Abstract
Objective
To evaluate the role of human macrophage metalloelastase (HME) in pancreatic cancer.
Summary Background Data
HME, a member of the human matrix metalloproteinase family, possesses elastolytic activity and is critical for the degradation of extracellular matrix proteins. Inasmuch as tumor invasion and metastasis formation require lysis of extracellular matrix, HME plays a critical role in both processes.
Methods
HME expression was analyzed by Northern blot analysis, reverse transcriptase–polymerase chain reaction, Western blot analysis, and immunohistochemistry in 39 pancreatic cancer tissues and 13 normal controls. The molecular data were related to clinicopathologic parameters and patient survival.
Results
In human pancreatic cancer, overexpression of HME mRNA was present in 25 of 39 pancreatic cancer tissues (64%) and in five pancreatic cancer cell lines. In contrast, low levels of HME mRNA expression were present in 13 normal pancreatic tissues samples. By Western blot analysis, high levels of HME were found in pancreatic cancer tissues and in the pancreatic cancer cell lines compared with the normal controls. Fifty-six percent of the cancer samples exhibited HME immunoreactivity in the cancer cells, and 63% in the stromal cells. Analysis of the survival data revealed that patients whose tumors exhibited HME mRNA overexpression lived significantly shorter compared with patients whose tumors did not overexpress HME. No relationship between HME expression and tumor stage, tumor grading, or presence of lymph node metastases was found.
Conclusions
These findings indicate that HME participates in pancreatic cancer progression and that its presence worsens the prognosis. These data suggest a benefit of its inhibition in the treatment of pancreatic cancer.
Matrix metalloproteinases (MMPs) make up a family of zinc-dependent endoproteinases that degrade extracellular matrix (ECM), a relatively stable structural material that lies under epithelia and surrounds connective tissue cells and is composed of collagen type I–V, elastin, proteoglycans, and basement membranes. 1–3 The balance between MMPs and their endogenous inhibitors (tissue inhibitors of MMP [TIMP]) is an important part of complex regulated ECM interactions in angiogenesis as well as in tissue remodeling. Human macrophage metalloelastase (HME, synonym: matrix metalloproteinase-12) is a member of the MMP family and was initially identified in human alveolar macrophages of cigarette smokers. 4 HME degrades elastin and a broad selection of matrix and nonmatrix substrates, including laminin-1, fibronectin, entactin, collagen type IV, heparan, and chondroitin sulfates. 5,6 Because of its great ability to degrade ECM, it is physiologically synthesized and released by macrophages to penetrate basement membranes and to invade normal and diseased tissue. 7–11 Up to this time, HME mRNA has been detected in normal human tissues only in macrophages, placenta, 12 and in low levels in the normal aorta. 13 The expression of HME in vivo in benign diseased tissues has so far been shown in abdominal aortic aneurysm, 13 intestinal ulceration, 14 cutaneous granulomas, 15 and synovial-like interface tissue between bone and prostheses. 16 However, HME is also expressed in malignant cells such as skin cancer, 17 astrocytomas, glioblastomas, 18 and hepatocellular cancer, in which its expression is associated with hypovascularization. 19,20 This might be caused by the ability of HME to cleave plasminogen to angiostatin and other kringle products, leading to the inhibition of angiogenesis in these tumors. 21,22 In comparison with other MMPs, HME is the most efficient angiostatin-producing MMP. 22,23 On the other hand, HME production and HME activity are closely related to the presence of serine proteinases. The serine proteinases plasminogen and thrombin can regulate HME activity through distinct mechanisms: posttranslational secretion of preformed HME protein, induction of protein secretion, and extracellular enzymatic activation of the HME proenzyme to its active form. 24,25 Tumor necrosis factor-alpha (TNF-α) and interleukin-1 (IL-1) are activated similar to HME through the protein kinase C pathway, and there is increasing evidence that TNF-α is partly activated through several MMPs. 26
Using DNA chip technology (Affymetrix, Santa Clara, CA), we simultaneously analyzed more than 5,600 human genes in pancreatic cancer, chronic pancreatitis, and the normal pancreas (unpublished data). HME was identified as a gene that is strongly upregulated in many human pancreatic cancers but not in chronic pancreatitis and in the normal pancreas. Therefore, to further investigate HME expression and its influence on the prognosis of patients with pancreatic cancer, Northern blot analysis, reverse transcription–polymerase chain reaction (RT-PCR) analysis, Western blot analysis, and immunohistochemistry were applied and the molecular data were related to clinical data.
METHODS
Patients and Tissue Collection
Normal human pancreatic tissue samples were obtained through an organ donor program from 13 persons (4 women, 9 men) individuals who were free of any pancreatic disease. The median age of the organ donors was 45 years (range 33–63). Pancreatic cancer tissue samples were obtained from 39 patients (19 women, 20 men) undergoing pancreatic resection. In all patients with cancer, the resection margins were tumor-free. The median age of the patients with pancreatic cancer was 67 years (range 49–79). According to the international classification of the UICC, there were 8 stage I, 7 stage II, 23 stage III, and 1 stage IV tumors. Tumor grading was well differentiated in 7 cases, moderately differentiated in 25 cases, and undifferentiated in 7 cases. As a second control group, chronic pancreatitis tissue samples were obtained from 16 patients (4 women, 12 men) undergoing a pancreatic head resection. The median age of the patients with chronic pancreatitis was 46 years (range 33–57).
Freshly removed tissue samples were cut in the operating room on surgical removal and randomly divided for histologic analysis (immediately fixed in paraformaldehyde solution for 12–24 hours and paraffin-embedded for immunohistochemistry) or were snap-frozen in liquid nitrogen and maintained at −80°C until use for RNA and protein extraction. Studies were approved by the Human Subjects Committee of the University of Bern.
