Abstract
Objective
To establish whether the prevalence of positive reverse transcriptase–polymerase chain reaction (RT-PCR) results decreased during the first 3 months after colorectal cancer excision, and to assess whether persistence of RT-PCR positivity after primary colorectal cancer excision was related to tumor stage or locally advanced and metastatic disease.
Methods
Systemic venous blood was collected from patients with colorectal cancer before and at intervals up to 12 weeks after surgery. RNA was extracted from the mononuclear cell fraction of the blood samples and subjected to RT-PCR using specific primers for carcinoembryonic antigen mRNA and cytokeratin-20 mRNA. Healthy individuals with no history of cancer were used as controls.
Results
The results of RT-PCR were positive in 81 of 116 patients with colorectal cancer before surgery, with no significant differences in preoperative prevalence by Dukes stage or presence of locally advanced or metastatic disease. There was a significant decrease in the prevalence of RT-PCR positivity at 24 hours after surgery compared with before surgery. On subgroup analysis by Dukes stage, only the decrease in Dukes A and B patients reached significance. Seven of the 143 controls were RT-PCR positive.
Conclusions
Circulating tumor cells were present before treatment in most patients with colorectal cancer regardless of tumor stage or metastases. Clearance of circulating tumor cells within 24 hours of colorectal cancer excision was greatest in tumors with the best prognosis.
Experimental in vivo studies suggest that circulating tumor cells (CTCs) can be detected early in the natural history of solid tumor growth, before the development of metastases. 1 Hematogenous metastasis is thought to be an inefficient process where fewer than 0.01% of CTCs establish metastases, 2 and most CTCs are cleared from the circulation within 24 hours. 3 Thus, if CTCs appear early in the presence of an invasive solid tumor and disappear rapidly after complete solid tumor removal, they should be detectable before and disappear soon after curative excision in patients with colorectal carcinoma.
Detection of CTCs in colorectal cancer was first reported more than 40 years ago. 4 Use of the reverse transcriptase–polymerase chain reaction (RT-PCR) to identify mRNA coding for tumor-associated proteins has increased the sensitivity of detecting tumor cells spiked into normal blood to one colorectal cancer cell in a million white blood cells. 5,6 Despite this, studies based on a single preresection peripheral blood sample have not shown that RT-PCR status is significantly associated with prognosis. 7,8 Using multiple blood sampling with two RT-PCR markers to increase the probability of CTC identification before primary tumor removal, positive RT-PCR results have been found in most (>70%) patients with colorectal carcinoma, with little variation in the prevalence of positivity by tumor stage. 9 Previous clinical studies relying on one RT-PCR marker within a single blood sample that have assessed changes in RT-PCR prevalence with colorectal cancer excision have not shown a consistent pattern of change after primary tumor removal. 10–13
There were two aims to the present study. First, we sought to establish whether the prevalence of RT-PCR positives decreased during the first 3 months after colorectal cancer excision, using triple blood sampling and two RT-PCR markers. Second, because the probability of cure after excision of colorectal cancer is affected by primary tumor stage and the presence of metastases, 14 we sought to assess whether persistence of RT-PCR positivity after primary colorectal cancer excision was related to primary tumor Dukes stage or metastatic disease.
PATIENTS AND METHODS
Patients
Patients with colorectal cancer undergoing treatment in two hospitals between November 1997 and September 2000 were studied. The diagnosis of primary colorectal carcinoma was confirmed in all patients by endoscopic biopsy, and the primary tumor stage was confirmed by histologic examination of the resected primary tumor. All patients with colorectal liver metastases had histologic evidence of primary colorectal carcinoma, computed tomography (CT) scan evidence of liver metastases, and a subsequent history of liver metastasis growth on serial CT scans associated with a progressive increase in the serum level of carcinoembryonic antigen (CEA). Healthy patients with no history of cancer who were undergoing intermediate surgical procedures such as herniorrhaphy or hemorrhoidectomy were recruited as controls.
The protocol was approved by the Chelsea and Westminster Hospital Research Ethics Committee, and written informed consent to be included was obtained from all patients in the study.
