Abstract
In Escherichia coli, there are multiple paralogous copies of the enzyme API [A5P (D-arabinose 5-phosphate) isomerase], which catalyses the conversion of the pentose pathway intermediate Ru5P (D-ribulose 5-phosphate) into A5P. A5P is a precursor of Kdo (3-deoxy-D-manno-octulosonate), an integral carbohydrate component of various glycolipids coating the surface of the OM (outer membrane) of Gram-negative bacteria, including LPS (lipopolysaccharide) and many group 2 K-antigen capsules. The K-antigen-specific API KpsF has been cloned from the uropathogenic E. coli strain CFT073 and its biochemical properties characterized. Purified recombinant KpsF [K-API (K-antigen API)] is tetrameric and has optimal activity at pH 7.8. The enzyme is specific for A5P and Ru5P, with Km (app) values of 0.57 mM for A5P and 0.3 mM for Ru5P. The apparent kcat in the A5P to Ru5P direction is 15 and 19 s−1 in the Ru5P to A5P direction. While most of the properties are quite similar to its LPS API counterpart KdsD, the catalytic constant is nearly 10-fold lower. K-API is now the second Kdo biosynthetic related gene that has been characterized from the kps group 2 capsule cluster.
Keywords: D-arabinose 5-phosphate isomerase (API), 3-deoxy-D-manno-octulosonate (Kdo), Escherichia coli, group 2 capsule biosynthesis, K-antigen, lipopolysaccharide
Abbreviations: A5P, D-arabinose 5-phosphate; API, A5P isomerase; BTP, Bis-Tris propane; Kdo, 3-deoxy-D-manno-octulosonate; K-API, K-antigen API; LPS, lipopolysaccharide; MALDI-MS, matrix-assisted laser-desorption ionization MS; OM, outer membrane; Ru5P, D-ribulose 5-phosphate
INTRODUCTION
The surface of the OM (outer membrane) in Gram-negative bacteria is the main interface through which the bacterium senses and responds to its environment. The surface of the OM of Escherichia coli is decorated with a number of diverse polysaccharides, including LPS (lipopolysaccharide), enterobacterial common antigen and various capsular polysaccharides. In E. coli alone, there are over 80 distinct capsular polysaccharides collectively known as K-antigens [1]. Capsular polysaccharides are the outermost cell-surface structure, and accordingly they have been implicated as key components in many of the initial host–pathogen interactions [2]. K-antigens have been postulated to protect the bacterium both directly by conferring resistance to non-specific host defence mechanisms (complement cascade) and indirectly by masking the presence of underlying molecules, which are more immunostimulatory, such as LPS (endotoxin), from triggering an immune response.
K-antigens are classified into four groups according to various genetic and biosynthetic criteria [3]. Most of the pathogenic extraintestinal isolates associated with infection belong to group 2. Group 2 capsules are encoded by the kps gene cluster located on a pathogenicity island near serA, and share a common genetic organization that is subdivided into three regions [4]. Region 1 consists of six co-transcribed genes (kpsFEDUCS), while region 3 contains two genes (kpsMT) convergently transcribed with respect to region 1. Region 3 contains the ATP-binding-cassette transporter and, together with region 1 genes, is believed to be involved in the export of the nascent polymer from the cytoplasmic face of the inner membrane to the OM surface [5]. Region 2 is flanked by regions 1 and 3 and contains a variable number of serotype-specific genes encoding protein products responsible for the synthesis and assembly of the saccharide portion of the K-antigen repeat unit. Much work has been done on the transcriptional regulation of the kps locus in the prototype group 2 capsule systems from E. coli K1 and K5 [6–9]. Expression is under the control of multiple global regulators, including IHF (integration host factor), the nucleoid-associated protein H-NS, BipA and the anti-terminator RfaH, resulting in the co-ordinated repression of capsule synthesis at temperatures below 20 °C and induction at 37 °C. Thermoregulation has clear implications with respect to virulence during infection.