Cell Culture Experiments
Human pancreatic cancer cells were routinely grown in DMEM (PANC-1, MIA-PaCa-2) or RPMI (T3M4, ASPC-1, and CAPAN-1) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained at 37°C in a humid chamber with 5% CO2 and 95% air atmosphere.
Probe Synthesis for Northern Blot Analysis
For Northern blot analysis, a 346-bp fragment of human HME cDNA was amplified by RT-PCR using the following HME primers: forward, 5′-TGGCCATTCAGGTCTTG-3′; reverse, 5′-AAGCAGCTTCAATGCCAGAT-3′. The purified PCR products were cloned into the pGEM-T vector (Promega Biotechnology, Madison, WI) according to the manufacturer’s instructions. The identity of the cDNA fragment was confirmed by sequence analysis using the dye terminator method (ABI 373A; Perkin Elmer, Rotkreuz, Switzerland). A 190-bp fragment of mouse 7S that cross-hybridizes with human 7S was used to verify equivalent RNA loading and transfer in Northern blot analysis. 27 The probes were radiolabeled with [α-32P]dCTP (NEN Life Science Products AG, Geneva, Switzerland) using a random primer labeling system (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK).
Northern Blot Analysis
Total RNA was isolated from all pancreatic tissue samples and the five pancreatic cancer cell lines (ASPC-1, CAPAN-1, MIAPaCa-2, PANC-1, and T3M4) by the guanidinium isothiocyanate-phenol-chloroform extraction method. 28 The procedures used have been described in detail previously. 29 Briefly, after electrophoresis of total RNA in 1.2% agarose/1.8 mol/L formaldehyde gels, the RNA was electrotransferred onto nylon membranes and cross-linked by UV irradiation. The filters were prehybridized for 5 hours at 42°C and hybridized for 20 hours at 42°C in the presence of the radiolabeled cDNA probe for HME (106 cpm/mL). Afterwards, blots were rinsed twice with 2× SSC at 50°C, and washed twice with 0.2× SSC/2% SDS at 55°C for 10 minutes. 30,31 All blots were exposed at −80°C to Kodak XAR-5 films with Kodak intensifying screens, and the intensity of the radiographic bands was quantified by video image analysis using the Image-Pro plus software (Media Cybernetics, Silver Spring, MD). To verify equivalent RNA loading on Northern blot membranes, filters were rehybridized with the 7S cDNA probe, as reported before. 29
RT-PCR of HME in Pancreatic Cancer Cell Lines
Because HME expression could not be detected by Northern blot analysis in the pancreatic cancer cell lines, RT-PCR analysis was applied.
All cell line RNA samples were normalized to total RNA along with controls for nonspecific DNA contamination (absence of RNA template). First strand cDNA synthesis was performed in a 20 μL reaction volume using 1 μg total RNA. Samples were incubated at 42°C for 60 minutes and the reaction was terminated by heating the samples to 99°C for 5 minutes. RT products served as the template for PCR amplification using hot start PCR. Reactions included a 5-minute incubation at 94°C for denaturation and 40 PCR cycles (1 minute at 94°C, 1 minute at 61°C, and 1 minute at 72°C). Afterwards the samples were incubated for 8 minutes at 72°C for final extension. Five-microliter aliquots of each sample were resolved by electrophoresis on 1.2% agarose gels in the presence of 5 ng/mL ethidium bromide, and the DNA fragments were visualized under UV light to detect the PCR amplification products at the anticipated size (346 bp). Loading control was done with glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Western Blot Analysis
Approximately 200 mg of frozen normal and pancreatic cancer tissues was powdered and thawed in an ice-cold suspension buffer (10 mmol/L Tris-HCl, pH 7.6, 100 mmol/L NaCl) containing a proteinase inhibitor cocktail (Roche Diagnostics, Rotkreuz, Switzerland). Cultured pancreatic cancer cells (1 × 106) were washed in ice-cold phosphate-buffered saline and collected and resuspended in 1 mL ice-cold suspension buffer, which was also used for protein extraction of the human tissue samples. Tissues and cells were homogenized for 5 minutes, then centrifuged (14,000 rpm, 20 minutes at 4°C). The supernatants were collected and the protein concentration was measured with the micro BCA protein assay (Pierce Chemical Co., Rockford, IL). In addition, after 48 hours of incubation with the five pancreatic cancer cell lines, serum-free medium was sucked out and used for Western blot analysis.
Forty micrograms of protein from each sample was diluted in sample buffer (250 mmol/L Tris-HCl, 4% SDS, 10% glycerol, 0.006% bromphenol blue, and 2% β-mercaptoethanol), boiled for 5 minutes, cooled on ice for 5 minutes, and size-fractionated on 12% SDS-polyacrylamide gels. Gels were transferred onto nitrocellulose membranes, and the membranes were incubated in blocking solution (5% nonfat milk in 20 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.1% Tween-20 [TBS-T]), followed by incubation with monoclonal mouse antihuman HME C-terminal domain antibody (R&D Systems, Inc., Minneapolis, MN; 1:500 dilution), or human polyclonal rabbit anti-HME, N-terminus antibody (Chemicon International, Inc., Temecula, CA; 1:1,000 dilution) in blocking solution for 12 hours at 4°C. The membranes were washed with TBS-T and incubated with a sheep antimouse Ig horseradish peroxidase linked secondary antibody (Amersham Pharmacia Biotech; 1:5,000 dilution), or a donkey antirabbit Ig horseradish peroxidase linked secondary antibody (Amersham Pharmacia Biotech; 1:3,000 dilution). Antibody detection was performed with the enhanced chemoluminescence (ECL) Western blot detection system (Amersham Pharmacia Biotech). Signals were quantified by video image analysis using the Image-Pro plus software.
