 and 10). These results show that the SXL-binding site/GU-rich enhancer element is important both for the default choice of the proximal poly(A) site in males and for sex-specific switching in females. Third, we switched the order of the proximal (PA) and distal (DA) polyadenylation sites. For DP transcripts, males exclusively used the DA site (now located upstream) (Figure 3, lane 5). Moreover, DP females showed no switching to the PA site (now located downstream) (Figure 3, lane 6). We can draw the following conclusions from these experiments: (i) The distal site is competent to support efficient polyadenylation in (DP) males, which argues against the possibility that poly(A) switching requires a female germline-specific activator (or activation) of the distal site. (ii) The natural order of the two poly(A) sites plays an important role in their sex-specific usage. (iii) Cis competition with the proximal poly(A) site contributes to why the distal site is normally not used.
Figure 3.
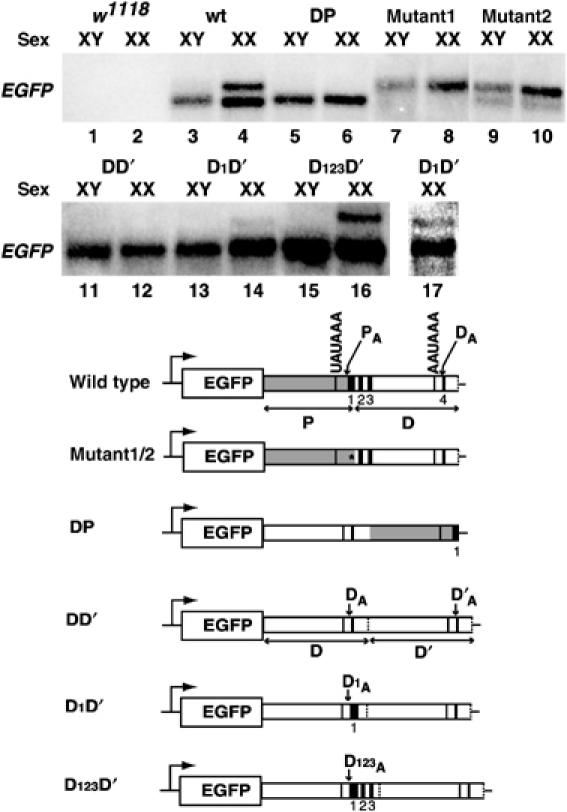
The SXL-binding site is important for the sex-specific poly(A) switching of e(r). (Top) The order of the two poly(A) sites and the proximal GU-rich element are important for default polyadenylation in males. Northern analysis was performed using an EGFP probe on three independent transgenic lines for each construct. w1118 is the parental line without the EGFP transgene. (Bottom) Schematics of reporter constructs with the e(r) promoter (site of transcription is indicated by an arrow upstream of EGFP) and different 3′UTRs of e(r) used to generate various transgenic lines. P and D represent the proximal (gray box) and distal (white box) portions of the 3′UTR, respectively. Locations of the proximal (PA), distal (DA), duplicated distal (DA′), and modified distal (D1A and D123A) poly(A) sites are shown. The GU-rich elements (1, 2, 3, and 4) are as in Figure 1A(ii). In the DP construct, the SXL-binding site is downstream of the P sequence and is indicated by the black box labeled 1. The asterisk indicates mutation of the proximal GU-rich element. The wt sequence of the proximal GU-rich element (Figure 1A(ii), 1) was changed to TGTTTTAGCACACACCACACGAATTCACACAACAC in mutant1 or to TGTGTGTGTTGAATTCGTG in mutant2. Lane 17 is an overexposure of lane 14.
Figure 4.
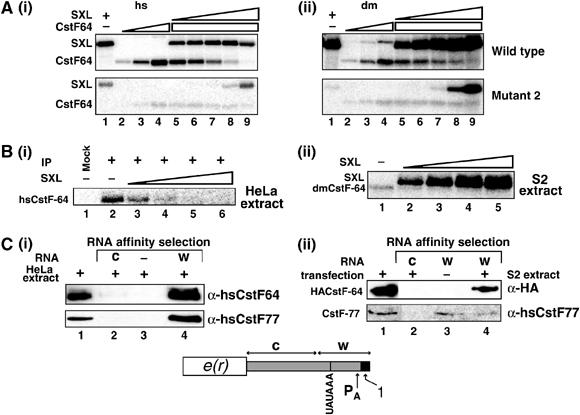
SXL competes with CstF-64 for binding to the proximal GU-rich element in vitro. (A) (i) Radiolabeled RNA containing the proximal GU-rich element was crosslinked to specific proteins and analyzed on an SDS–polyacrylamide gel. The concentrations (ng/μl) for SXL and hsCstF-64 were as follows: SXL (i and ii): lane 1, 150.0; lane 5, 1.85; lane 6, 5.5; lane 7, 16.5; lane 8, 50.0; lane 9, 150.0; hsCstF-64 (i): lane 2, 13.8; lane 3, 41.6; lanes 4–9, 125.0; and dmCstF-64 (ii): lane 2, 16.6; lane 3, 50.0; lanes 4–9, 150.0. The mutant2 is as described in Figure 3. Longer exposure is shown because of the weaker signal from dmCstF-64. (B) SXL competes with the human CstF-64 (i) and the putative Drosophila CstF-64 (ii) in the nuclear extract for binding to the proximal GU-rich element. Identity of the human hsCstF-64 protein was confirmed by immunoprecipitation with anti-CstF64 antibodies. (C) RNA affinity selection confirms that the CstF complex assembles on the proximal GU-rich element. The proximal GU-rich element (w), but not a control RNA (c), pulls down both CstF-64 and CstF-77 from the HeLa (i) and S2 (ii) nuclear extracts. A schematic of the 3′UTR and the positions of the RNAs used for affinity selection are shown.
