Abstract
Characterization of AtMLH3, the Arabidopsis homologue of the prokaryotic MutL mismatch repair gene, reveals that it is expressed in reproductive tissue where it is required for normal levels of meiotic crossovers (COs). Immunocytological studies in an Atmlh3 mutant indicate that chromosome pairing and synapsis proceed with normal distribution of the early recombination pathway proteins. Localization of the MutS homologue AtMSH4 occurs, suggesting that double Holliday junctions (dHjs) are formed, but the MutL homologue AtMLH1, which forms a heterocomplex with AtMLH3, fails to localize normally. Loss of AtMLH3 results in an ∼60% reduction in COs and is accompanied by a substantial delay of ∼25 h in prophase I progression. Analysis of the chiasma distribution in Atmlh3 suggests that dHj resolution can occur, but in contrast to wild type where most or all dHjs are directed to form COs the outcome is biased in favour of a non-CO outcome by a ratio of around 2 to 1. The data are compatible with a model whereby the MutL complex imposes a dHj conformation that ensures CO formation.
Keywords: Arabidopsis, meiosis, mismatch repair, recombination
Introduction
The eukaryotic homologues of the Escherichia coli MutS and MutL mismatch repair (MMR) proteins play important roles in maintaining genome stability during both mitosis and meiosis (Kolodner and Marsischky, 1999; Hoffmann and Borts, 2004; Svetlanov and Cohen, 2004). Studies in yeast have identified four MutL homologues that form functionally distinct heterodimers. Two of these, Mlh1/Pms1 and Mlh1/Mlh2, are proposed to have roles in the correction of different classes of DNA mismatch, whereas the Mlh1/Mlh3 heterodimer appears to play an important role in promoting meiotic crossovers (COs) (Wang et al, 1999). Studies in Mlh1-deficient yeast have revealed that they exhibit reduced spore viability and a reduction in COs such that map distance in an interval on chromosome III is reduced by 21–33% (Wang et al, 1999). Mouse knockouts in MLH1 disrupt meiotic recombination in both male and female animals, resulting in the formation of unpaired univalent chromosomes at the first meiotic division (Baker et al, 1996; Woods et al, 1999). The early stages of prophase I appear normal in the knockout mice, but as the synaptonemal complex disassembles at the onset of diplotene the absence of chiasmata is revealed. An mlh3 mouse knockout has a similar, although not identical effect on meiotic progression to that of the mlh1 knockout (Lipkin et al, 2002). Chromosome synapsis is normal, but very few bivalents persist beyond pachytene. However, in contrast to the mlh1 knockout where spermatocyte apoptosis is induced swiftly at diplotene, a substantial proportion of mlh3 knockout spermatocytes progress through to metaphase I/anaphase I, where following chromosome missegregation apoptosis occurs. Female mice are infertile, failing to complete meiosis I after fertilization.
In accord with these findings, immunolocalization studies using light and electron microscopy have revealed that MLH1 and MLH3 proteins colocalize as foci on mouse chromosomes during pachytene and that their distribution is consistent with each being a component of the late recombination nodules (RNs) (Moens et al, 2002; Marcon and Moens, 2003). Moreover a recent study, in which the phosphatase inhibitor okadaic acid was used to induce the precocious onset of diplotene in mouse spermatocytes, succeeded in demonstrating that the MLH1/MLH3-containing RNs are localized to the chiasmata (Marcon and Moens, 2003).
Studies of MMR genes in Arabidopsis thaliana have identified several homologues of the E. coli MutS gene, namely AtMSH2, AtMSH3, AtMSH4, AtMSH5, AtMSH6-1, AtMSH6-2 and AtMSH7 (Ade et al, 1999; Culligan and Hays, 2000). Of the Arabidopsis MutL homologues, AtMLH1 has been studied to a limited extent (Jean et al, 1999). The predicted protein shares 37% identity, 55% similarity over its entire length to MLH1 from mouse and 31% identity, 48% similarity to the yeast protein. However, a corresponding mutant was not isolated, hence a functional analysis of this gene was not conducted. In addition to AtMLH1, the study revealed two other MutL homologues in the Arabidopsis genome. Phylogenetic analysis indicated that one of these is likely to be the Arabidopsis homologue of PMS1. Although the other, originally designated AtMLHx, was also considered to be a member of the PMS1 family, it appeared somewhat distinct, and was placed in an intermediate position between PMS1 and MLH1 (Jean et al, 1999). Recently, AtMLHx has been redesignated as AtMLH3 (Figure 1A) (Alou et al, 2004). The AtMLH3 protein contains a predicted MutL domain in the NH2-terminal region of the protein, which exhibits 32% amino-acid identity over a region of 228 aa to that present in the Arabidopsis AtMLH1 protein. However, AtMLH3 (1151 aa) is considerably larger than MLH1 (737 aa) and contains an additional MutL domain. This does not exhibit any significant homology to the other MutL domain in the protein or with that in AtMLH1. However, it shares significant (37%) similarity with the MutL domain found in the mouse MLH3 protein. Phylogenetic analysis clearly places AtMLH3 within the MLH3 group (Alou et al, 2004). Despite discrepancies in their sizes, the mouse and yeast MLH3 proteins (192 and 715 aa, respectively) are thought to perform a similar role in meiotic recombination. Hence, this suggests that although AtMLH3 is larger than either, it might also function in meiosis. The work described below has revealed that AtMLH3 is specifically expressed in reproductive tissues of Arabidopsis and that the protein localizes to foci associated with the chromosome axes during prophase I of meiosis. Analysis of two independent T-DNA insertion mutants of the gene confirms a role in the formation of meiotic COs and provides new insight into the role played by the MutL homologues in CO/non-CO resolution of double Holliday junctions (dHjs).
Figure 1.