Immunohistochemistry
Immunohistochemical analysis was performed with the streptavidin alkaline phosphatase technique. Briefly, consecutive 3- to 5-μm paraffin-embedded tissue sections were dewaxed and rehydrated. Antigen retrieval was achieved by boiling the sections in citrate buffer in an 850-watt microwave oven for 8 minutes, followed by two similar treatments for 5 minutes each. Afterwards, the sections were washed in Tris-buffered saline and incubated at room temperature for 30 minutes with 10% normal goat serum before 20 hours of incubation at 4°C with the primary polyclonal rabbit antihuman HME antibody (Chemicon International; dilution 1:150) diluted in 10% normal goat serum. After incubation with the secondary biotinylated goat antirabbit antibody (Kirkegaard & Perry Laboratories Inc., Gaithersburg, MD), the sections were incubated with a streptavidin-biotin/alkaline phosphatase complex (Kirkegaard & Perry Laboratories Inc.). The color reaction was developed with new fuchsin-naphthol AS-BI (Sigma Chemical Co., St. Louis, MO) followed by counterstaining with hematoxylin. 32 To ensure specificity of the primary antibody, consecutive tissue sections were incubated in the absence of the primary antibody with normal goat serum. In these cases no immunostaining was detected.
Statistical Analysis
Data are expressed as median and range. The clinical and histopathologic characteristics of the HME-positive and HME-negative patient groups were compared with univariate analysis using the Mann-Whitney test. Survival curves were plotted according to the Kaplan-Meier method; the statistical differences were analyzed using the log-rank test. To determine independent prognostic values with respect to overall survival, the grouping of the patients according to HME mRNA expression level, patient age, gender, tumor size, presence of lymph node metastases, tumor grading, and tumor stage were included in the Cox proportional regression analysis. Further, Spearman correlation analysis was performed. P < .05 was defined as significant. All the statistical analyses were performed using the SPSS 8.0 software (SPSS Inc., Chicago, IL).
RESULTS
Detection of HME mRNA Expression in Normal Pancreas, Chronic Pancreatitis, and Pancreatic Cancer
Thirteen normal, 16 chronic pancreatitis, and 39 pancreatic cancer samples were investigated by Northern blot analysis to detect HME mRNA expression. The transcript size of HME mRNA in pancreatic tissue samples was approximately 1.8 kb and is in accordance with previous reports, and no aberrant mRNA moieties were found. 4 In the normal pancreas only very low levels of HME mRNA were detectable; in chronic pancreatitis, HME mRNA expression levels higher than those of the normal pancreas were detected in 3 of 16 (19%) tissue samples. In contrast, in the pancreatic cancer samples, HME mRNA levels were markedly increased in 25 of 39 samples (64%) (Fig. 1). Quantification of the mRNA signals revealed that HME mRNA levels were 41-fold increased in pancreatic cancer tissues when all cancer samples were compared with the normal samples. When only cancer samples with increased expression were evaluated, the increase in comparison to normal controls was 50-fold.
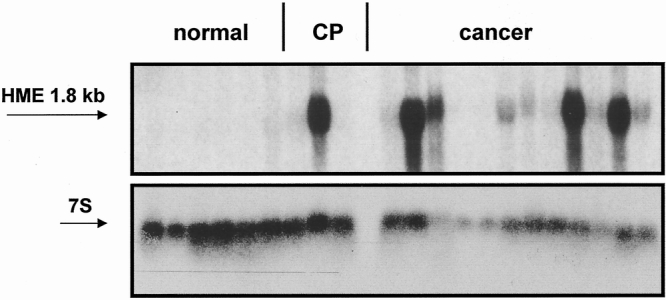
Figure 1. Northern blot analysis of human macrophage metalloelastase (HME) mRNA in normal pancreas, chronic pancreatitis (CP), and pancreatic cancer. 7S hybridization was used to verify equivalent RNA loading and transfer.
HME mRNA Expression in Pancreatic Cancer Cell Lines
In the five human pancreatic cancer cell lines T3M4, ASPC-1, CAPAN-1, PANC-1, and MIA-PaCa-2, no HME mRNA expression signals could be detected by standard Northern blot analysis. However, by RT-PCR analysis, HME mRNA expression was detectable in all five pancreatic cell lines at moderate levels (Fig. 2).
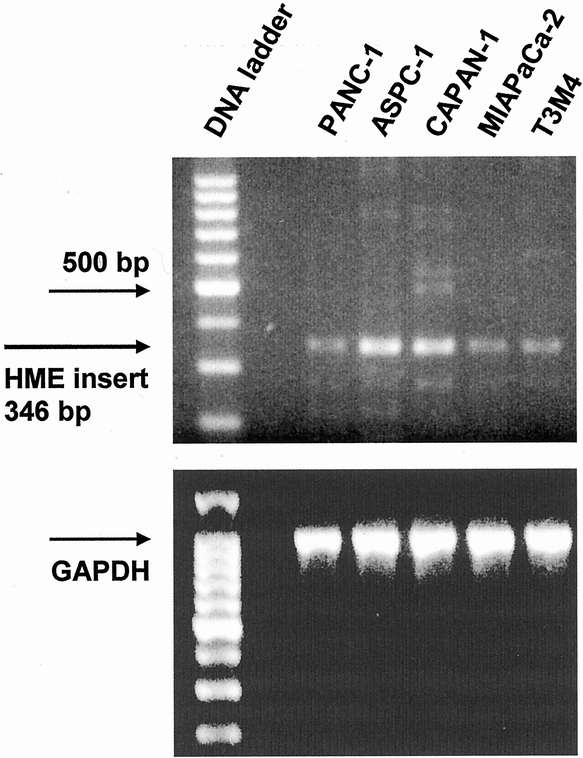
Figure 2. Human macrophage metalloelastase reverse transcriptase–polymerase chain reaction analysis in cultured pancreatic cancer cell lines. GAPDH was used to verify equivalent loading.