Blood Sampling
Blood was sampled within 24 hours before and at 24 hours, 1 week, 6 weeks, and 12 weeks after laparotomy for primary colorectal cancer excision. To reduce the false-positive risk from venesection needle-cored epithelial cells entering the venesection needle lumen, 15 an intravenous cannula was inserted and 5 mL blood was aspirated before sample collection. Three 14-mL samples of systemic venous blood were then collected at 1-minute intervals from each patient into 7-mL Vacutainers (two 7-mL aliquots per 14-mL sample) containing sodium EDTA.
Three blood samples were taken via a cannula (as above) in every fourth control subject before surgery. In all remaining control subjects, a single blood sample was taken via a cannula. The first blood sample result in control subjects undergoing triple sampling was used with single blood sample results when deriving control subject positivity for a single blood sample. Control subjects were recruited regularly throughout the course of the study to ensure that control results did not vary during the course of the study.
RNA Extraction From Blood
Blood samples were processed within 15 minutes of venesection to prevent RNA degradation. The two 7-mL Vacutainer aliquots were combined and made up one sample. Each was centrifuged at 1,400 rpm (200 g) for 10 minutes and the resulting supernatant containing plasma was removed. This volume was replaced by phosphate-buffered saline (PBS). Cell fractionation was achieved by layering the resulting suspension onto a density gradient separation medium (Histopaque-1077, Sigma Diagnostics, St. Louis, MO) using Accuspin tubes (Sigma Diagnostics) followed by centrifugation at 1,400 rpm (200 g) for 30 minutes. Mononuclear cells, including tumor cells, contained in the upper chamber were then harvested by centrifugation at 1,400 rpm (200 g) for 15 minutes at 4°C, and the supernatant was discarded. Total RNA was extracted from the residual cell pellets by a modified method of Chomczynski and Sacchi, 16 using RNAzol Stat-50 (Biogenesis, Bournemouth, UK), which is optimized to avoid any DNA contamination during the extraction. The extracted RNA was washed in diethyl pyrocarbonate (DEPC)-75% ethanol before being dissolved in DEPC-water and stored at −70°C until further analysis. The RNA yield was maximized by the addition of 10 μL polyinosinic acid (an RNA carrier) solution (16 g/L) to each sample. All glassware was rinsed in DEPC-water and autoclaved, and solutions were made up in DEPC-water.
Reverse Transcriptase–Polymerase Chain Reaction
Two micrograms of total RNA was used for reverse transcription to cDNA in a final volume of 40 μL made up from 400 units Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL, Paisley, UK), 40 units RNAse inhibitor (RNAsine, Promega, Madison, WI), 8 μL reaction buffer (250 mmol/L Tris-HCl, pH 8.3, 375 mmol/L KCl, and 15 mmol/L MgCl2), and a 2-μL mix of 10 mmol/L of each dNTP (Promega, Southampton, UK). One hundred picomoles of random hexamers (Perkin-Elmer, Foster City, CA) was added, and primer annealing was performed by incubation for 2 minutes at 70°C followed by rapid quenching on ice for 5 minutes. Reverse transcription was performed by incubating the samples for 1 hour at 42°C, and then 95°C for 5 minutes to inactivate the reverse transcriptase. The final volume of the cDNA reaction was then diluted by adding 40 μL DEPC-water. Ten microliters of this cDNA solution was then used for the final PCR reaction after making up to 50 μL by adding 5 μh PCR buffer (10× Tris-HCl, KCl, (NH4)2SO4, 15 mmol/L MgCl2 [pH 8.7] [20°C] [Qiagen, Crawley, UK]), 2 mmol/L MgCl2 (Qiagen), 250 μmol/L dNTP (Promega, UK), 2.5 units Taq Normal DNA Polymerase (Qiagen), and 0.5 μmol/L of both sense and antisense primers (MWG-Biotech, Milton Keynes, UK), and DEPC = water.