While much is known about the gene functions of the kps cluster and of the regulatory network controlling their expression, details concerning the mechanism of ligation of the K-antigen polysaccharide to the lipid anchor acceptor remain obscure [3]. Schmidt and Jann [10] originally reported the identity of the lipid anchor as a single Kdo (3-deoxy-D-manno-octulosonate) residue attached to a 1,2-dipalmitoylphosphatidic acid, suggesting that Kdo may serve a universal role in group 2 capsule biosynthesis by linking the polysaccharide region to the lipid domain, reminiscent of Kdo's well-studied essential role in LPS biosynthesis [11]. Accordingly, there is an increase in specific activity levels of CMP-Kdo synthetase (KpsU) in extracts from group 2 capsule strains only when grown at permissive temperatures [12]. In addition to KpsU, a second potential Kdo biosynthetic gene from the kps cluster (kpsF) was initially identified based on its ability to restore LPS biosynthesis in a Neisseria meningitidis kdsD construct [13], which was then successfully repeated in a kdsD mutant of Yersinia pestis [14]. KdsD (formerly YrbH) is an API [A5P (D-arabinose 5-phosphate) isomerase] from the LPS pathway that catalyses the 1,2-aldo/keto isomerization of Ru5P (D-ribulose 5-phosphate) to A5P, a precursor of Kdo [15]. Despite the conservation of both KpsU and KpsF among all group 2 isolates, E. coli K1 (and K92) strains do not have Kdo substituted at the reducing end of their polysaccharides [3]. In order to begin to understand better the role of Kdo in capsule biogenesis, kpsF from the group 2 capsule strain E. coli CFT073 (O6:K2:H1) [16] was cloned, and the biochemical properties of recombinant capsular K-API (K-antigen API) are reported for the first time. The properties of K-API differ in comparison with its LPS (KdsD, L-API) counterpart, suggesting that K-API may have additional or alternative roles besides producing A5P for Kdo synthesis.
EXPERIMENTAL
Materials
E. coli strain CFT073 (O6:K2:H1) was obtained from Professor Harry L. T. Mobley (University of Michigan). PCR primers were synthesized by Invitrogen. Thermal cycling was performed using an MJ Research PTC-200 Peltier thermal cycler. Failsafe™ PCR PreMix Selection kit was purchased from Epicentre (Madison, MI, U.S.A.). The Promega Wizard DNA purification kit was utilized for plasmid purification. Chemically competent E. coli XL1-Blue (Stratagene) or E. coli BL21(DE3) (Novagen) cells were used for plasmid transformations. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. DNA sequencing was performed by the University of Michigan Biomedical Resources Core Facility. Hi Load™ Q-Sepharose (16/10) fast flow for anion-exchange chromatography, Fast Desalting (HR 10/10) columns for metal analysis studies and Superose 12 (HR 10/30) columns for gel-filtration native protein molecular mass analysis were all obtained from Amersham Biosciences. The gel-filtration protein molecular mass marker kit (12–200 kDa) was from Sigma.
Cloning of E. coli CFT073 kpsF
The kpsF gene was directly amplified from the E. coli CFT073 genome using standard whole-cell PCR methodologies using the following forward (GGTGCTAGAATTCATATGTCTGAAAGACATTTACC) and reverse (GAATTCGGATCCAAGTTAGTCGAAAATGCGCACAAGGCC) primers in Failsafe™ PCR 2X premix buffer ‘G’ (Epicentre). Primers were designed to incorporate NdeI and BamHI sites respectively and used to ligate the PCR product in the similarly restricted expression vector pT7-7 that had been pretreated with calf intestinal alkaline phosphatase. The ligation mixture was used to transform chemically competent E. coli XL1-Blue cells, and a transformant harbouring the correct pT7-kpsF plasmid was identified by restriction analysis and confirmed by DNA sequencing. The plasmid was transformed into E. coli BL21(DE3) cells for protein expression.