Immunohistochemistry
HME immunoreactivity was absent in the normal pancreas (Fig. 3), and in chronic pancreatitis samples only the three samples that exhibited enhanced HME mRNA expression on Northern blot analysis showed HME immunoreactivity. In these chronic pancreatitis samples HME immunoreactivity was present in macrophages, most ductal cells, some acinar cells, and some nerves. In contrast, in chronic pancreatitis samples that had not exhibited enhanced HME mRNA expression, HME immunoreactivity was absent.
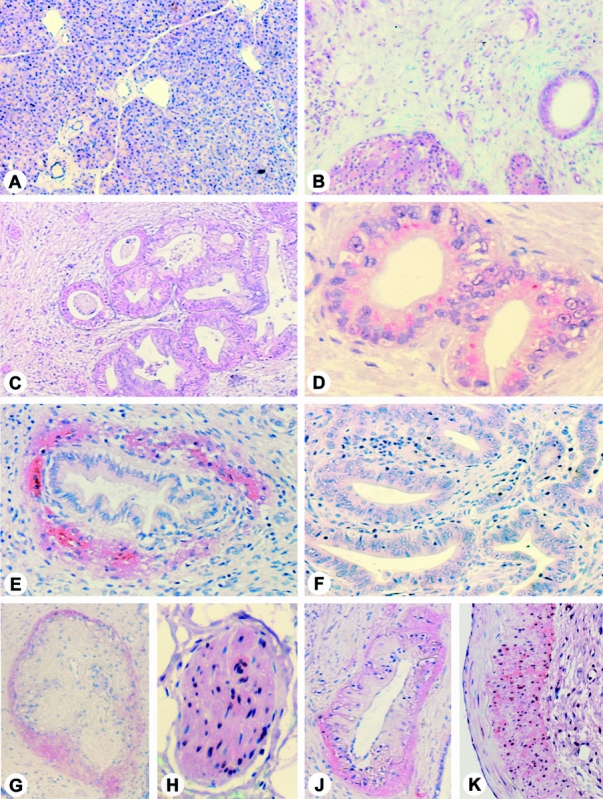
Figure 3. Human macrophage metalloelastase (HME) immunohistochemistry in the normal pancreas (A), in chronic pancreatitis (B), and in pancreatic cancer sections (C–K). In the normal pancreas, only occasionally faint HME immunoreactivity was present, whereas in the chronic pancreatitis samples with enhanced HME mRNA expression levels, strong immunoreactivity was found by Northern blot analysis in remaining acinar and ductal cells and in macrophages. In pancreatic cancer, cytoplasmic HME immunoreactivity was present in the cancer cells (C, D); however, there were also cancer samples in which the tumor cells were HME negative (F). Some cancer samples exhibited strong stromal HME immunoreactivity around the cancer cell ducts (E), nerves (G), and vessels (J). Some enlarged pancreatic nerves and blood vessels (H) also exhibited HME immunoreactivity. (A, B, C, F, original magnification ×50; G, J, original magnification ×100; E, original magnification ×150; D, H, K, original magnification ×200)
In pancreatic cancer samples HME immunoreactivity was predominantly present at moderate intensity in the cancer cells and in a focal pattern in stromal cells surrounding the cancer cells. Some cancer cells with ductal formation exhibited no HME immunoreactivity. In the pancreatic cancer tissues some HME immunoreactivity was also present in enlarged pancreatic nerves, arterial walls, and in a circular pattern in stromal tissue adjacent to arteries and nerves. Of 32 pancreatic cancer tissues, 18 (56%) exhibited HME immunoreactivity in the cancer cells and 20 (63%) in the stromal cells. In eight samples (25%), HME immunoreactivity was simultaneously present in cancer cells and in surrounding stromal cells. HME immunostaining was comparable in primary tumor samples of stage I/II cancers (no metastases present) and stage III/IV cancers (lymph node or distal metastases present). In addition, when HME immunostaining was simultaneously analyzed in primary tumor samples and in corresponding lymph node metastases, no difference in HME immunoreactivity was observed.
Western Blot Analysis of HME in Normal Pancreas and Pancreatic Cancer
Western blot analysis of HME was performed in normal human pancreas and pancreatic cancer samples to quantify differences in protein levels. Incubation with the monoclonal and polyclonal antibodies revealed a strong 54-kd HME protein band in all cancer tissue samples, whereas in the normal controls only a weak band was present (Fig. 4). Quantification of the protein signals by video image analysis revealed that the HME protein levels were 4.7-fold increased in pancreatic cancer samples compared with the normal controls.
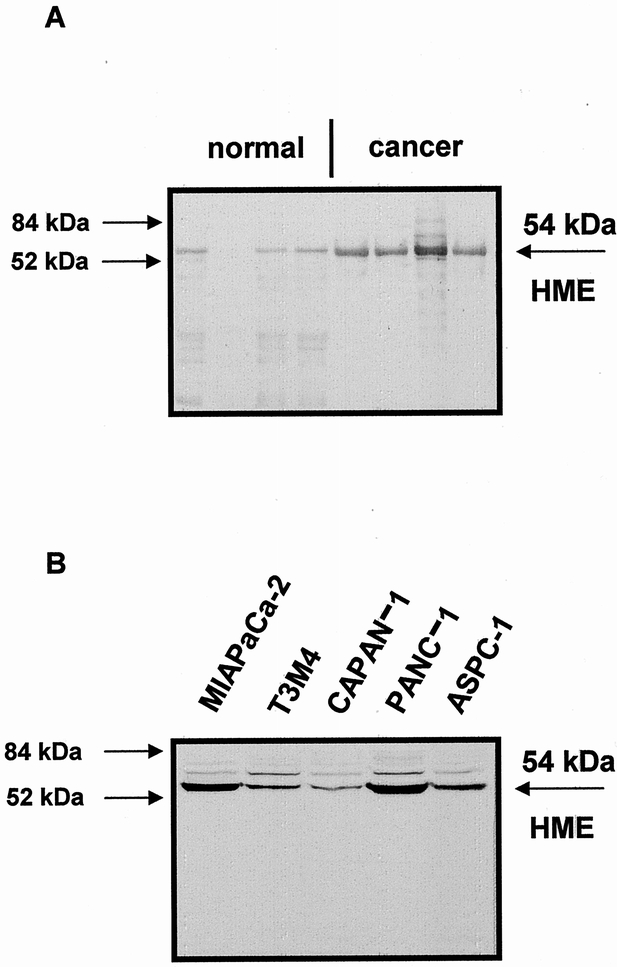
Figure 4. (A) Western blot analysis of human macrophage metalloelastase (HME) in normal pancreas and in pancreatic cancer. (B) Western blot analysis of HME in pancreatic cancer cell lines. A 54-kd HME protein band was detectable in the pancreatic tissues and in the cancer cell lines.