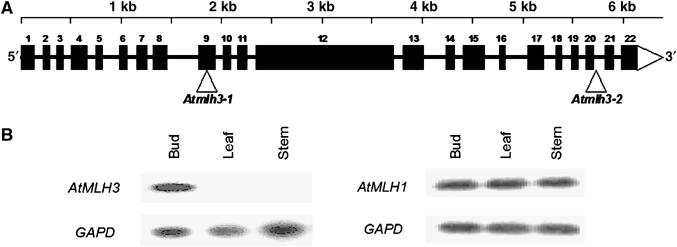
(A) Map of the 6.3 kb At4g35520 locus showing the exon/intron organization of AtMLH3. The exons are shown as numbered black boxes. The triangles indicate the T-DNA insertion sites in Atmlh3-1 and Atmlh3-2. (B) RT–PCR expression analysis revealing that in contrast to AtMLH1, expression of AtMLH3 is restricted to reproductive tissue.
Results
AtMLH3 is expressed in reproductive tissue
Studies in yeast and mouse indicate that MLH3 and MLH1 function as a heterocomplex during meiosis, but unlike MLH1, the MLH3 protein is thought to have only a limited role in DNA MMR in mitotic cells (Flores-Rozas and Kolodner, 1998; Kolas and Cohen, 2004). This suggested that expression of an Arabidopsis homologue of MLH3 may be restricted to or more abundant in reproductive tissue. To explore this possibility, we carried out RT–PCR using AtMLH3- and AtMLH1-specific primers with mRNA from a range of plant tissues (Figure 1B). This clearly revealed that AtMLH3 was expressed in bud tissue but was not detectable in vegetative tissues, whereas expression of AtMLH1 was detected in all the tissues tested. This finding is consistent with a role for AtMLH3 during meiosis in Arabidopsis, but cannot be absolutely definitive, as buds are comprised of a mixture of reproductive and vegetative cells.
AtMLH3 localizes to meiotic chromosome axes in mid–late prophase I
An antibody was raised to a 347 aa region comprising residues 804–1151 of the AtMLH3 protein. FITC-labelled anti-AtMLH3 Ab was used to immunolocalize AtMLH3 on meiotic chromosome spreads prepared from Arabidopsis pollen mother cells (PMCs) at different stages of meiosis (Figure 2A–C). In order to accurately establish when AtMLH3 is initially detectable, dual immunolocalization was performed with antibodies that recognize the meiotic proteins ASY1, AtMSH4 and AtMLH1, which may be used to monitor prophase I progression from early leptotene through to pachytene (Armstrong et al, 2002; Higgins et al, 2004, 2005). ASY1 is associated with the chromosome axes and its first appearance identifies the onset of leptotene (Armstrong et al, 2002). At this stage, AtMLH3 was not detectable. As the meiocytes progressed to zygotene, AtMLH3 foci gradually appeared. At pachytene, the mean number of foci per nucleus was 9.4 (n=42), which is in close agreement with the mean chiasmata frequency of 9.86 previously reported for Arabidopsis (Higgins et al, 2004). The AtMLH3 foci continued to be detectable throughout pachytene, where they were found to colocalize with AtMLH1 (Figure 2D–F). The foci remained present as the chromosomes began to desynapse during diplotene, but by metaphase I they could no longer be detected. Thus, the timing and frequency of the AtMLH3 foci are consistent with a role for the protein in the later stages of meiotic recombination.
Figure 2.
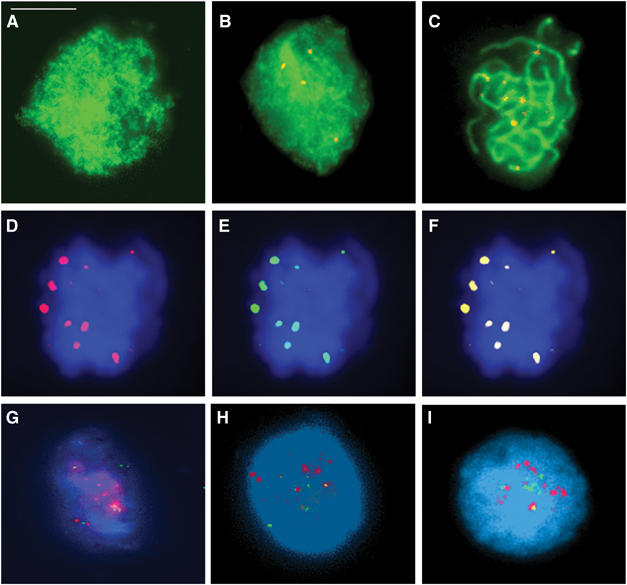
(A–C) Dual immunolocalization of AtMLH3 (red) and ASY1 (green). (A) At leptotene, ASY1 is localized to the developing chromosome axes. (B) AtMLH3 foci first become detectable during zygotene. (C) At pachytene 9–10, AtMLH3 foci are found in association with the chromosome axes. (D–F) Colocalization of the MutL homologues at pachytene. (D) AtMLH3 (red), (E) AtMLH1 (green) and (F) merged image. (G–I) Limited colocalization of AtMLH3 and AtMSH4. At early zygotene, AtMSH4 foci (red) are abundant with few AtMLH3 foci (green) detectable (G). At mid-zygotene (H) through to late zygotene/early pachytene (I), there is an increase in AtMLH3 foci and a reduction in AtMSH4 foci. Colocalization between the foci is consistently observed, but is limited to only few foci (1–2) per nucleus. Bar=10 μm.
Dual-immunolocalization studies were conducted to establish the relationship of AtMLH3 and AtMSH4 during zygotene (Figure 2G–I). At early zygotene, numerous AtMSH4 foci are detected with few if any AtMLH3 foci present. During mid-zygotene through to late zygotene/early pachytene, there was a continual decrease in the number of AtMSH4 foci, whereas the number of AtMLH3 foci increased to ∼10 per nucleus. Although most AtMLH3/AtMSH4 foci did not appear to colocalize, we consistently observed colocalization of 1–2 foci in each nucleus (N=20).