Primer Sequences
To amplify fragments of CEA cDNA 5,6 and cytokeratin-20 cDNA, 5,6 we designed the following specific primers to avoid amplification of any other related gene members. They were also designed to bridge intronic sequences to prevent the risk of genomic DNA yielding a product of the same size. CEA primer sequences were: sense primer 5′ CCATGGGAGTCTCCCTCG 3′; antisense primer 5′ GTAGCTTGCTGTGTCAT TTC 3′. These CEA primer sequences in the presence of cDNA CEA amplify a 641-bp fragment (Fig. 1). Cytokeratin-20 primer sequences were: sense primer 5′CAGACACACGGTGAACTATGG 3′; antisense primer 5′GATCAGCTTCCACTGTTAGACG 3′. These cytokeratin-20 primer sequences in the presence of cDNA CEA amplify a 370-bp fragment.
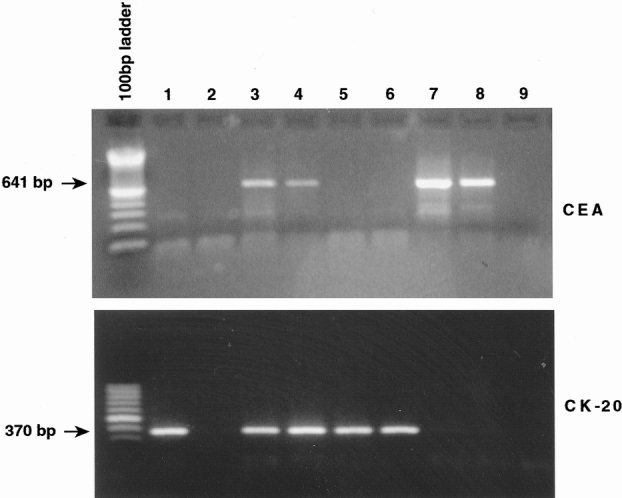
Figure 1. Reverse transcriptase–polymerase chain reaction identified 641- and 370-bp fragments indicative of cDNA coding for carcinoembryonic antigen (CEA) and cytokeratin-20 cDNAs, respectively. Electrophoretic gel (top): Lanes 1 to 6 and lane 8, blood from seven patients with colorectal cancer showing CEA cDNA positive results for patients 3, 4, and 8; lane 7, CEA cDNA positive control (HT115 colorectal cell line); lane 9, CEA cDNA negative control (H2O plus reagents). Electrophoretic gel (bottom): Lanes 1 to 3 and lanes 5 to 8, blood from seven patients with colorectal cancer showing cytokeratin-20 cDNA positive results for patients 1, 3, 5, and 6; lane 4, cytokeratin-20 cDNA positive control (HT115 colorectal cell line); lane 9, cytokeratin-20 cDNA negative control (H2O plus reagents).
Polymerase Chain Reaction
Polymerase chain reactions were performed using a thermocycler Primus G Plus (MWG-Biotech). The conditions for both CEA cDNA PCR amplification and cytokeratin-20 cDNA PCR amplification were 94°C for 10 minutes. This single cycle was followed by 29 cycles at 94°C for 30 seconds, 60°C for 1 minute, and 74°C for 1 minute. The final elongation step was 72°C for 10 minutes.
Identification of Polymerase Chain Reaction Products
Polymerase chain reaction products were separated by electrophoresis on a 1.5% agarose gel stained with ethidium bromide to enable DNA visualization under UV light. Samples that had not been reverse transcribed did not give rise to product. The identity of the two products had previously been confirmed by Southern blotting 6 and direct sequencing. 17
When samples were processed for PCR, a positive control produced from either the HT29 or HT115 colorectal cancer cell line and a negative control where cDNA was substituted by nuclease free water were included. The quality of the cDNA was assessed using primers for the ubiquitous housekeeping gene β-microglobulin (data not shown). The RT-PCR method was standardized by loading equal volumes (20 μL) of PCR product onto agarose gels to ensure that the intensity of the bands reflected equivalent amounts of detectable product. To assess RNA integrity and quality, formaldehyde RNA gels were run at differing intervals on randomly selected samples. The operator was unaware of the source of each sample while performing the RT-PCR.