HME Protein in Pancreatic Cancer Cell Lines
Western blot analysis was also performed in the five pancreatic cancer cell lines. The 54-kd HME form was present in all cell lines, with highest levels in PANC-1 and MiaPaCa-2 and similar lower levels in T3M4, CAPAN-1, and ASPC-1 cells. Further, HME was present in the supernatant of serum-free medium T3M4, CAPAN-1, PANC-1, and MIAPaCa-2 cells, but not in the supernatant of ASPC-1 cells.
Relationship of HME mRNA Expression to Clinical and Histopathologic Data
Three patients were excluded from the survival analysis because they died early after surgery (two died of postoperative complications and one died of apoplexy). The cancer samples were grouped according to their HME mRNA expression levels. Samples that exhibited enhanced HME expression in comparison with the normal controls were summarized as positive samples, and samples without enhanced HME expression were summarized as negative samples. There was no significant difference between the two HME groups when patient age (P = .4), gender (P = .25), tumor size (P = .2), presence of lymph node metastasis (P = .69), tumor stage (P = .99), and tumor grading (P = .22) were compared by univariate analysis.
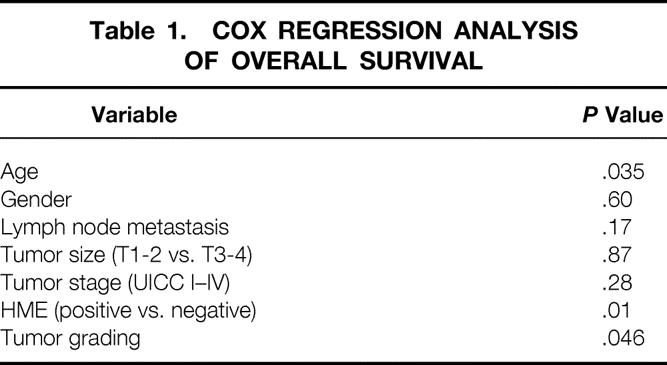
The median duration of follow-up of the patients was 12.5 months. In the HME mRNA-positive group (n = 22), the median survival was 11.5 months (lower quartile 8.0 months, upper quartile 16.5 months). In the HME mRNA-negative group (n = 14), the median survival was 22 months (lower quartile 9.0 months, upper quartile 43.2 months). Four patients in the HME-negative group are still alive 43, 44, 56, and 67 months after tumor resection. Kaplan-Meier analysis and log-rank analysis of the postoperative survival curves of patients whose tumors were HME positive or negative revealed a significantly poorer survival for patients with HME-positive tumors (P < .01) (Fig. 5). Cox proportional regression analysis revealed that the HME status, tumor grading, and age are independent prognostic values (Table 1).
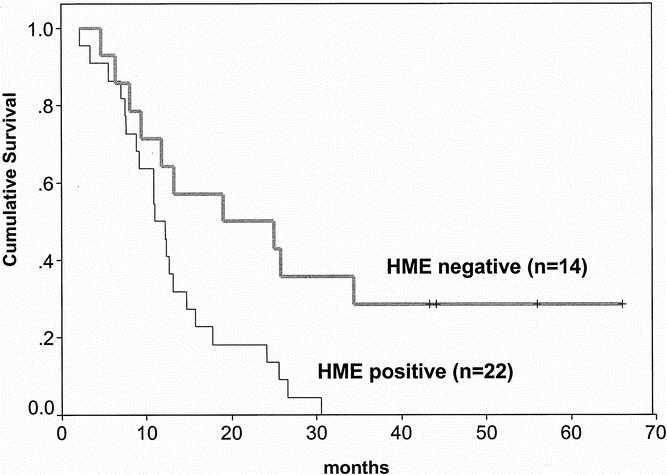
Figure 5. Overall survival of 36 patients with pancreatic adenocarcinoma who underwent tumor resection (by the log-rank test). Patients whose tumors expressed human macrophage metalloelastase (HME) mRNA showed a poorer survival compared with patients whose tumors were HME mRNA-negative.
Table 1. COX REGRESSION ANALYSIS OF OVERALL SURVIVAL
Spearman correlation analysis revealed no relationship between the HME status and age, gender, the tumor size, presence of lymph node metastases, tumor stage, or tumor grading.