AtMLH3-deficient Arabidopsis exhibit reduced fertility and meiotic defects
To determine if AtMLH3 was required for meiosis, we identified two Atmlh3 mutant lines among the Salk Institute T-DNA insertion collection (Figure 1A). The position of the T-DNA within AtMLH3 was determined in each case using PCR and nucleotide sequencing. The first line, Atmlh3-1, was found to carry a T-DNA insert 114 bp into exon 9 of the gene. Cytological analysis using fluorescence in situ hybridization (FISH) with a T-DNA probe indicated that in addition to the insertion on chromosome 4 in AtMLH3 the line possessed a second insertion on chromosome 5. Crosses were therefore made to obtain a single insertion line, which was confirmed by FISH (Supplementary Figure 1). As it is predicted that an insertion in exon 9 would result in a truncation of the AtMLH3 protein to an NH2-terminal peptide of just 262 aa of the total 1151 aa, it seems likely that Atmlh3-1 is a null mutant. This is supported by the absence of the corresponding RNA transcript and protein, as determined by RT–PCR (data not shown) and immunolocalization respectively (Supplementary Figure 2A). Analysis of the second line (Atmlh3-2) revealed that the T-DNA was inserted within the coding region, 279 bp from the 3′ end of the gene. Although this mutation could have resulted in the production of a truncated AtMLH3 protein, immunolocalization using anti-MLH3 Ab failed to detect any evidence of residual protein during prophase I (Supplementary Figure 2B).
Vegetative growth of both T-DNA insertion mutants was indistinguishable from wild-type plants, with no apparent somatic abnormalities. However, differences became apparent when the plants began to set seed. The wild-type plants developed normal length siliques of a more or less consistent length containing an average of 52.8 seeds (n=32 siliques), whereas Atmlh3-1 and Atmlh3-2 siliques contained means of 26 (n=50 siliques) and 22.8 (n=32) seeds, respectively. A reduced fertility phenotype is typical of that found for other meiotic mutants in Arabidopsis (Caryl et al, 2003), although in those characterized to date the reduction in seed set is more dramatic than that observed for the Atmlh3 mutant lines. Nevertheless, this observation further suggested a role for AtMLH3 in meiosis.
Cytological analysis of Atmlh3-deficient lines reveals defects in recombination
The reduced fertility phenotype of the mutant lines is consistent with a role for AtMLH3 in meiosis. To investigate this further, meiotic chromosome spreads were prepared using PMCs isolated from the mutants Atmlh3-1 and Atmlh3-2 and a wild-type control. These were then examined using fluorescence microscopy. Figure 3 shows a cytological profile of Atmlh3-1. Early prophase I from leptotene through to pachytene was indistinguishable from wild type with apparently normal chromosome pairing, leading to full synapsis at pachytene (Figure 3A). However, as the chromosomes desynapsed towards the end of prophase I and began to condense during late diplotene/diakinesis, it became clear that a proportion (see later for details) of the homologous chromosome pairs lack chiasmata and are present as univalents at metaphase I (Figure 3B and C). This resulted in missegregation at the first meiotic division, leading to the formation of dyads containing aberrant chromosome numbers. This in turn resulted in aneuploid tetrads following the second meiotic division (Figure 3D). Analysis of mlh3-2 revealed an identical effect on meiosis with a reduction in the average number of meiotic COs leading to aneuploidy at the second division (Supplementary Figure 3). These observations clearly demonstrate that AtMLH3 is required for normal progression through meiosis and thus plays a crucial role in meiosis.
Figure 3.
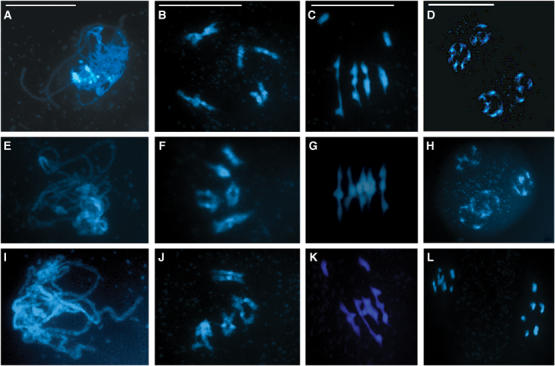
(A–D) Cytological analysis of Atmlh3-1. Prophase I appears normal with fully synapsed homologous chromosomes at pachytene (A). However, as the chromosomes desynapse at the end of prophase I and begin to condense during diplotene/diakinesis (B) before aligning at metaphase I (C), the presence of univalents becomes clear. This leads to subsequent missegregation and the formation of unbalanced tetrads (D). (E–H) Complementation of Atmlh3-1 with a genomic fragment encoding AtMLH3 results in the restoration of normal meiosis. Fully synapsed chromosomes are observed at pachytene (E) with five bivalents at diplotene/diakinesis and metaphase I (F, G) leading to the formation of balanced tetrads containing five chromosomes (H). (I–L) Crossing Atmlh3-1 and Atmlh3-2 fails to restore normal meiosis, indicating that the mutants are allelic. Cytologically, the line is indistinguishable from the parental single knockout lines, apparently normal at pachytene (I) but with univalents at diplotene/diakinesis and metaphase I (J, K) leading to missegregation at the dyad stage. Bar=10 μm.
Complementation of Atmlh3-1 and allelism with Atmlh3-2
To confirm that mutation of the AtMLH3 gene was responsible for the reduced fertility phenotype, an 11.42 kb genomic XbaI fragment encoding the full-length gene was cloned from BAC F8D20 into pCAMBIA 1302. The construct was then transformed into Atmlh3-1. Transformed lines were selected on hygromycin medium and transferred to soil. Analysis of 50 independent transformants revealed that fertility was fully restored (50–55 seeds/silique) (Supplementary Figure 4). Cytological analysis of two lines revealed that meiosis was entirely normal (Figure 3E–H).