Definition of Positive Result
A patient with colorectal cancer was regarded as RT-PCR positive for that time point if any of the three blood samples taken at that time point was positive for either CEA or cytokeratin-20 mRNA. A control subject was regarded as having a positive test if any blood sample was positive for either CEA or cytokeratin-20 mRNA.
Statistical Analysis
Comparisons of the prevalence of tumor cell positivity between groups were by contingency table analysis using the chi-square distribution. Comparisons of CTC prevalence within a group at different time points were by binomial test.
RESULTS
Patients
One hundred sixteen patients with colorectal cancer (71 men, 45 women; mean age 68 ± 10 years) were studied. Eighty-six patients underwent primary tumor excision with curative intent (primary tumor stage: Dukes A, 17; B, 48; C, 21), and 30 patients underwent palliative primary tumor resection or bypass because of metastatic (liver metastases, 26 patients) or locally unresectable disease (4 patients). One hundred forty-three control subjects (75 men, 68 women; mean age 39 ± 16 years) were studied.
Prevalence of Polymerase Chain Reaction Positivity
The preoperative prevalence of RT-PCR positivity in the patients with colorectal cancer (81/116 patients [69%]) was significantly (P < 10−6) greater than in the control subjects undergoing triple blood sampling (4/36 patients [11%]). Seven of 143 (4.9%) control subjects were RT-PCR positive (CEA mRNA, 3 patients; cytokeratin-20, 4 patients) on a single preoperative blood sample (RT-PCR numbers 186, 532, 544, 958, 1,287, 1,804, and 2,647 in the total series of 3,310 cancer and control RT-PCRs).
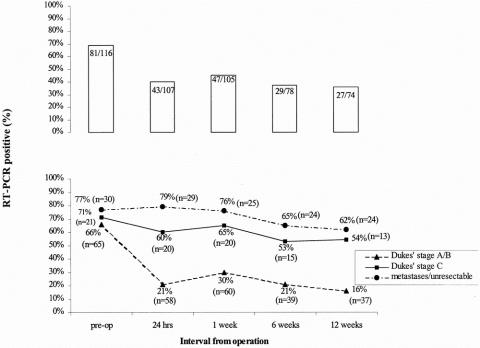
The prevalence of RT-PCR positivity 24 hours to 12 weeks after surgery in the colorectal cancer group is shown in Figure 2. There was a significant decrease (P < 10−4) in the prevalence of RT-PCR positivity 24 hours after surgery compared with before surgery. Sixty-one percent of patients who were RT-PCR positive before surgery had a negative result by 24 hours after surgery; in contrast, 14.8% of patients who were RT-PCR negative before surgery developed a positive result by 24 hours after surgery (Table 1).
Figure 2. Top: Prevalence of a reverse transcriptase–polymerase chain reaction (RT-PCR) positive test (%) in patients with colorectal cancer before and at intervals after surgery (number of positive patients and total patients studied in each group are shown within each bar). There was a significant decrease (P = 10−4) in the prevalence of RT-PCR positivity result at 24 hours after surgery, but no additional change at up to 12 weeks after surgery. Bottom: Prevalence of an RT-PCR positive test (%) in patients with colorectal cancer before and at intervals after surgery. A significant decrease compared with the preoperative RT-PCR prevalence was seen at 24 hours after “curative” resection of node-negative tumors (Dukes A and B) but not node-positive ones (Dukes C). Numbers in parentheses represent number of patients available for blood sampling at each time point.