DISCUSSION
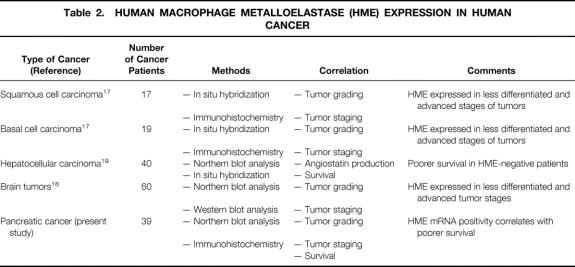
This study analyzed for the first time HME mRNA expression in a comparative manner in the normal pancreas, chronic pancreatitis, and pancreatic cancer. The rationale for studying HME in human pancreatic cancer was its identification as a markedly upregulated gene by DNA chip technology screening, a technology by which the expression of more than 5,600 human genes could be analyzed simultaneously (unpublished data). In our study, HME mRNA expression was low in the normal pancreas. In contrast, HME mRNA expression was enhanced in 64% of the pancreatic cancers, and overexpression was associated with poorer survival. HME expression analysis has also been conducted previously in other malignancies. In contrast to our findings, in hepatocellular carcinomas a better prognosis was reported for HME-expressing tumors. 19,20 Patients with hepatocellular carcinomas whose tumors overexpressed HME mRNA had less frequent portal vein invasion and less frequent intrahepatic metastasis than those without HME expression. These histopathologic associations were in accordance with the survival analysis, in which patients with hepatocellular carcinoma whose tumors did not express HME mRNA had poorer survival rates than those whose tumors expressed HME mRNA. Based on these data it was speculated that HME might be associated with angiostatin generation, which indirectly might possess inhibitory effects on tumor growth. 19,20 Inasmuch as angiostatin can be generated in vitro from its precursor plasminogen by several other MMPs and enzymes, factors other than HME might possess a stronger influence on the growth behavior of hepatocellular cancer cells. 22,33–36 However, the impact of HME on cancer prognosis seems to vary, and at present HME seems to be a positive prognostic factor only in hepatocellular carcinomas; in other nongastrointestinal malignancies, such as squamous cell carcinomas, basal cell carcinomas, and brain tumors, HME is associated with a more aggressive disease status and worse tumor differentiation. 17,18 Further, anaplastic astrocytomas and grade 4 glioblastomas expressed significantly higher HME levels than grade 1 meningiomas, 18 indicating that HME mRNA expression is associated with advanced tumor stages in brain tumors (Table 2).
Table 2. HUMAN MACROPHAGE METALLOELASTASE (HME) EXPRESSION IN HUMAN CANCER
Western blot analysis in the pancreatic tissue samples and the cancer cell lines visualized a 54-kd HME protein band. The detected protein is most likely a precursor of active HME because in macrophages different active forms weighing between 22 and 29 kd have previously been described. 4,12,25 However, it is surprising that one of these active HME forms could not be detected in the pancreatic cancer tissues. HME is activated from its precursor by autoactivation 4,12 and by serine proteinases, 25 which are present in high levels in pancreatic tumors. 37 This observation cannot readily be explained: in other tumors, such as colon cancer and hepatocellular carcinoma, active HME is clearly detectable (unpublished own data). We detected active HME in colon cancer tissue samples by Western blot analysis in a 22-kd form and in hepatocellular carcinoma tissue samples in a 29-kd form, but in the same blots there was no active form for pancreatic cancer samples, but rather only an intermediate form at approximately 36 kd. Immunohistochemical analysis localized HME in pancreatic cancer cells in 56% of the tumor samples, and in 63% of the tumors HME immunoreactivity was present in stromal cells. Inasmuch as HME immunostaining signals were mainly localized in cytoplasm, it is likely that inactive HME is released from the cancer cells in the interstitial space, where it becomes activated by serine proteinases and other enzymes. 37 This hypothesis is supported by the experiments in the pancreatic cancer cell lines, in which HME could be detected intracellularly but also was released in the tissue culture medium.
Previously several other MMPs have been analyzed in pancreatic carcinomas, and in addition to HME, enhanced MMP-1 mRNA levels, obtained by in situ hybridization, were also associated with poorer survival. 38,39 However, in contrast to HME, the expression levels of MMP-1 seem to be quite low because detection by Northern blot analysis was not possible, suggesting that they may not be of predominant biologic relevance. 38,39 Similar to MMP-1, which was not associated with tumor metastasis, 39 HME expression in primary pancreatic cancer tissues was not different in metastatic and nonmetastatic primary tumors. Further, no difference in HME expression was found in cancer cells of the primary tumor and metastatic cancer cells of the same tumors, suggesting that HME seems not to have an important function in the development of metastases in pancreatic cancer, as had been reported before for other genes. 40,41 MMP-2, -7, -9, and -11 are also upregulated in pancreatic cancer, and their expression influences the extent of the desmoplastic reaction, tumor invasiveness, and metastatic potential. 42–49 Inhibition of MMP-2 and -9 positively influences the prognosis in a murine model of human pancreatic cancer. 50 A number of MMP inhibitors have been developed, and some are being tested in clinical studies in pancreatic cancer therapy. 51 First results of clinical and experimental studies suggest that the inhibition of MMP activity is more beneficial for patients with early pancreatic cancer than in advanced disease. 52,53 In recent years more selective inhibitors of MMPs associated with tumor progression were developed to avoid the side effects caused by the disturbances in the balance of MMP–TIMP–extracellular matrix interactions. 54 Our findings of marked HME overexpression in many pancreatic tumors and their correlation with poorer survival suggest that inhibition of HME activity might provide a therapeutic benefit. However, the effects of HME inhibition have to be evaluated in clinical trials.
Acknowledgments
The authors thank Dr. J. Kleeff for his assistance in the HME primer design and Mr. P. Pavek, Technopol a.s., Slovakia, and Mr. A. Masar, Slovenske Elektrarne a.s., Slovakia, for their financial support of Dr. Balaz.
Footnotes
Correspondence: Helmut Friess, MD, Department of General Surgery, University of Heidelberg, Im Neuenheimer Feld 110, D-69120 Heidelberg, Germany.
E-mail: helmut_friess@med.uni-heidelberg.de
Accepted for publication October 1, 2001.
References
- 1.Okada Y, Nagase H, Harris ED Jr. Matrix metalloproteinases 1, 2, and 3 from rheumatoid synovial cells are sufficient to destroy joints. J Rheumatol 1987; 14: 41–42. [PubMed] [Google Scholar]
- 2.Bejarano PA, Noelken ME, Suzuki K, et al. Degradation of basement membranes by human matrix metalloproteinase 3 (stromelysin). Biochem J 1988; 256: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay ED, ed. Cell Biology of Extracellular Matrix, 2d ed. New York, London: Plenum Press; 1991.