In addition to the complementation test, an allelism test was carried out by reciprocally crossing heterozygous Atmlh3-1 and Atmlh3-2 lines. All the progeny genotyped as Atmlh3-1/Atmlh3-2 (N=12) exhibited a reduced fertility phenotype and univalent chromosomes were present at metaphase I (Figure 3I–L and Supplementary Figure 4)
Immunolocalization of meiotic proteins in Atmlh3 mutants
To further characterize the effect of the Atmlh3 mutation on meiosis, fluorescence immunolocalization studies were carried out on spread preparations of PMCs at prophase I from the Atmlh3 mutants. The localization of ASY1 and the synaptonemal complex transverse filament protein ZYP1 (Higgins et al, 2005) in Atmlh3-1 was indistinguishable from that in wild-type PMCs, leading us to believe that axis formation, chromosome pairing and synapsis are unaffected in the mutant (Figure 4A and B; the results for Atmlh3-2 were identical and are presented in Supplementary Figure 2C and D). This finding suggested that early steps in the recombination pathway proceed as normal in Atmlh3-1. This was supported by the observation that distribution of AtRAD51, a component of early RNs, was unaltered in the mutant line. Immunolocalization using anti-AtRAD51 Ab revealed that the protein is first detectable as numerous punctate foci associated with the developing chromosome axes during early leptotene (Figure 4C). These remain throughout zygotene/early pachytene until mid-pachytene when they rapidly decline in number. The disappearance of the AtRAD51 foci is associated with the removal of the early RNs and the emergence of the late RNs that correspond to sites of the chiasmata (Moens et al, 2002). Although not proven, it is postulated that the late RNs represent a subset of early RNs that have been ‘marked' as the future sites of COs/chiasmata.
Figure 4.
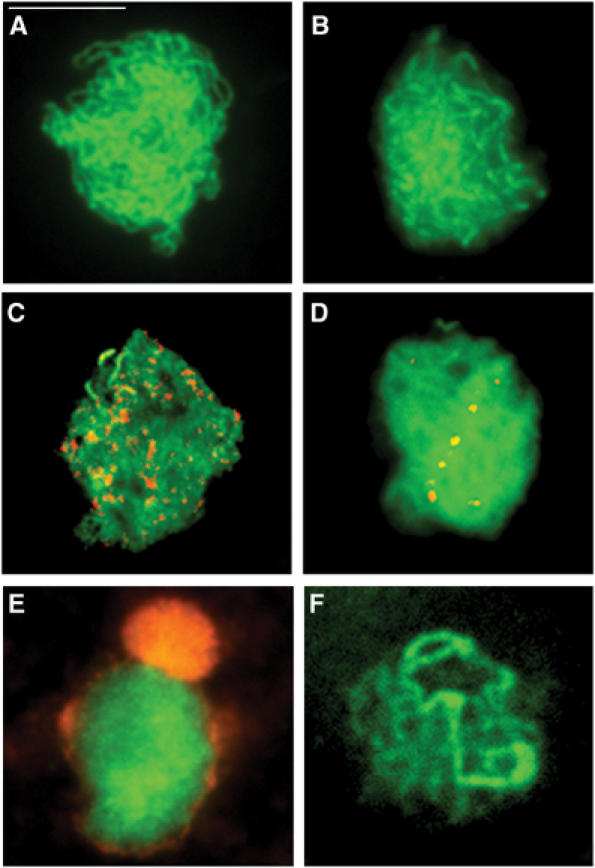
Immunolocalization of meiotic proteins in spread preparations of PMCs in an Atmlh3-1 mutant. Localization of the axis-associated protein ASY1 (green) at zygotene (A) and the SC protein ZYP1 (green) at pachytene (B) appears normal, indicating that chromosome pairing and synapsis occur in the absence of AtMLH3. Dual localization of ASY1 (green) with the recombination protein RAD51 (red) (C) and ZYP1 (green) with AtMSH4 (red) (D) suggests that recombination progresses to the dHj stage. However, normal localization of AtMLH1 fails to occur. Dual localization with ASY1 (green) at leptotene reveals aberrant nucleolar localization of AtMLH1 (red) (E). At pachytene, ZYP1 (green) is present but there is a complete absence of AtMLH1 foci (F). Bar=10 μm.
The MutS homologue, AtMSH4, is proposed to play a crucial role in this transitional period (Moens et al, 2002). We therefore used an anti-AtMSH4 antibody to investigate localization of the protein in Atmlh3-1. This was found to be indistinguishable from that previously reported in wild-type Arabidopsis (Higgins et al, 2004). Numerous MSH4 foci were first detectable at late leptotene. As prophase I progressed, the number of foci gradually reduced in number, with a few (<10) persisting through to mid-pachytene (Figure 4D). However, in contrast to the wild-type meiocytes, we were unable to detect any evidence of AtMLH1 foci in Atmlh3-1; instead, the protein was found to accumulate in the nucleolus at leptotene before disappearing before pachytene (Figure 4E and F). This strongly suggests that AtMLH1 localization is dependent on AtMLH3.
Prophase I progression is delayed in Atmlh3
Studies in yeast and more recently in Arabidopsis suggest a role for an intra-prophase I surveillance system that maintains coordination between prophase I progression and the recombination pathway (Boerner et al, 2004; Higgins et al, 2005). We therefore investigated prophase I progression in Atmlh3 using bromo-deoxyuridine pulse labelling of meiocytes (Armstrong et al, 2003). This revealed that as a result of the mutation, prophase I was extended substantially such that the first meiotic division was delayed by 25 h compared to a wild-type control (Supplementary Figure 5). Based on this observation, it seems conceivable that in the absence of the AtMLH3/AtMLH1 complex, the recombination defect is somehow detected and as a result prophase I is delayed until resolution of the dHjs.