Table 1. POLYMERASE CHAIN REACTION POSITIVITY BEFORE AND AFTER SURGERY
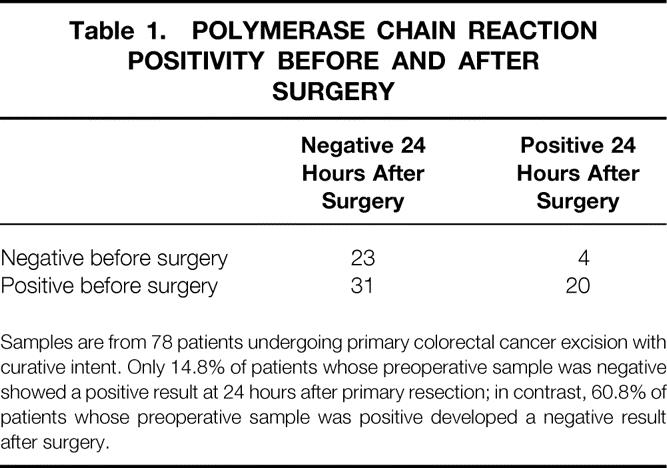
Samples are from 78 patients undergoing primary colorectal cancer excision with curative intent. Only 14.8% of patients whose preoperative sample was negative showed a positive result at 24 hours after primary resection; in contrast, 60.8% of patients whose preoperative sample was positive developed a negative result after surgery.
There were no significant differences between the prevalence of positivity in samples at 24 hours after surgery and subsequently. Assuming the random chance of a subsequent negative result was 54/78 (RT-PCR negative at 24 hours/all RT-PCR results at 24 hours), then the probability of tests at further time points also being negative would be 0.33 (negative at three subsequent time points), 0.44 (negative at two time points), 0.20 (negative at one time point), and 0.03 (negative at no subsequent time point). Of the 51 patients with colorectal cancer who were RT-PCR negative at 24 hours after primary tumor excision and in whom complete follow-up was available, 43 were negative at three subsequent time points (P < 10−4 compared with random), 5 were negative at two (P < 10−4), and 3 were negative at one (P = .001), whereas no patient who was RT-PCR negative at 24 hours was RT-PCR positive at all subsequent time points. Therefore, the association between a negative RT-PCR result at 24 hours after primary tumor excision and subsequent negative RT-PCR results at up to 12 weeks from primary tumor excision was unlikely to be random. Similarly, significantly different associations from random were also found for RT-PCR positivity after primary tumor excision with curative intent, and for RT-PCR positive and negative analyses that included patients known to have incurable disease.
There was no significant difference in the prevalence of preoperative RT-PCR positivity by tumor stage or presence of metastases (Dukes A, 12/17 patients; B, 31/48; C, 15/21; metastases/unresectable 23/30). For patients undergoing primary tumor resection with curative intent (patients with metastases or locally unresectable disease excluded), there was a significant reduction in the prevalence of RT-PCR positivity after surgery in those with node-negative primary tumors (Dukes A/B, P < 10−4) but not node-positive primary tumors (Dukes C, P = .659; see Fig. 2). There was no significant decrease in the prevalence of RT-PCR positivity after laparotomy among patients with metastatic or unresectable disease.
The prevalence of RT-PCR positivity at 24 hours after resection with curative intent was significantly lower (P = .0027) in patients with node-negative primary tumors (Dukes A/B) compared with node-positive (Dukes C) primary tumors. This significant difference by node status in patients undergoing resection with curative intent persisted in RT-PCR analyses at subsequent intervals after surgery (1 week, P = .012; 6 weeks, P = .042; 12 weeks, P = .02).
No significant differences were found in the prevalence of RT-PCR positivity between different times after surgery, for any tumor stage after resection with curative intent, and for patients with metastases or locally unresectable disease.
DISCUSSION
Although the validity of the present RT-PCR methods has been confirmed by in vitro studies 5,6 in which blood samples from healthy individuals without cancer were spiked with colorectal cancer cells, confirmatory results in patients with cancer using tests for CTCs based on other methods 18,19 are needed. We used healthy young (mean age 39 years) individuals undergoing intermediate surgery as controls throughout the study, and it is unlikely that RT-PCR-positive results in this age group were produced by occult carcinomas. It is thought that false-positive results can arise from needle coring of epithelial cells, expressing epithelial antigens such as CEA or cytokeratin-20 into the blood, 15 and it has also been reported that cells expressing cDNA for CEA can be identified in the blood of patients without cancer who have inflammatory conditions such as colitis. 13,20 We identified a positive rate of approximately 5% per single blood sample and 11% per patient for triple blood sampling for CEA and cytokeratin-20 combined. The prevalence of these noncancer-related positive results in control subjects was of roughly sixfold less magnitude than that observed before primary tumor resection in patients with colorectal cancer. Thus, it seems likely that 85% to 90% of the positive results in our patients with colorectal cancer were produced by mRNA from CTCs.