The above experiments suggested that the SXL-binding site adjacent to PA was important for sex-specific poly(A) switching. To complement results from the mutagenesis of the GU-rich element, we asked whether the GU-rich sequence(s) downstream of PA was able to confer female-specific switching onto an otherwise nonresponsive site in vivo. Therefore, we duplicated the distal site (DD′), and analyzed it for sex-specific splicing. As would be expected from the DP construct, the first distal site (DA) was used in males (lane 11). Furthermore, there was no activation of the second distal site (DA′) in females (lane 12), indicating that DA is nonresponsive to female-specific switching. However, insertion of three GU-rich elements (or the SXL-binding site(s)) (1, 2, and 3; see Figure 1A(ii)) downstream of D (D123D′) supported efficient poly(A) switching in the female germline (lane 16), but not in males (lane 15). Incorporation of the first GU-rich element alone (D1D′) caused a smaller but reproducibly detectable poly(A) switching (lanes 14 and 17). Lack of poly(A) switching in DP and DD′ females served as a negative control for the specificity of sex-specific poly(A) switching. We conclude that GU-rich elements downstream of the proximal poly(A) site confer female-specific poly(A) site repression onto an otherwise nonresponsive site, causing sex-specific poly(A) switching.
SXL competes for the binding of CstF-64 to the proximal GU-rich element in vitro
The experiments described above show that both SXL and the putative SXL-binding site(s) are important for the female germline-specific poly(A) switching in e(r). Next, we investigated the molecular mechanism for the poly(A) switching of e(r). The cleavage stimulatory factor 64 (CstF-64), a component of the CstF complex of the poly(A) machinery, is known to directly bind to GU-rich enhancer elements and stimulate polyadenylation by facilitating the binding of the cleavage and polyadenylation specificity factor to the AAUAAA signal (Zhao et al, 1999). Therefore, we reasoned that SXL could compete with CstF-64 for binding to the proximal GU-rich element in e(r), resulting in the activation of the distal poly(A) site in females.
To test this hypothesis, we performed a competition assay. In an in vitro RNA-binding experiment using photochemical crosslinking to radiolabeled RNA and analysis on an SDS–polyacrylamide gel, we found that recombinant SXL and the RNA-binding domain of the human (hsCstF64) and Drosophila (dmCstF-64) CstF-64 proteins crosslinked to the proximal GU-rich element (Figure 4A(i) and (ii), lanes 1–4, wt). Most importantly, SXL competed in vitro, in a concentration-dependent manner, for the crosslinking of both the human (Figure 4A(i), lanes 5–9, wt) and the Drosophila (Figure 4A(ii), lanes 5–9, wt) CstF-64 to the proximal GU-rich element, indicated by the disappearance of the crosslinked CstF-64 band. However, both SXL and CstF-64 showed significantly reduced crosslinking to a GU-rich element mutant (mutant2), which is a reflection of their specificity for GU-rich sequences and is consistent with the known RNA-binding properties of these proteins (Singh et al, 1995; Takagaki and Manley, 1997). In addition, at these concentrations, SXL showed no detectable competition for the low-level binding of CstF-64 to the mutant sequence (Figure 4A(i) and (ii), lanes 5–9, mutant2). We conclude that the proximal GU-rich element is a specific, high-affinity SXL-binding site, and that SXL directly competes in vitro for the binding of CstF-64 to this element.