Overexpression and purification of recombinant KpsF
E. coli BL21(DE3) cells harbouring the pT7-kpsF plasmid were grown in 1 litre of 2YT medium [1.6% (w/v) tryptone, 1% yeast extract and 1% NaCl] containing ampicillin (100 μg/ml) at 37 °C with shaking (250 rev./min). Once the culture reached the mid-exponential growth phase (D600∼0.7–0.9), the culture was allowed to cool to 18 °C before being induced with isopropyl β-D-thiogalactoside at a final concentration of 0.4 mM. After 16 h of growth at 18 °C, the cells were harvested by centrifugation (6500 g, 15 min, 4 °C). The cell pellet (11.2 g wet weight) was suspended in 35 ml of buffer A (20 mM Tris/HCl and 1 mM dithiothreitol; pH 8.0) and then sonicated on ice (5×30 s pulses with 2 min pauses in between). Cellular debris was removed by centrifugation (40000 g, 30 min, 4 °C) and the supernatant was decanted (40 ml; 640 mg of protein). The solution was loaded on to a Hi Load™ (16/10) Q-Sepharose fast flow column that had been pre-equilibrated with buffer A. The protein was eluted using a 0–600 mM gradient of NaCl in buffer A over 120 min. Fractions containing primarily KpsF (∼35 kDa), as judged by SDS/PAGE, were pooled (24 ml; 315 mg of protein) and precipitated by the slow addition of an ammonium sulphate-saturated buffer A solution with stirring at room temperature (22 °C) until 35% saturation was reached. The solution was clarified by centrifugation (40000 g, 45 min, 22 °C), and the protein pellet was resuspended in buffer A. The solution was dialysed against two changes of 2 litres of buffer A for 24 h each at 4 °C. The preparation was greater than ∼95% homogeneous as judged by SDS/PAGE analysis. The final total yield of recombinant KpsF protein was 230 mg/l of cell culture. Purified enzyme was divided into aliquots and stored at −80 °C.
Assay of API activity
A 96-well microplate assay using the cysteine-carbazole colorimetric assay was used for all assays as previously described [15]. Briefly, a solution containing 25 μl of enzyme at various concentrations in buffer (100 mM Tris/HCl; pH 7.8) was incubated at 37 °C for 3 min in a 96-well reaction PCR plate using a Peltier thermal cycler. The reaction was initiated with 25 μl of an A5P solution and subsequently quenched after various time intervals with 50 μl of 12.5 M H2SO4. One unit of enzyme activity is defined as the conversion of 1 μmol of sugar phosphate per minute at 37 °C.
Molecular mass of recombinant KpsF
Protein samples were prepared by buffer exchange via overnight dialysis against 2 litres of 5 mM Hepes (pH 8.0) at 4 °C. The subunit mass of KpsF was determined by MALDI (matrix-assisted laser-desorption ionization)-MS on a VESTEC-2000 instrument using a sinipinic acid matrix by the University of Michigan Protein Structure Facility. The native molecular mass was determined by gel filtration utilizing a Superose 12 column (HR 10/30) with 20 mM Tris/HCl buffer (pH 8.0, 150 mM NaCl) as eluent. The elution volume was determined in triplicate for all samples and protein standards (12–200 kDa).
Metal content analysis
Enzyme samples as isolated above were prepared for metal analysis by extensive dialysis (48 h) against 2 litres of metal-free buffer (20 mM Trizma/HCl; pH 7.5) at 4 °C. EDTA-treated protein samples were prepared by first incubating enzyme in the presence of 10 mM EDTA for 2 h at 4 °C, and were subsequently desalted using a Fast Desalting column (HR 10/10) with 20 mM Tris/HCl (pH 7.5) as eluent. Bivalent metal content of the untreated or EDTA-treated samples was determined by high-resolution inductively coupled plasma-MS on a Finnigan MAT ELEMENT instrument at the Department of Geology, University of Michigan.
Effect of metals and pH on the activity of KpsF
Samples of KpsF as isolated and described above were diluted in 100 mM Tris/HCl buffer (pH 7.8) and incubated with various bivalent metals or EDTA at 20 μM for 30 min at 4 °C. Activity was assayed at 37 °C under saturating substrate conditions in triplicate with a 3 min reaction time (50 mM Tris/HCl, 80 nM KpsF, 10 mM A5P and 10 μM bivalent metal or EDTA).