Chiasmata are binomially distributed in the Atmlh3 T-DNA mutants
The initial cytological examination of the AtMLH3-deficient lines revealed that the protein is required for wild-type levels of meiotic COs. It was notable however that a significant number of chiasmata remained, suggesting that CO events are not entirely dependent on AtMLH3 in Arabidopsis. One alternative explanation might be that the COs were due to residual AtMLH3 activity. However, in view of the studies outlined above, which indicate a complete absence of the protein in the mutant lines, this does not seem likely. We therefore decided to determine the numerical distribution of residual COs in Atmlh3-1 in more detail. By using FISH with probes recognizing the 45S and 5S ribosomal subunit repeat sequences combined with chromosome morphology, it is possible to accurately identify each of the Arabidopsis chromosomes and chromosome arms (Fransz et al, 1998; Sanchez-Moran et al, 2002). This enabled the number of chiasmata per meiocyte and per chromosome to be determined. A total of 60 metaphase I nuclei were analysed for each mutant (Figure 5A–C). The mean chiasma frequency in Atmlh3-1 is 3.92 compared to 9.86 for wild-type Arabidopsis (Columbia), a reduction of around 60%. Essentially identical data were obtained for Atmlh3-2, which has a mean chiasma frequency of 3.84.
Figure 5.
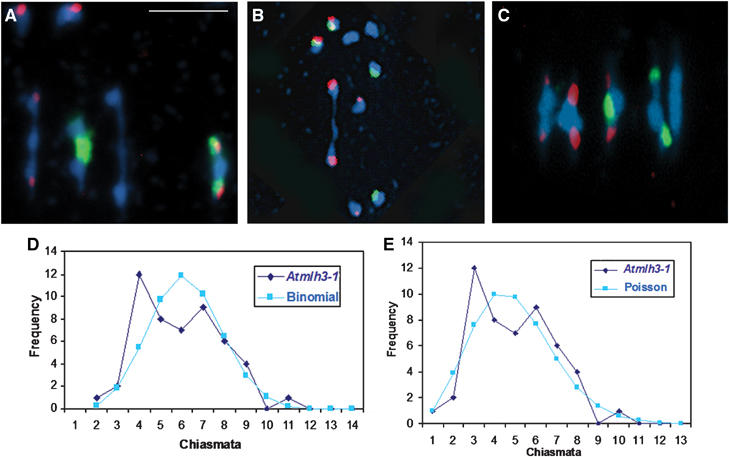
(A, B) Representative metaphase I nuclei of Atmlh3-1 cells after FISH to mark the positions of 5S rDNA (red) and 45S rDNA (green) loci. (A) A nucleus with a total of four chiasmata, comprising a single distal chiasma in chromosomes 1, 2, 3 and 4. (B) A nucleus with a single distal chiasma in chromosome 5. (C) An example of a typical wild-type nucleus with nine chiasmata is shown for comparison. (D) Observed (diamonds) and binomial-predicted (squares) distributions of chiasma numbers per cell for Atmlh3-1. (E) Observed data (diamonds) and Poisson-predicted (squares) distribution.
Comparing the numerical distribution of chiasmata in Atmlh3 mutant(s) to one or more theoretical distributions may give some insights into the mode of action of the AtMLH3 gene in the wider context of chiasma/CO regulation. For this purpose, we focused our attention on the Atmlh3-1 mutant. Despite having a relatively low mean chiasma frequency per cell (3.92), this line showed an unusually wide range of cell chiasma frequencies (0–9). This contrasts with the situation in wild-type meiosis where, typically, the range of cell chiasma frequencies is much narrower (8–12), and suggests that chiasma formation is much less controlled in the mutant(s).
We initially compared the mutant cell chiasma frequencies to the binomial distribution. The rationale behind this is that the AtMLH3 gene may be responsible for driving a preselected set of recombination intermediates (RIs) towards COs, and hence chiasmata. It is proposed that in the absence of AtMLH3, these dHjs can resolve independently either as COs or as non-COs with probability p of resolving as COs and probability q (=1−p) of resolving as non-COs at each of k RIs per cell. If these conditions are obtained, then CO numbers per cell should fit the binomial distribution (p+q)k. However, the binomial distribution is usually applied when the values of p and k are known, which is not the case here. Nevertheless, we can determine, by iteration, the values that give the closest fit of our mutant cell chiasma data to the binomial distribution. We found that the best fit to the binomial distribution was obtained with a p-value of 0.35 and k-values of 11 (χ2(6)=9.46; P>0.1) or 12 (χ2(7)=11.15; P>0.1) (Figure 5D). As up to 12 chiasmata per cell are observed in wild-type Arabidopsis, these values of k are quite reasonable. Of course, this is likely a gross simplification because, for example, k (the number of RIs) may differ from cell to cell. Additionally, it seems highly probable that Arabidopsis has a second recombination pathway that accounts for an average of 1.5 chiasmata per cell (Higgins et al, 2004) that would be unaffected by the Atmlh3 mutation. However, this subset of COs do not exhibit interference and thus may occur too close to other COs to be distinguished separately as chiasmata.
An alternative approach that avoids some of these difficulties is to compare the chiasma distribution to the Poisson distribution that is related to the binomial distribution. The practical advantage of this is that there is no necessity to find values for p and k, as the Poisson distribution is specified by the mean (μ) that is derived directly from the data. When this comparison was carried out, the Atmlh3 cell chiasma frequency distribution did not differ significantly from a Poisson distribution (χ2(6)=5.84; P>0.3) (Figure 5E). In this context, the possible presence of a subset of interference free (random) chiasmata originating from a second recombination pathway, which also follow a Poisson distribution (Higgins et al, 2004), is not a problem because when two Poisson distributions are combined the resulting distribution is itself Poissonian (http://mathworld.wolfram.com/PoissonDistribution.html; Eric W Weisstein ‘Poisson distribution').
Discussion
MLH1 and MLH3 are eukaryotic homologues of the bacterial MMR gene MutL. A previous investigation in Arabidopsis resulted in the identification of three MutL homologues, AtMLH1, AtPMS1 and AtMLHx (Jean et al, 1999). Subsequently, the same authors redesignated AtMLHx as AtMLH3 on the basis of sequence homology to the yeast and mammalian genes (Alou et al, 2004). The investigation reported here provides strong functional evidence that the AtMLH3 protein is required for normal meiotic recombination in Arabidopsis. Immunolocalization studies indicate that in an Atmlh3 mutant, recombination progresses to the later stages. Based on this, we propose that resolution of dHjs can occur in the absence of the MLH3 protein, but that resolution is biased in favour of a non-CO outcome.