To confirm that the competition seen with recombinant SXL and CstF64 is also relevant in the context of crude nuclear extract and in the context of endogenous CstF complex, we performed the following experiments. First, SXL competed with the endogenous human CstF64 in HeLa nuclear extract (Figure 4B(i), lane 2 versus lanes 3–6). The identity of the human CstF64 was confirmed using immunoprecipitation with α-CstF64 antibodies (Figure 4B(i), lane 1 versus 2). In addition, SXL competed with the putative Drosophila CstF64 protein (the only crosslinked band was of the expected molecular mass), in the nuclear extract from S2 cells (Figure 4B(ii), lanes 2–5), although antibodies were unavailable to independently confirm the identity of the Drosophila ortholog. Second, to determine whether intact CstF complex assembled onto the proximal poly(A) site, we performed an RNA affinity selection, using RNAs corresponding to the proximal polyadenylation elements (w) or an upstream control sequence (c) immobilized onto agarose beads. CstF64 from nuclear extract prepared from HeLa cells (Figure 4C(i), lane 4) and from S2 cells transfected with HA-tagged CstF64 (Figure 4C(ii), lane 4) was specifically recruited to the appropriate RNA substrate (w; see Schematics). As negative controls, CstF64 was not pulled down by the control RNA (c) (Figure 4C(i) and (ii), lane 2), by beads only (i, lane 3), or from an extract prepared from untransfected S2 cells (ii, lane 3). Moreover, the affinity-selected CstF64 co-immunoprecipitated with CstF77 in both HeLa and S2 nuclear extracts (Figure 4C(i) and (ii), lane 4, bottom panel), indicating that intact CstF complex is recruited to the proximal GU-rich element; a weaker signal for the Drosophila extract (Figure 4C(ii), lane 4, bottom panel) reflects poor crossreactivity of anti-hsCstF77 antibodies to the Drosophila ortholog. We conclude that SXL competes with the CstF64 subunit of the intact CstF complex in nuclear extract for binding to the proximal GU-rich element.
We determined the binding affinities of both SXL and CstF64 for RNAs containing single and multiple potential SXL-binding sites (sites 1 or 1–3). The binding affinity of SXL for site 1 (Kd∼10 nM) (Figure 5A, lanes 8–12) was comparable to that of the well-characterized high-affinity SXL-binding site in tra (lanes 2–6) (Valcarcel et al, 1993), within 2- to 3-fold experimental variation. SXL formed a single complex on the RNA substrate with a single binding site. However, on the RNA with three sites, we observed up to three or four supershifted RNA–protein complexes in an SXL concentration-dependent manner (lanes 14–19), suggesting that SXL can bind to each of these sites. However, CstF64 bound to both RNAs in a similar manner, with a binding affinity (Kd) of ∼350 nM for site 1 based on the disappearance of free RNA in a gel mobility shift (Takagaki and Manley, 1997) and on UV crosslinking approaches (Figure 5B(i) and (ii), lanes 2–7 and 9–14). This affinity is within the range (∼100 nM to 1.5 μM) reported for CstF64-selected RNA aptamers (Takagaki and Manley, 1997). Thus, both SXL and CstF64 bind to the GU-rich elements with high affinity. Moreover, occupancy of multiple sites by SXL could at least in part explain why the construct with three SXL sites (D123D′) is more efficient than that with a single site (D1D′) in conferring female-specific poly(A) switching in a heterologous context (Figure 3).
Figure 5.
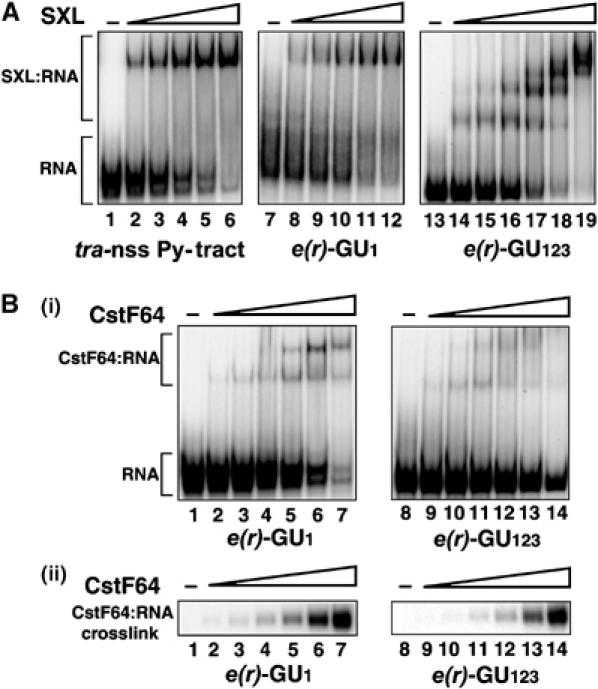
SXL binds to multiple binding sites in e(r). Gel mobility shift analysis with SXL (A) and CstF64 (B, i) using RNAs containing either one (e(r)-GU1) or three (e(r)-GU123) SXL-binding sites, shown in Figure 1A(ii). The transformer non-sex-specific Py-tract (tra-nss Py-tract), a known SXL-binding site (Valcarcel et al, 1993), was used as a positive control. (B, ii) The same RNA-binding reaction as in panel (B, i) except that the RNA–protein complex was crosslinked with UV light and separated on a denaturing SDS–polyacrylamide gel. The concentrations (ng/μl) for SXL and hsCstF-64 were as follows. (A) SXL: lanes 1, 7, and 13, no protein; lanes 2, 8, and 15, 0.34; lanes 3, 9, and 16, 0.68; lanes 4, 10, and 17, 1.37; lanes 5, 8, and 18, 2.75; lanes 6, 12, and 19, 5.5; lane 14, 0.17; and (B, i and ii) hsCstF-64: lanes 1 and 8, no protein; lanes 2 and 9, 1.7; lanes 3 and 10, 5.1; lanes 4 and 11, 15.3; lanes 5 and 12, 46.0; lanes 6 and 13, 140.0; lanes 7 and 14, 420.0.