- 4.Shapiro SD, Kobayashi DK, Ley TJ. Cloning and characterization of a unique elastolytic metalloproteinase produced by human alveolar macrophages. J Biol Chem 1993; 268: 23824–23829. [PubMed] [Google Scholar]
- 5.Chandler S, Cossins J, Lury J, et al. Macrophage metalloelastase degrades matrix and myelin proteins and processes a tumour necrosis factor-alpha fusion protein. Biochem Biophys Res Commun 1996; 228: 421–429. [DOI] [PubMed] [Google Scholar]
- 6.Gronski TJ Jr, Martin RL, Kobayashi DK, et al. Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J Biol Chem 1997; 272: 12189–121894. [DOI] [PubMed] [Google Scholar]
- 7.Shipley JM, Wesselschmidt RL, Kobayashi DK, et al. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA 1996; 93: 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Normann SJ. Macrophage infiltration and tumor progression. Cancer Metastasis Rev 1985; 4: 277–291. [DOI] [PubMed] [Google Scholar]
- 9.Whitworth PW, Pak CC, Esgro J, et al. Macrophages and cancer. Cancer Metastasis Rev 1990; 8: 319–351. [DOI] [PubMed] [Google Scholar]
- 10.Mantovani A, Bottazzi B, Colotta F, et al. The origin and function of tumor-associated macrophages. Immunol Today 1992; 13: 265–270. [DOI] [PubMed] [Google Scholar]
- 11.Emmrich J, Weber I, Nausch M, et al. Immunohistochemical characterization of the pancreatic cellular infiltrate in normal pancreas, chronic pancreatitis and pancreatic carcinoma. Digestion 1998; 59: 192–198. [DOI] [PubMed] [Google Scholar]
- 12.Belaaouaj A, Shipley JM, Kobayashi DK, et al. Human macrophage metalloelastase. Genomic organization, chromosomal location, gene linkage, and tissue-specific expression. J Biol Chem 1995; 270: 14568–14575. [DOI] [PubMed] [Google Scholar]
- 13.Curci JA, Liao S, Huffman MD, et al. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J Clin Invest 1998; 102: 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaalamo M, Karjalainen-Lindsberg ML, Puolakkainen P, et al. Distinct expression profiles of stromelysin-2 (MMP-10), collagenase-3 (MMP-13), macrophage metalloelastase (MMP-12), and tissue inhibitor of metalloproteinases-3 (TIMP-3) in intestinal ulcerations. Am J Pathol 1998; 152: 1005–1014. [PMC free article] [PubMed] [Google Scholar]
- 15.Vaalamo M, Kariniemi AL, Shapiro SD, et al. Enhanced expression of human metalloelastase (MMP-12) in cutaneous granulomas and macrophage migration. J Invest Dermatol 1999; 112: 499–505. [DOI] [PubMed] [Google Scholar]
- 16.Takei I, Takagi M, Santavirta S, et al. Messenger ribonucleic acid expression of 16 matrix metalloproteinases in bone-implant interface tissues of loose artificial hip joints. J Biomed Mater Res 2000; 52: 613–620. [DOI] [PubMed] [Google Scholar]
- 17.Kerkela E, Ala-Aho R, Jeskanen L, et al. Expression of human macrophage metalloelastase (MMP-12) by tumor cells in skin cancer. J Invest Dermatol 2000; 114: 1113–1119. [DOI] [PubMed] [Google Scholar]
- 18.Kachra Z, Beaulieu E, Delbecchi L, et al. Expression of matrix metalloproteinases and their inhibitors in human brain tumors. Clin Exp Metastasis 1999; 17: 555–566. [DOI] [PubMed] [Google Scholar]
- 19.Rivas MJ, Arii S, Furutani M, et al. Expression of human macrophage metalloelastase gene in hepatocellular carcinoma: correlation with angiostatin generation and its clinical significance. Hepatology 1998; 28: 986–993. [DOI] [PubMed] [Google Scholar]
- 20.Gorrin-Rivas MJ, Arii S, Mori A, et al. Implications of human macrophage metalloelastase and vascular endothelial growth factor gene expression in angiogenesis of hepatocellular carcinoma. Ann Surg 2000; 231: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Reilly MS, Holmgren L, Shing Y, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 1994; 79: 315–328. [DOI] [PubMed] [Google Scholar]
- 22.Cornelius LA, Nehring LC, Harding E, et al. Matrix metalloproteinases generate angiostatin: effects on neovascularization. J Immunol 1998; 161: 6845–6852. [PubMed] [Google Scholar]
- 23.Dong Z, Kumar R, Yang X, et al. Macrophage-derived metalloelastase is responsible for the generation of angiostatin in Lewis lung carcinoma. Cell 1997; 88: 801–810. [DOI] [PubMed] [Google Scholar]
- 24.Carmeliet P, Moons L, Lijnen R, et al. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet 1997; 17: 439–444. [DOI] [PubMed] [Google Scholar]
- 25.Raza SL, Nehring LC, Shapiro SD, et al. Proteinase activated receptor-1 regulation of macrophage elastase secretion by serine proteinases. J Biol Chem 2000; 275: 41243–41250. [DOI] [PubMed] [Google Scholar]
- 26.St-Denis A, Chano F, Tremblay P, et al. Protein kinase C-alpha modulates lipopolysaccharide-induced functions in a murine macrophage cell line. J Biol Chem 1998; 273: 32787–32792. [DOI] [PubMed] [Google Scholar]
- 27.di Mola FF, Friess H, Scheuren A, et al. Transforming growth factor-betas and their signaling receptors are coexpressed in Crohn’s disease. Ann Surg 1999; 229: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156–159. [DOI] [PubMed] [Google Scholar]