AtMLH3 localizes to homologous chromosomes during meiotic prophase I
Studies in mouse have revealed that MLH3 protein localizes as foci to the chromosome axes at early to mid-pachytene, persisting until early diplotene (Lipkin et al, 2002). At mid- to late pachytene, one to two foci were observed per chromosome, colocalizing with MLH1. It has been proposed that these foci mark the positions of meiotic COs that will subsequently appear as chiasmata in diplotene, diakinesis and metaphase I. This has recently been confirmed in mouse where both MLH1 and MLH3 have been shown to localize to the sites of chiasmata precociously induced using okadaic acid treatment of spermatocytes (Marcon and Moens, 2003).
Our immunolocalization studies have revealed that AtMLH3 localizes as discrete foci during prophase I of meiosis. The protein is first detected at mid-zygotene. By pachytene, the number of foci detected is ∼10 and corresponds closely to the number of COs. In the absence of AtMLH3, the early recombination pathway remains, as far as can be judged, unaltered. However, in agreement with the mouse, the loading of AtMLH1 is dependent on AtMLH3, as AtMLH1 foci are not observed in the Atmlh3 mutants. Instead, the protein remains associated with the nucleolus. The basis of the nucleolar accumulation is unknown, but it has previously been noted in the case of the meiotic protein SWI1 and it has been suggested that nucleolus may be a reservoir that somehow regulates protein availability (Visintin and Amon, 2000; Mercier et al, 2003). Additionally, AtMLH3 foci are not detected in an Atmsh4 background (Higgins et al, 2004), whereas the number and distribution of AtMSH4 foci in Atmlh3 is indistinguishable from that observed in wild-type meiocytes. Thus, the chronology of loading of the MMR proteins appears similar in yeast, mammals and Arabidopsis, with MSH4/MSH5 loading early in prophase I to be followed at a later stage by MLH3/MLH1.
The role of AtMLH3 in CO formation
Evidence from budding yeast has confirmed that Msh4/Msh5 and Mlh1/Mlh3 function in the same recombination pathway (Hunter and Borts, 1997). In addition, mutant analysis of Mlh1 in budding yeast has indicated that the role of the protein in CO formation is independent of its role in MMR (Argueso et al, 2003). Hence, it has been proposed that the MutL homologues combine along with the MutS homologues Msh4 and Msh5 in some structural role to promote meiotic COs. However, analysis reveals that msh4/msh5 mutants exhibit a greater defect in recombination frequency than their mlh1/mlh3 counterparts (Wang et al, 1999). Our analysis of the chiasma frequency in the Atmlh3 mutants reveals a reduction in chiasma frequency from around ∼10 in wild type to ∼3.9. In comparison, an Atmsh4 mutant has only ∼1.6 residual chiasmata (Higgins et al, 2004). This suggests that the two classes of MMR proteins have differing roles in CO formation.
This is also supported by a difference in the effect of the mutations on prophase I progression. Mutation of both AtMSH4 and AtMLH3 results in a delay in prophase I. This is consistent with studies in budding yeast and more recently in Arabidopsis that suggest an intra-prophase surveillance system that detects recombination defects and then imposes an appropriate response that maintains normal coordination of cellular processes (Boerner et al, 2004). Importantly however, the delay in prophase I progression is significantly greater in Atmlh3 (+25 h) than in Atmsh4 (+8 h) (Higgins et al, 2004). One possible explanation is that the CO-designated double-strand breaks (DSBs) are directed to different fates in the two mutants. We propose that in Atmsh4, all interference-sensitive COs lose their CO designation such that all the DSBs, aside from the ∼15% that are processed via the second recombination pathway, are repaired before dHj formation, possibly by the synthesis-dependent strand annealing pathway (Paques and Haber, 1999). In Atmlh3, this route may not be available because the CO-designated DSBs probably progress to dHjs. Although it appears that in the absence of the MutL complex, these may still be substrates for resolution, the rate and outcome of this process is compromised. This interpretation is consistent with a recent in vitro study of the human hMSH4/hMSH5 heterodimer. This has led to the suggestion that the MutS complex forms a clamp that embraces the homologous chromosomes at the site of a progenitor dHj, thereby stabilizing the RI before its conversion to a mature dHj and subsequent resolution to form a CO (Snowden et al, 2004). Hence, in the absence of the MutS proteins, stable mature dHjs are not formed.
The same authors speculate that the role of the MutL homologues may be to promote the release of the hMSH4/hMSH5 heterodimers from the DNA. However, our data cannot readily be reconciled with this hypothesis. It appears that the localization of AtMSH4 in the Atmlh3 mutants is identical to that in the wild type. In particular, there is no evidence to suggest that the turnover of the AtMSH4 foci at late prophase is delayed, as might be expected if the AtMLH3/AtMLH1 heterocomplex was required for release of the MutS proteins from the homologous chromosomes. However, we cannot exclude the possibility that in the absence of the MutL proteins, the MutS proteins are removed from the DNA by some other mechanism.
An alternative suggestion, based on the proposed role of the prokaryotic MutL protein, is that the Mlh3/Mlh1 heterocomplex has a signal transduction role possibly between Msh4/Msh5 complexes bound at adjacent dHjs, or recruits the Hj resolvase to the RI (Hoffmann and Borts, 2004). Co-immunoprecipitation using mouse testes extracts suggests that MLH3 and MSH4 do interact, but corresponding attempts from meiotic budding yeast cultures have failed to establish evidence of an interaction (Santucci-Darmanin et al, 2002; Wang and Kung, 2002). Cytological studies in mouse support a direct interaction between the two classes of MMR complexes, but indicate that this is transient in nature (Santucci-Darmanin et al, 2000). Similarly, in Arabidopsis, colocalization of AtMSH4 and AtMLH3 at a small proportion of RIs, together with the absence of AtMLH3 foci in an Atmsh4 mutant (Higgins et al, 2004), is indicative of a direct, yet transient association. Nevertheless, studies have also shown that by late pachytene the chromosomes are devoid of AtMSH4 foci (Higgins et al, 2004), whereas the AtMLH1/AtMLH3 foci persist through to diplotene. This suggests that the role of the MLH1/MLH3 complex is not simply restricted to providing a molecular link between two MSH4/MSH5 heterodimers, as suggested.