Poly(A) switching provides a means for translation regulation in the female germline
To determine the biological significance of the female germ-line-specific poly(A) switching of e(r) on gene expression, we fused portions of the 3′UTR of e(r) (proximal (P), distal (D), or both (P+D)) or of control 3′UTRs (Adh and K10) downstream of the coding region of an EGFP reporter that was expressed from the e(r) promoter, and introduced these constructs into flies. Expression of the EGFP reporter was determined by fluorescence microscopy in isolated ovaries. Nontransgenic flies showed barely detectable signal (Figure 6A, panel w1118), whereas control 3′UTRs allowed efficient expression of the EGFP reporter (panels Adh and K10). The non-sex-specific portion of e(r) showed an extremely high level of reporter expression (panel P). However, the female-specific portion of e(r) showed significantly reduced reporter expression (panel D) in germ cells. There was a detectable signal in the surrounding follicle cells, which are of somatic origin, suggesting that this 3′UTR is otherwise competent to support translation. The entire 3′UTR (P+D) of e(r) showed reporter expression level much lower than that in the P line (panel P+D), consistent with the extent of poly(A) switching in this line (Figure 6B). These results were also confirmed using immunostaining with anti-GFP antibodies (middle panels). To exclude the possibility that the level of EGFP expression was due to differences in levels of RNA synthesis or stability, we performed Northern analysis. The level of EGFP poly(A)+ RNA was comparable in P, D, and P+D flies (Figure 6B, lanes 3–5). Thus, we argue that the EGFP signal reflects primarily differences in reporter translation. We conclude that the female-specific portion of e(r) represses reporter translation in vivo in the female germline.
Figure 6.
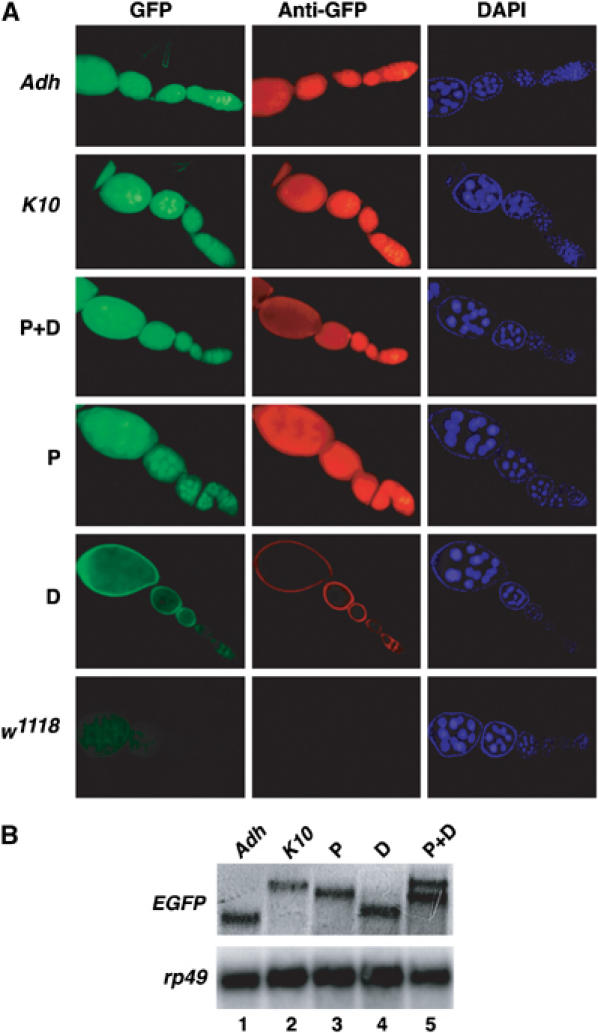
The female-specific portion of e(r) represses translation in vivo. (A) Different portions of the 3′UTR of e(r) (P, D, P+D) (see Materials and methods for details) and control 3′UTRs (Adh, K10) were placed downstream of an EGFP reporter under the control of the e(r) promoter, and analyzed for reporter translation in vivo using fluorescence microscopy (left panels) and immunostaining with anti-GFP antibodies (middle panels) in isolated ovaries. DAPI staining (right panels) is also shown. (B) Northern analysis of RNA from the transgenic lines.
Discussion
In this study, we demonstrate that SXL regulates e(r) expression in vivo by a novel mechanism—polyadenylation switching—which allows translation regulation in the female germline.
Model for the sex-specific polyadenylation of e(r)
A model that explains the SXL-mediated regulation of e(r) must answer the following questions. First, what is the molecular basis for the default polyadenylation pattern in male flies? Second, how does SXL mediate poly(A) switching in females? Third, why does the poly(A) switching of e(r) occur primarily in the female germline?
Male-specific polyadenylation. We show that three key factors account for the default poly(A) pattern of e(r) in male flies: differences in the binding affinities of CstF-64 for the two alternative GU-rich elements; order of the two polyadenylation sites; and cis competition between the two poly(A) signals. First, the longer GU-rich enhancer element downstream of the proximal cleavage/polyadenylation site provides at least in part a basis for the increased apparent binding affinity for CstF-64, most likely by providing multiple registers for binding (Banerjee et al, 2003), and thus confers competitive advantage to the proximal poly(A) site.