As previously reported, 9 we found a high prevalence of RT-PCR positivity in patients with untreated colorectal cancer that was not significantly influenced by tumor stage. It is unlikely that the 69% of patients with colorectal cancer (including 71% of patients with Dukes A cancers) found to be RT-PCR positive before surgery were all destined to develop recurrent disease. This is consistent with experimental 1 and clinical 8 studies indicating that CTC positivity in association with colorectal cancer does not imply that metastasis has occurred.
The prevalence of RT-PCR positivity was almost halved at 24 hours after colorectal cancer surgery. It is unlikely that this was an effect of the anesthetic, surgical manipulation, or blood transfusion because a similar statistically significant reduction was not observed after operations where metastatic or locally unresectable tumor was present. It was also unlikely that RT-PCR positivity at 24 hours after surgical excision could be explained by patient variations in the rate of CTC clearance after complete tumor excision, because the prevalence of RT-PCR positivity at 24 hours after surgery did not significantly decrease in subsequent assessments over 12 weeks. The results were consistent with the experimental observation 3 that clearance of tumor cells from the circulation occurs within 24 hours of tumor removal.
A positive RT-PCR test after colorectal cancer excision might be a random occurrence arising from a needle-cored epithelial cell or a blood sample taken from a patient with inflammation. The finding of a significantly different association from random between either negative or positive RT-PCR results at 24 hours after surgery with subsequent results up to 12 weeks after surgery suggests that RT-PCR results at different time points after surgery were causally associated. The association of a positive postoperative RT-PCR result with persisting disease is supported by the finding of more postoperative RT-PCR positivity in patients known to have unresectable or metastatic disease than in those undergoing apparently curative resections (see Fig. 2). In addition, no significant reduction in RT-PCR positivity was detected after apparently complete tumor excision among patients with lymph node-positive tumors (Dukes C) in whom there was a high probability of recurrence, compared with the significant reduction detected in node-negative tumors (Dukes A or B), where the risk of recurrence was lower. These findings suggest that RT-PCR after primary tumor excision may be capable of identifying patients destined to develop recurrence after apparently curative removal of colorectal cancer.
The results also indicate that use of the present RT-PCR technique to predict recurrence of colorectal cancer is likely to be associated with errors. These include a false-positive rate of 10% to 15%, because the noncancer-related sources of positivity observed in our control subjects would be expected to occur also in patients with colorectal cancer. In addition, the false-negative rate was 25% or more: roughly 25% of patients known to have metastatic or unresectable colorectal cancer had a negative RT-PCR test (see Fig. 2). A longitudinal study assessing the accuracy of RT-PCR positivity after colorectal cancer excision in predicting treatment outcome is now in progress.
In summary, CTCs ceased to be detectable in 30% of patients with colorectal cancer within 24 hours of apparently complete tumor excision, and the persistence of cancer cells in the circulation was more likely when tumor excision was incomplete or associated with adverse tumor stage. Although the present RT-PCR techniques are unlikely to be 100% accurate in predicting an adverse treatment outcome, RT-PCR of blood taken at 24 hours after primary tumor excision may provide prognostic information after resection of colorectal cancer.
Footnotes
H.P. and R.Q.W. were Stefan Galeski Research Fellows. N.LeM., Z.A.J.K., and C.G. were supported by Colon Cancer Concern, London, UK. R.A. was supported by the E. B. Moller Charitable Trust, London, U.K.
Correspondence: Timothy G. Allen-Mersh, MD, FRCS, Department of Surgery, Imperial College School of Medicine, Chelsea and Westminster Hospital, 369 Fulham Road, London SW10 9NH, UK.