Thus, the current balance of evidence suggests that the MMR complexes perform different, sequential roles, and that any direct interaction may be relatively transient. One possibility is that the MutS complex establishes the dHj in a CO configuration and that this is then maintained by the action of MLH3/MLH1 until CO resolution occurs. However, for the reasons outlined above, we favour a model where the role of MSH4/MSH5 may be to enable the establishment of a mature dHj, with the MutL complex then imposing a CO configuration before resolution. In addition, it may also influence the recruitment/activity of the, as yet unidentified, dHj resolvase.
CO versus non-CO resolution in Atmlh3 mutants
It now seems likely that the distribution of CO sites is determined early in the recombination pathway. Mutant analysis of the ZMM proteins, which comprise Zip1, Zip2, Zip3, Msh4/5 and Mer3, indicates that this may be as early as or before the single-end invasion stage (SEI) during early prophase I (Boerner et al, 2004). Evidence suggests the SEIs that are designated to form dHj intermediates, mostly or all, resolve as COs (Allers and Lichten, 2001; Boerner et al, 2004). Hence, it is the resolution of dHjs as COs, rather than their distribution, that is influenced by the activity of AtMLH3. It therefore follows that the numbers and distribution of the residual COs in the Atmlh3 mutant lines reflect, in part, the probability of a designated CO resolving as a CO or non-CO event. Analysis of the chiasma frequency data from the Atmlh3 mutants revealed that it was possible to fit them to both a binomial distribution and a Poisson distribution. The binomial distribution is consistent with a model where designated RIs can resolve in one of two ways (CO/non-CO). The best-fit value of p (0.35) suggests that about 35% of dHjs proceed to COs in the absence of AtMLH3, whereas when AtMLH3 is present they all form COs. As mentioned earlier, this is undoubtedly an oversimplification because of the existence of a second recombination pathway that accounts for an average of 1.6 COs per cell (Higgins et al, 2004). Nevertheless, in the absence of AtMLH3 protein, the default resolution of dHjs to COs is insufficient to ensure the obligate CO that is essential for accurate chromosome disjunction at the first meiotic division. The Poisson distribution is normally taken as indicating a random numerical distribution of events. However, in this case, even though the distribution does not differ significantly from a Poisson distribution, it is probably not completely random. From other evidence, it seems that potential CO sites are selected from a much larger number of early RIs (DSBs, etc.) through the establishment of recombination complexes, which exhibit CO interference (Boerner et al, 2004). It appears likely that this is also the case in wild-type Arabidopsis, but in the Atmlh3 mutants they can be resolved as COs (35%) or non-COs (65%), thereby generating a quasirandom distribution that nevertheless fits a Poisson distribution.
The numerical distribution of AtMLH3 and AtMSH4 foci during prophase I
One observation that remains to be resolved is the relationship between the number of AtMLH3 foci and AtMSH4 foci detected by immunocytochemistry. Previous studies have shown that at mid-leptotene there are around 80–100 AtMSH4 foci. This decreases such that by zygotene there are around 25 AtMSH4 foci associated with RIs (Higgins et al, 2004). This is more than twice the number of AtMLH3 foci, which is closely correlated with the number of chiasmata. Hence, it seems that AtMLH3 does not interact with all RIs. The mechanism whereby AtMLH3 recognizes and interacts with CO-designated RIs remains to be determined. Equally, the significance of the additional AtMSH4 foci is unclear. There appears to be a correlation between the number of AtMSH4 foci and early RIs. Nevertheless, it seems unlikely that AtMSH4 has a recombination role in the non-CO pathway, as studies in budding yeast indicate that non-CO formation is unaffected in zmm mutants at high temperature (Boerner et al, 2004). Therefore, it is conceivable that the AtMSH4 foci at non-CO sites are performing some yet to be defined structural role.
Materials and methods
Plant material and nucleic acid extraction
A. thaliana ecotype Columbia (0) was used in this study for wild-type analysis. The T-DNA insertion lines SALK_015849 (Atmlh3-1) and SALK_041465 (Atmlh3-2) were obtained from the SALK Institute via NASC for mutant analysis (Alonso, 2003). Plants were grown, material harvested and nucleic acid extractions were performed as previously described by Higgins et al (2004).
Semiquantitative RT–PCR for AtMLH3 and AtMLH1 transcript expression
RT–PCR was carried out as previously described (Higgins et al, 2004). The gene-specific primers for AtMLH3 were 5′-TGTGCACTCAGAGACCCAAGATG-3′ (forward) and 5′-ACACCCAGAATGCATGGAACCGC-3′ (reverse). For AtMLH1, the primers were 5′-CCAGGTATGCTGGAGACTGTAAG (forward) and 5′-GAACTGAATACCGTCACCCGATG (reverse). The primers for the GAPD control were 5′-CTTGAAGGGTGGTGCCAAGAAGG-3′ (forward) and 5′-CCTGTTGTCGCCAACGAAGTCAG-3′ (reverse).
T-DNA insertion site mapping
The T-DNA insertion site of SALK_015849 was mapped with primers LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′ and Atmlh3-1 T-DNA 5′-AACTCTTTGAGTGGTGAACATTTGG-3′. The T-DNA insertion site of SALK_041465 was mapped with LBa1 and Atmlh3-2 T-DNA 5′-TGACATCCAACTCATCTTTCAATCC-3′. The PCR products were cloned into pDrive (Qiagen) and sequenced. Pairs of primers were used to determine if the plants were homozygous or heterozygous for the T-DNA insertion. Primers Atmlh3-1 T-DNA and Atmlh3-1WT 5′-TGTGCACTCAGAGACCCAAGATG-3′ were used to amplify the wild-type genomic region of SALK_015849 and primers Atmlh3-2 T-DNA and Atmlh3-2WT 5′-TTGAGTGTGCGGATGACTGG-3′ for SALK_041465.