Second, the order of the two poly(A) sites is also important. Contrary to our expectation that the proximal site is inherently stronger than the distal site, DP males (Figure 3, lane 5) exclusively use the DA site. This demonstrates that the arrangement of the two sites is important for the default poly(A) site choice and excludes the possibility that the distal site is inherently weak. The usage of the DA site is consistent with the known RNA-binding properties of CstF-64 (Takagaki and Manley, 1997) and sequences of natural poly(A) sites (MacDonald et al, 1994). Moreover, in this system, the promoter-proximal site is always used by default in males (Figure 3; PA in wt, DA in DP, and D123A in D123D). Both transcription and splicing influence polyadenylation (Niwa and Berget, 1991; Hirose and Manley, 2000; Vagner et al, 2000; Proudfoot et al, 2002). As our EGFP reporter, unlike the endogenous transcript, lacks an intron, coupling between the transcription and polyadenylation machineries best explains why the promoter-proximal site is always preferred in males.
Third, mutation of the SXL-binding site or the proximal GU-rich element compromises CstF64 binding and activates the distal site in males. This is what would be expected if the SXL-binding site is also a polyadenylation element (or the CstF64-binding site) and if SXL blocks the proximal poly(A) site, which is consistent with the SXL blockage model proposed for splicing regulation (Sosnowski et al, 1989; Inoue et al, 1990; Valcarcel et al, 1993). As CstF-64 binds directly to the GU-rich enhancer element to facilitate recruitment of the polyadenylation machinery to the poly(A) signal (Zhao et al, 1999), reduced affinity of CstF-64 to the mutant proximal GU-rich element provides a basis for the activation of the distal site in males. This shows that the longer GU-rich element associated with the proximal site is also important for the default poly(A) site choice. It is possible that the non-canonical poly(A) element (UAUAAA instead of AAUAAA) and its sequence context (Figure 1A(ii)) confer increased dependence on the long GU-rich element for polyadenylation at the proximal site. This feature could make the proximal GU-rich element and CstF-64 binding an attractive target for SXL regulation.
Female-specific poly(A) switching. The simplest explanation for our combined results is that in the female germline SXL competes for the binding of CstF-64 to the proximal GU-rich element and represses polyadenylation at the proximal site, leading to activation of the distal site (Figure 7); SXL does not bind to the distal GU-rich element (data not shown). The proximal GU-rich element is much longer than is necessary for polyadenylation or CstF-64 binding (MacDonald et al, 1994; Takagaki and Manley, 1997), most likely to confer competitive advantage (by providing multiple registers for CstF-64 binding; Banerjee et al, 2003) to the proximal site in males and to accommodate SXL binding in females. Moreover, SXL and CstF-64 have similar sequence preference (GU-rich sequences devoid of cytidines) (Singh et al, 1995; Takagaki and Manley, 1997). Therefore, it is not surprising that we could not uncouple the sequence requirement of the proximal GU-rich element for polyadenylation in males and for poly(A) switching in females (Figure 3), as was performed using a three nucleotide U-to-C substitution for SXL-mediated splicing regulation of tra (Sosnowski et al, 1989; Inoue et al, 1990), which involves competition between SXL and U2AF (Valcarcel et al, 1993); U2AF prefers uridine- and cytidine-rich sequences devoid of guanosines (Singh et al, 1995, 2000). Specificity of SXL for the proximal but not the distal GU-rich element and our ability to convert a poly(A) site that is nonresponsive to poly(A) switching to a site that supports female-specific switching provide an explanation for why switching occurs in wt and D123D′ females, but not in DP and DD′ females (Figure 3). Moreover, SXL-dependent poly(A) site switching of e(r) (Figure 3) as well as splice site switching of Sxl and msl2 transcripts displays a need for multiple SXL-binding sites for efficient regulation (Forch and Valcarcel, 2003). The ability of SXL to discriminate between alternative GU-rich sequences is the crux of the sex-specific poly(A) switching. GU-rich sequences belong to a class of regulatory RNA motifs that are defined by base composition rather than an exact sequence. Such simple repetitive sequences offer several advantages for gene regulation: for example, binding affinity but not specificity for RNA-binding proteins can change as a function of the length of the sequence (Banerjee et al, 2003); two proteins (e.g., SXL and CstF-64) with different binding site length requirements can recognize the same sequence (Singh et al, 1995; Takagaki and Manley, 1997); a regulatory protein such as SXL can regulate multiple processes, polyadenylation and splicing, by antagonizing CstF-64 and U2AF65, respectively, because of the possibility of overlapping binding specificities; and single nucleotide changes are less disruptive.
Figure 7.