E-mail: t.allenmersh@ic.ac.uk
Accepted for publication June 15, 2001.
References
- 1.Glaves D. Correlation between circulating cancer cells and incidence of metastases. Br J Cancer 1983; 48: 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur J Cancer 1973; 9: 223–227. [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ. Metastasis. Quantitative analysis of distribution and fate of tumor emboli labelled with 125 I-5-iodo-2′-deoxyuridine. J Natl Cancer Inst 1970; 45: 773–782. [PubMed] [Google Scholar]
- 4.Engell H. Cancer cells circulating in circulating blood. Acta Chir Scand 1955; 201. [PubMed] [Google Scholar]
- 5.Burchill SA, Bradbury MF, Pittman K, et al. Detection of epithelial cancer cells in peripheral blood by reverse transcriptase-polymerase chain reaction. Br J Cancer 1995; 71: 278–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonas S, Windeatt S, O-Boateng A, et al. Identification of carcinoembryonic antigen-producing cells circulating in the blood of patients with colorectal carcinoma by reverse transcriptase polymerase chain reaction. Gut 1996; 39: 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Takagi Y, Aoki S, et al. Significant detection of circulating cancer cells in the blood by reverse transcriptase–polymerase chain reaction during colorectal cancer resection. Ann Surg 2000; 232: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardingham JE, Kotasek D, Sage RE, et al. Detection of circulating tumor cells in colorectal cancer by immunobead: PCR is a sensitive prognostic marker for relapse of disease. Mol Med 1995; 1: 789–794. [PMC free article] [PubMed] [Google Scholar]
- 9.Wharton RQ, Jonas SK, Glover C, et al. Increased detection of circulating tumor cells in the blood of colorectal carcinoma patients using two reverse transcription-PCR assays and multiple blood samples. Clin Cancer Res 1999; 5: 4158–4163. [PubMed] [Google Scholar]
- 10.Weitz J, Kienle P, Lacroix J, et al. Dissemination of tumor cells in patients undergoing surgery for colorectal cancer. Clin Cancer Res 1998; 4: 343–348. [PubMed] [Google Scholar]
- 11.Mori M, Mimori K, Ueo H, et al. Molecular detection of circulating solid carcinoma cells in the peripheral blood: the concept of early systemic disease. Int J Cancer 1996; 68: 739–743. [DOI] [PubMed] [Google Scholar]
- 12.Funaki NO, Tanaka J, Sugiyama T, et al. Perioperative quantitative analysis of cytokeratin 20 mRNA in peripheral venous blood of patients with colorectal adenocarcinoma. Oncol Rep 2000; 7: 271–276. [DOI] [PubMed] [Google Scholar]
- 13.Castells A, Boix L, Bessa X, et al. Detection of colonic cells in peripheral blood of colorectal cancer patients by means of reverse transcriptase and polymerase chain reaction. Br J Cancer 1998; 78: 1368–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dukes C, Bussey H. The spread of rectal cancer and its effect on prognosis. Br J Surg 1958; 12: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wharton R, Patel H, Jonas S, et al. Venesection needle coring increases positive results with RT-PCR for detection of circulating cells expressing CEA mRNA. Clin Exp Metastasis 2000; 18: 291–294. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156–159. [DOI] [PubMed] [Google Scholar]
- 17.Wyld DK, Selby P, Perren TJ, et al. Detection of colorectal cancer cells in peripheral blood by reverse transcriptase polymerase chain reaction for cytokeratin 20. Int J Cancer 1998; 79: 288–293. [DOI] [PubMed] [Google Scholar]
- 18.Rehse MA, Corpuz S, Heimfeld S, et al. Use of fluorescence threshold triggering and high-speed flow cytometry for rare event detection. Cytometry 1995; 22: 317–322. [DOI] [PubMed] [Google Scholar]
- 19.Racila E, Euhus D, Weiss AJ, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA 1998; 95: 4589–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardingham JE, Hewett PJ, Sage RE, et al. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int J Cancer 2000; 89: 8–13. [PubMed] [Google Scholar]