Construction of the AtMLH3 complementation plasmid
An XbaI (11 242 bp) fragment of BACF8D20.11 (between sites 4729 and 15 971) encoding the entire coding sequence of AtMLH3 was cloned into the binary vector pCAMBIA1302 (http://www.cambia.org) for Agrobacterium-mediated transformation of mutant line Atmlh3-1.
Plant transformation
The binary plasmid construct was introduced into Agrobacterium tumefaciens (GV3101 pMP90) (Koncz and Schell, 1986) and plants transformed as previously described (Higgins et al, 2004). Transformed plants were selected using hygromycin selection (20 μg/ml).
Antibody production
A 1041 bp fragment encoding a 347 aa region comprising residues 804–1151 of AtMLH3 was amplified from Arabidopsis bud cDNA. An NdeI site was designed into primer AtMLH3expF 5′-CGCATATGCATCTTAAGTGCAGCATCC-3′ and an XhoI site into primer AtMLH3expR 5′-GGCTCGAGACTTTTAGCGTTGTCTAAGCGTG-3′. The PCR fragment was digested with NdeI/XhoI and ligated into the expression vector pET21b (Novagen). The construct was transferred to E. coli BL21 (Novagen) cells and purified refolded recombinant protein prepared as described previously (Kakeda et al, 1998). Rabbit and rat polyclonal antisera were produced against the recombinant protein (ISL, Paignton, UK).
Nucleic acid sequencing
Nucleotide sequencing was carried out by the genomics laboratory, Biosciences, University of Birmingham, UK.
Cytological procedures
The cytological methods were carried out as previously described (Higgins et al, 2004), except that the images presented in the figures are Z-stacks. The following antibodies were used in the study: anti-MLH3 (rabbit/rat) anti-ZYP1 (rabbit/rat), anti-AtRAD51 (rabbit), anti-ASY1 (rabbit/rat), anti-MSH4 (rabbit) and anti-AtMLH1 (rabbit) (Mercier et al, 2003; Higgins et al, 2004, 2005). FISH on metaphase I chromosomes was carried using the 45S rDNA and 5S rDNA probes. To identify single insert lines of Atmlh3-1, FISH was carried out with a T-DNA probe from pROK2.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Acknowledgments
We thank Dr Anthony Caryl and Dr James Higgins for useful discussions during the course of this work. We also thank undergraduate students Orestis Laszos, Cheng Fang, Darren Arbon and Tom Butts for their contribution to the initial analysis of AtMLH3. We are also grateful to Stephen Price, the horticulture staff and the Biosciences genomics unit for technical support. Finally, we thank the reviewers for their insight, which has enabled us to significantly improve the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council, UK.
References
- Ade J, Belzile F, Philippe H, Doutriaux MP (1999) Four mismatch repair paralogues coexist in Arabidopsis thaliana: AtMSH2, AtMSH3, AtMSH6-1 and AtMSH6-2. Mol Gen Genet 262: 239–249 [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Alonso JM (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana (vol 301, pg 653, 2003). Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alou AH, Jean M, Diomingue FJ, Belzile F (2004) Structure and expression of AtPMS1, the Arabidopsis ortholog of the yeast DNA repair gene PMS1. Plant Sci 167: 447–456 [Google Scholar]
- Argueso JL, Kijas AW, Sarin S, Heck J, Waase M, Alani E (2003) Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol Cell Biol 23: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FCH (2002) Asy1, a protein required for melotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FCH, Jones GH (2003) A meiotic time-course for Arabidopsis thaliana. Sexual Plant Reprod 16: 141–149 [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13: 336–342 [DOI] [PubMed] [Google Scholar]
- Boerner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Caryl AP, Jones GH, Franklin FCH (2003) Dissecting plant meiosis using Arabidopsis thaliana mutants. J Exp Botany 54: 25–38 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Hays JB (2000) Arabidopsis MutS homologs—AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7—form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell 12: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H, Kolodner RD (1998) The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA 95: 12404–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 13: 867–876 [DOI] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann ER, Borts RH (2004) Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res 107: 232–248 [DOI] [PubMed] [Google Scholar]
- Hunter N, Borts RH (1997) Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev 11: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Jean M, Pelletier J, Hilpert M, Belzile F, Kunze R (1999) Isolation and characterization of AtMLH1, a MutL homologue from Arabidopsis thaliana. Mol Gen Genet 262: 633–642 [DOI] [PubMed] [Google Scholar]
- Kakeda K, Jordan ND, Conner A, Ride JP, Franklin-Tong VE, Franklin FCH (1998) Identification of residues in a hydrophilic loop of the Papaver rhoeas S protein that play a crucial role in recognition of incompatible pollen. Plant Cell 10: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Cohen PE (2004) Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet Genome Res 107: 216–231 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT (1999) Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9: 89–96 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, Schwartzberg P, Collins FS, Cohen PE (2002) Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31: 385–390 [DOI] [PubMed] [Google Scholar]
- Marcon E, Moens P (2003) MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FCH (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B (2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci 115: 1611–1622 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH (2002) Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin S, Neyton S, Lespinasse F, Saunieres A, Gaudray P, Paquis-Flucklinger V (2002) The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet 11: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V (2000) MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J 14: 1539–1547 [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Svetlanov A, Cohen PE (2004) Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp Cell Res 296: 71–79 [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A (2000) The nucleolus: the magician's hat for cell cycle tricks (vol 12, pg 372, 2000). Curr Opin Cell Biol 12: 372–377 [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N (1999) Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kung WM (2002) Supercomplex formation between Mlh1–Mlh3 and Sgs1–Top3 heterocomplexes in meiotic yeast cells. Biochem Biophys Res Commun 296: 949–953 [DOI] [PubMed] [Google Scholar]
- Woods LM, Hodges CA, Baart E, Baker SM, Liskay M, Hunt PA (1999) Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J Cell Biol 145: 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5