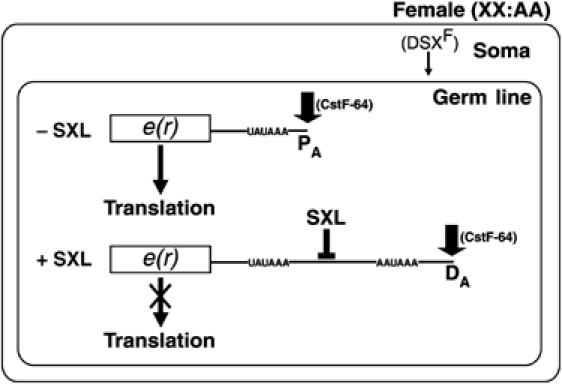
A model for sex-specific polyadenylation site switching of e(r) in the female germline. (−SXL), CstF-64 binds to the proximal GU-rich element and promotes assembly of the polyadenylation machinery preferentially at the proximal site, producing the e(r)-nss isoform. (+SXL), SXL competes with CstF-64 for binding to the proximal GU-rich element, leading to the activation of the distal poly(A) site. The e(r)-nss transcript is translated in the female germline, but e(r)-fs isoform is not; SXL may also be involved in translation repression. Thick arrows indicate the preferred (proximal (PA) or distal (DA)) cleavage/polyadenylation sites.
It is possible that future development of an in vitro poly(A) switching assay may provide additional evidence for a direct role of SXL or formally exclude a less likely possibility of involvement of another factor that is regulated by SXL, is an RNA-binding protein, is female germline specific, and has RNA-binding properties identical to those of SXL. Nonetheless, several lines of evidence presented here (Figures 2, 3 and 4) strongly argue that SXL has a direct role in female germline-specific poly(A) switching of e(r).
Female germ-line specificity of poly(A) switching. Intriguingly, the female germline-specific e(r)-fs transcript is barely detectable in the female soma where SXL is present and functional. Our experiments have ruled out that female germline-specific activation of the distal site contributes to poly(A) switching. Several factors, which are consistent with the repression of the proximal site, could account for why SXL-dependent poly(A) switching is seen in a female germline-specific manner. For example, levels of SXL change during oogenesis (Vied and Horabin, 2001), female germline-specific SXL isoforms are known (Bopp et al, 1991; Samuels et al, 1991), and changes in the nuclear translocation of SXL are observed during oogenesis (Bopp et al, 1993). Finally, SXL may associate with a germline-specific cofactor for efficient poly(A) switching, as described for SXL-dependent translation repression of msl2 in embryonic extract (Grskovic et al, 2003).
Comparison with other examples of poly(A) site switching
CstF-64 plays an important role in constitutive and regulated polyadenylation. The levels of CstF-64 change significantly during B-lymphocyte differentiation and an increase in the levels of CstF-64 is necessary and sufficient for the poly(A) site switching of the immunoglobulin heavy-chain (IgM) mRNA (Takagaki et al, 1996; Takagaki and Manley, 1998; Proudfoot et al, 2002). However, there are important differences between the IgM and e(r) systems. For example, in IgM, there are competing splicing and polyadenylation sites, the two poly(A) sites are separated by >2 kb, and changes in relative concentrations/activities of ubiquitously expressed GU-rich element-binding proteins (CstF64 and hnRNPs) contribute to poly(A) site choice (Takagaki et al, 1996; Takagaki and Manley, 1998; Zhao et al, 1999; Veraldi et al, 2001; Castelo-Branco et al, 2004). In contrast, the sex-specific poly(A) site choice for e(r) is unaffected by splicing, the two sites are separated by only 221 nucleotides, and the absence or presence of the female-specific protein SXL correlates with poly(A) site choice. Whereas poly(A) switching produces different protein isoforms (secreted versus membrane bound) of IgM, it affects the 3′UTR rather than the coding sequence of e(r) and influences protein synthesis. Finally, unlike most other examples of alternative polyadenylation, which often involve skipping of a weaker proximal poly(A) site to allow use of a stronger distal site by default (Perez Canadillas and Varani, 2003), a promoter-proximal site is always preferred by default in e(r).
Biological significance
The poly(A) site switching described here provides a new mechanism for the retention of an SXL-binding site(s) (in 3′UTR) in a female germline-specific manner for translation repression, although it remains to be determined whether the elements that mediate translation regulation are similar to or different from those required for poly(A) switching. SXL-mediated alternative splicing (intron retention in the 5′UTR) of msl2 is another mechanism for retention of SXL-binding sites for translation repression in somatic cells in females (Forch and Valcarcel, 2003). Thus, it would not be surprising if SXL were also involved in translation repression. Comparison of the two mRNAs should provide important insights into the molecular mechanism of translation regulation.