- 29.Friess H, Lu Z, Graber HU, et al. bax, but not bcl-2, influences the prognosis of human pancreatic cancer. Gut 1998; 43: 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friess H, Yamanaka Y, Büchler M, et al. A subgroup of patients with chronic pancreatitis overexpress the c-erbB-2 protooncogene. Ann Surg 1994; 220: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanaka Y, Friess H, Büchler M, et al. Synthesis and expression of transforming growth factor beta-1, beta-2, and beta-3 in the endocrine and exocrine pancreas. Diabetes 1993; 42: 746–756. [DOI] [PubMed] [Google Scholar]
- 32.Mazzucchelli L, Blaser A, Kappeler A, et al. BCA-1 is highly expressed in Helicobacter pylori-induced mucosa-associated lymphoid tissue and gastric lymphoma. J Clin Invest 1999; 104: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson BC, Sang QA. Angiostatin-converting enzyme activities of human matrilysin (MMP-7) and gelatinase B/type IV collagenase (MMP-9). J Biol Chem 1997; 272: 28823–28825. [DOI] [PubMed] [Google Scholar]
- 34.Lijnen HR, Ugwu F, Bini A, et al. Generation of an angiostatin-like fragment from plasminogen by stromelysin-1 (MMP-3). Biochemistry 1998; 37: 4699–4702. [DOI] [PubMed] [Google Scholar]
- 35.O’Mahony CA, Seidel A, Albo D, et al. Angiostatin generation by human pancreatic cancer. J Surg Res 1998; 77: 55–58. [DOI] [PubMed] [Google Scholar]
- 36.Morikawa W, Yamamoto K, Ishikawa S, et al. Angiostatin generation by cathepsin D secreted by human prostate carcinoma cells. J Biol Chem 2000; 275: 38912–38920. [DOI] [PubMed] [Google Scholar]
- 37.Cantero D, Friess H, Deflorin J, et al. Enhanced expression of urokinase plasminogen activator and its receptor in pancreatic carcinoma. Br J Cancer 1997; 75: 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imamura T, Ohshio G, Mise M, et al. Expression of membrane-type matrix metalloproteinase-1 in human pancreatic adenocarcinomas. J Cancer Res Clin Oncol 1998; 124: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito T, Ito M, Shiozawa J, et al. Expression of the MMP-1 in human pancreatic carcinoma: relationship with prognostic factor. Mod Pathol 1999; 12: 669–674. [PubMed] [Google Scholar]
- 40.Guo XZ, Friess H, Maurer C, et al. KAI1 is unchanged in metastatic and nonmetastatic esophageal and gastric cancers. Cancer Res 1998; 58: 753–758. [PubMed] [Google Scholar]
- 41.Friess H, Guo XZ, Berberat P, et al. Reduced KAI1 expression in pancreatic cancer is associated with lymph node and distant metastases. Int J Cancer 1998; 79: 349–355. [DOI] [PubMed] [Google Scholar]
- 42.Gress TM, Muller-Pillasch F, Lerch MM, et al. Expression and in-situ localization of genes coding for extracellular matrix proteins and extracellular matrix degrading proteases in pancreatic cancer. Int J Cancer 1995; 62: 407–413. [DOI] [PubMed] [Google Scholar]
- 43.Koshiba T, Hosotani R, Wada M, et al. Involvement of matrix metalloproteinase-2 activity in invasion and metastasis of pancreatic carcinoma. Cancer 1998; 82: 642–650. [DOI] [PubMed] [Google Scholar]
- 44.Ellenrieder V, Alber B, Lacher U, et al. Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int J Cancer 2000; 85: 14–20. [DOI] [PubMed] [Google Scholar]
- 45.Bramhall SR, Stamp GWH, Dunn J, et al. Expression on collagenase (MMP2), stromelysin (MMP3), and tissue inhibitor of the metalloproteinases (TIMP1) in pancreatic and ampullary disease. Br J Cancer 1996; 73: 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bramhall SR, Neoptolemos JP, Stamp GW, et al. Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J Pathol 1997; 182: 347–355. [DOI] [PubMed] [Google Scholar]
- 47.Satoh K, Ohtani H, Shimosegawa T, et al. Infrequent stromal expression of gelatinase A and intact basement membrane in intraductal neoplasms of the pancreas. Gastroenterology 1994; 107: 1488–1495. [DOI] [PubMed] [Google Scholar]
- 48.Gong YL, Xu GM, Huang WD, et al. Expression of matrix metalloproteinases and the tissue inhibitors of metalloproteinases and their local invasiveness and metastasis in Chinese human pancreatic cancer. J Surg Oncol 2000; 73: 95–99. [DOI] [PubMed] [Google Scholar]
- 49.Maatta M, Soini Y, Liakka A, et al. Differential expression of matrix metalloproteinase (MMP)-2, MMP-9, and membrane type 1-MMP in hepatocellular and pancreatic adenocarcinoma: implications for tumor progression and clinical prognosis. Clin Cancer Res 2000; 6: 2726–2734. [PubMed] [Google Scholar]
- 50.Haq M, Shafii A, Zervos EE, et al. Addition of matrix metalloproteinase inhibition to conventional cytotoxic therapy reduces tumor implantation and prolongs survival in a murine model of human pancreatic cancer. Cancer Res 2000; 60: 3207–3211. [PubMed] [Google Scholar]
- 51.Heath EI, Grochow LB. Clinical potential of matrix metalloproteinase inhibitors in cancer therapy. Drugs 2000; 59: 1043–1055. [DOI] [PubMed] [Google Scholar]
- 52.McCawley LJ, Matrisian LM. Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol Med Today 2000; 6: 149–156. [DOI] [PubMed] [Google Scholar]
- 53.Bergers G, Javaherian K, Lo KM, et al. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science 1999; 284: 808–812. [DOI] [PubMed] [Google Scholar]
- 54.Erlichman C, Adjei AA, Alberts SR, et al. Phase I study of the matrix metalloproteinase inhibitor, BAY12–9566. Ann Oncol 2001; 12: 389–395. [DOI] [PubMed] [Google Scholar]