Although SXL regulates distinct steps along the gene expression pathway (splicing, translation, and now polyadenylation) and targets different factors (e.g., U2AF, TIA-1, and CstF-64) (Forch and Valcarcel, 2003), it uses a common underlying mechanism, that is, competition for the binding of factors that recognize GU- or U-rich sequences and play non-overlapping roles in the assembly of appropriate multi-ribonucleoprotein or -protein complexes. Thus, overlapping RNA-binding specificities and the possibility of combinatorial control from association with cofactors could allow SXL to specifically regulate distinct processes (splicing, translation, and polyadenylation) in different tissues. ER is a potential transcription repressor (Pogge von Strandmann et al, 2001), interacts with components of the transcription machinery (Amente et al, 2005), and may be subject to cell cycle regulation (Gelsthorpe et al, 1997). As loss of SXL in the female germline results in ovarian tumors, it is tempting to speculate that e(r) may act as an anti-oncogene and provide the missing link in the pathway connecting SXL to germ cell proliferation/differentiation and oncogenesis. Loss of e(r) from microinjection of double-stranded RNA into embryos results in non-sex-specific embryonic lethality, which could at least in part explain why the sex-specific regulation of the e(r) transcript escaped previous genetic screens (see Supplementary data).
Conclusions
Identification of e(r) represents the first known example of SXL-dependent poly(A) switching, and opens up the possibility that other genes may also be regulated in this manner. Poly(A) switching provides a new mechanism to allow subsequent translation regulation. As e(r) is suggested to be a transcription regulator, it appears to be a potent downstream target of the sex determination pathway and may help unravel the mysterious role of SXL in the female germline.
Materials and methods
Materials
The Sxl stocks were obtained from the Bloomington Stock Center, the snf stocks from Dr Helen Salz, the suppressor stocks from Drs Jamila Horabin and Daniel Bopp, and the mago and tudor stocks from Dr Robert Boswell. Flies were raised on standard corn meal food at 25°C. The cDNA clones and ESTs were obtained from Research Genetics, CA, and the e(r) genomic DNA was obtained from Dr Stuart Tsubota. The 3′UTR e(r)-fs probe (fs-UTR) was generated by PCR from the EST clone LD36385 using primers A and B. Recombinant GST-fusion proteins were as described: SXL (Valcarcel et al, 1993); hsCstF-64 (Takagaki and Manley, 1997); Drosophila CstF-64 cDNA was PCR amplified from the EST clone RE27227 using primers C and D and cloned into pGEX-6P-1 vector. The anti-HA antibody (12CA5) was purchased from Roche, and the anti-hsCstF-64 and anti-hsCstF-77 antibodies were obtained from Dr Christine Milcarek and Dr David Bentley.
Plasmids, DNA templates, and primers
The EGFP reporter constructs, transcription templates, EGFP translation reporter constructs, and primers are described in Supplementary data.
RNase protection assay
The RNase protection was performed essentially as per the manufacturer's instructions for the RPA III kit (Ambion Inc.).
Northern analysis
Northern analysis was performed as previously described (Robida and Singh, 2003).
Generation of transgenic lines
Transgenes were introduced into the Drosophila genome by P-element-mediated transformation (Serano et al, 1994).
Gel shift, RNA–protein UV crosslinking assay, and immunoprecipitation
The gel mobility shift and RNA–protein UV crosslinking assays were performed as previously described (Valcarcel et al, 1993; Singh et al, 1995). For immunoprecipitation, anti-hsCstF64 antibodies or preimmune serum were crosslinked to protein A beads, incubated with the crosslinked RNA–protein complexes, washed, and separated on an SDS–PAGE gel (see Supplementary data).
RNA affinity selection and Western analysis
RNA was synthesized from relevant templates using T7 RNA polymerase, immobilized on adipic acid dihydrazine agarose beads, incubated with nuclear extract prepared from untransfected S2 cells, from S2 cells transfected with HA-CstF-64, or from HeLa cells, and subjected to Western analysis using appropriate antibodies (see Supplementary data).
Whole-mount fluorescence and immunological detection of reporter translation
Female flies were fed on yeast paste for 2–4 days after eclosion. Ovaries were dissected in PBST (1 × PBS, 0.1% Triton X-100), fixed for 15 min in PBST with 4% formaldehyde, stained with DAPI, and examined for EGFP signal by florescence microscopy and by immunostaining using a Zeiss 2.2.1 microscope (Mohr et al, 2001).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Drs Tom Cech, Tom Blumenthal, Jens Lykke-Anderson, Mark Winey, Helen Salz, Fatima Gebauer, Juan Valcarcel, and members of the Singh laboratory for critical comments on the manuscript. We thank Drs Helen Salz, Jamila Horabin, Daniel Bopp, and the Bloomington Drosophila Stock Center for providing the fly stocks, Daniel Bopp for sharing unpublished results, Dr Bob Boswell and PJ Bennett for advice on fly genetics and fly stocks, and Indranil Dey for technical help with RNase protection. MDR thanks Kellie Hazell for her support. This work was supported in part by a training grant from the National Institutes of Health to MDR, and by grants from the National Institutes of Health (GM58576) and American Cancer Society (RSG-04-177-01) to RS.
References
- Amente S, Napolitano G, Licciardo P, Monti M, Pucci P, Lania L, Majello B (2005) Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett 579: 683–689 [DOI] [PubMed] [Google Scholar]
- Banerjee H, Rahn A, Davis W, Singh R (2003) Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA 9: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bopp D, Bell LR, Cline TW, Schedl P (1991) Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev 5: 403–415 [DOI] [PubMed] [Google Scholar]
- Bopp D, Horabin JI, Lersch RA, Cline TW, Schedl P (1993) Expression of the Sex-lethal gene is controlled at multiple levels during Drosophila oogenesis. Development 118: 797–812 [DOI] [PubMed] [Google Scholar]
- Bopp D, Schutt C, Puro J, Huang H, Nothiger R (1999) Recombination and disjunction in female germ cells of Drosophila depend on the germline activity of the gene sex-lethal. Development 126: 5785–5794 [DOI] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP (1985) tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43: 97–104 [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N (2004) Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P, Valcarcel J (2003) Splicing regulation in Drosophila sex determination. Prog Mol Subcell Biol 31: 127–151 [DOI] [PubMed] [Google Scholar]
- Fujii S, Amrein H (2002) Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J 21: 5353–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsthorpe M, Pulumati M, McCallum C, Dang-Vu K, Tsubota SI (1997) The putative cell cycle gene, enhancer of rudimentary, encodes a highly conserved protein found in plants and animals. Gene 186: 189–195 [DOI] [PubMed] [Google Scholar]
- Grskovic M, Hentze MW, Gebauer F (2003) A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J 22: 5571–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager J, Cline T (1997) Induction of female Sex-lethal RNA splicing in male germ cells: implications for Drosophila germline sex determination. Development 124: 5033–5048 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y (1990) Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature 344: 461–463 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877 [DOI] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14: 6647–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP, Wei G (1994) Sex determination of germ cells in Drosophila. Ciba Found Symp 182: 193–202 [DOI] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE (2001) The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev 15: 2886–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagengast AA, Stitzinger SM, Tseng CH, Mount SM, Salz HK (2003) Sex-lethal splicing autoregulation in vivo: interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development 130: 463–471 [DOI] [PubMed] [Google Scholar]
- Niwa M, Berget SM (1991) Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095 [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM, Varani G (2001) Recent advances in RNA–protein recognition. Curr Opin Struct Biol 11: 53–58 [DOI] [PubMed] [Google Scholar]
- Perez Canadillas JM, Varani G (2003) Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J 22: 2821–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogge von Strandmann E, Senkel S, Ryffel GU (2001) ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor. Biol Chem 382: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Robida MD, Singh R (2003) Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J 22: 2924–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Swain A (1995) Sex in the '90s. Cell 81: 483–493 [DOI] [PubMed] [Google Scholar]
- Sakashita E, Sakamoto H (1994) Characterization of RNA binding specificity of the Drosophila sex-lethal protein by in vitro ligand selection. Nucleic Acids Res 22: 4082–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK (1992) The genetic analysis of snf: a Drosophila sex determination gene required for activation of Sex-lethal in both the germline and the soma. Genetics 130: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK, Cline TW, Schedl P (1987) Functional changes associated with structural alterations induced by mobilization of a P element inserted in the Sex-lethal gene of Drosophila. Genetics 117: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Bopp D, Colvin RA, Roscigno RF, Garcia-Blanco MA, Schedl P (1994) RNA binding by Sxl proteins in vitro and in vivo. Mol Cell Biol 14: 4975–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Schedl P, Cline TW (1991) The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol Cell Biol 11: 3584–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T (1985) Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex-lethal in Drosophila melanogaster. Genetics 109: 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt C, Nothiger R (2000) Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127: 667–677 [DOI] [PubMed] [Google Scholar]
- Serano TL, Cheung HK, Frank LH, Cohen RS (1994) P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene 138: 181–186 [DOI] [PubMed] [Google Scholar]
- Singh R, Banerjee H, Green MR (2000) Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA 6: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176 [DOI] [PubMed] [Google Scholar]
- Sosnowski BA, Belote JM, McKeown M (1989) Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58: 449–459 [DOI] [PubMed] [Google Scholar]
- Steinmann-Zwicky M (1992) How do germ cells choose their sex? Drosophila as a paradigm. BioEssays 14: 513–518 [DOI] [PubMed] [Google Scholar]
- Steinmann-Zwicky M, Schmid H, Nothiger R (1989) Cell-autonomous and inductive signals can determine the sex of the germline of Drosophila by regulating the gene Sxl. Cell 57: 157–166 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL (1997) RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 17: 3907–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL (1998) Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Valcarcel J, Singh R, Zamore PD, Green MR (1993) The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362: 171–175 [DOI] [PubMed] [Google Scholar]
- Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C (2001) hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol 21: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vied C, Halachmi N, Salzberg A, Horabin JI (2003) Antizyme is a target of sex-lethal in the Drosophila germline and appears to act downstream of hedgehog to regulate sex-lethal and cyclin B. Dev Biol 253: 214–229 [DOI] [PubMed] [Google Scholar]
- Vied C, Horabin JI (2001) The sex determination master switch, Sex-lethal, responds to Hedgehog signaling in the Drosophila germline. Development 128: 2649–2660 [DOI] [PubMed] [Google Scholar]
- Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI (1994) Enhancer of rudimentary e(r) a highly conserved enhancer of the rudimentary gene. Genetics 138: 1163–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