The activity of KpsF in BTP (Bis-Tris propane) buffer (pH 6–9.75 in 0.25 pH unit intervals) at 37 °C was measured using the standard cysteine-carbazole colorimetric assay conditions. Activity was measured in triplicate with a 3 min reaction time (100 mM BTP, 80 nM KpsF, 1 mM EDTA and 10 mM A5P).
Substrate specificity
The substrate specificity of KpsF was tested with a panel of naturally abundant carbohydrates (50 mM Tris/HCl, pH 7.8, 1 mM EDTA, 90 nM KpsF and 10 mM substrate). After 10 min at 37 °C, reaction mixtures containing the potential alternative substrates D-arabinose, D-ribose 5-phosphate, D-glucose 6-phosphate, D-glucose 1-phosphate, D-glucosamine 6-phosphate or D-mannose 6-phosphate were quenched, and the presence of ketose was determined as outlined above using the colorimetric cysteine-carbazole assay, which detects both pentuloses and hexuloses [17]. Each carbohydrate was assayed in triplicate, along with appropriate controls that did not contain enzyme. The lower limit of detection for this assay is estimated to be less than 1% of ketose formation when using an initial 10 mM substrate concentration.
Determination of kinetic parameters
Reactions were performed at 37 °C using the end-point microplate assay with substrate concentrations typically ranging from 0.2 Km to 10 Km. After 2 min, reactions (50 mM Tris/HCl, pH 7.8, 1 mM EDTA and 90 nM KpsF) were quenched, at which point approx. <10% of substrate had been consumed. Initial rates (v0) were determined in triplicate and fitted to the standard Michaelis–Menten equation using non-linear least-squares regression to determine values for Km and kcat.
The equilibrium constant was determined using 31P NMR as previously described [15]. Solutions containing either 5 mM A5P or 5 mM Ru5P in 50 mM Tris/HCl (pH 7.5) with 1 mM EDTA and 10% (v/v) 2H2O were incubated in the presence of 900 nM KpsF at 37 °C. Reactions were periodically monitored by 31P NMR using a Bruker Avance DRX-300 instrument with WALTZ16 proton decoupling. A 10 s delay (>3 times the T1A5P/Ru5P) was used during the acquisition to ensure complete relaxation of the phosphorus nucleus, allowing for direct integration and comparison of peak areas for the two different sugar phosphates under observation. Chemical shifts were referenced to an external phosphoric acid standard (0 p.p.m.). Once there was no change in peak ratios for both samples, spectra were acquired (64 scans). Keq is reported for API in the direction of Ru5P product formation from A5P substrate ([Ru5P]/[A5P]).
Miscellaneous methods
Protein concentrations were determined using the Bio-Rad Protein Assay Reagent assay with BSA serving as the standard. One-dimensional SDS/PAGE was performed under reducing conditions with a 12% gel using a Mini-PROTEAN II electrophoresis unit (Bio-Rad).
RESULTS
Cloning, overexpression and purification of recombinant KpsF
The protein sequence deposited at the NCBI database for KpsF from E. coli CFT073 is 339 amino acids. The gene was initially cloned according to the deposited reading frame with the alternative start codon GTG, but MALDI-MS analysis revealed two recombinant protein peaks (m/z 35323 and 36579; results not shown) corresponding to a difference of 1256 Da. The first 12 N-terminal amino acids upstream of an internal methionine have a similar mass (1252 Da), suggesting that the methionine was actually the correct start codon. Further, there is a canonical ribosomal binding sequence [18] (AAGGAG) optimally spaced 5 base-pairs upstream of the internal methionine. Accordingly, the kpsF gene was re-cloned from the internal methionine in order to encode a 327-amino-acid open reading frame.
Recombinant KpsF was highly overexpressed using the pT7-7 expression vector. The vast majority of protein was localized in the cytoplasm, though a small fraction remained associated with the membrane fraction. Efficient purification to homogeneity was achieved in two steps using anion-exchange chromatography followed by ammonium sulphate precipitation (Figure 1). KpsF migrated as single band when analysed by SDS/PAGE under reducing conditions at approx. 35 kDa, and was estimated to be 95% homogeneous. The specific API activity after dialysis was 25 units/mg.
Figure 1. Purification of recombinant KpsF.
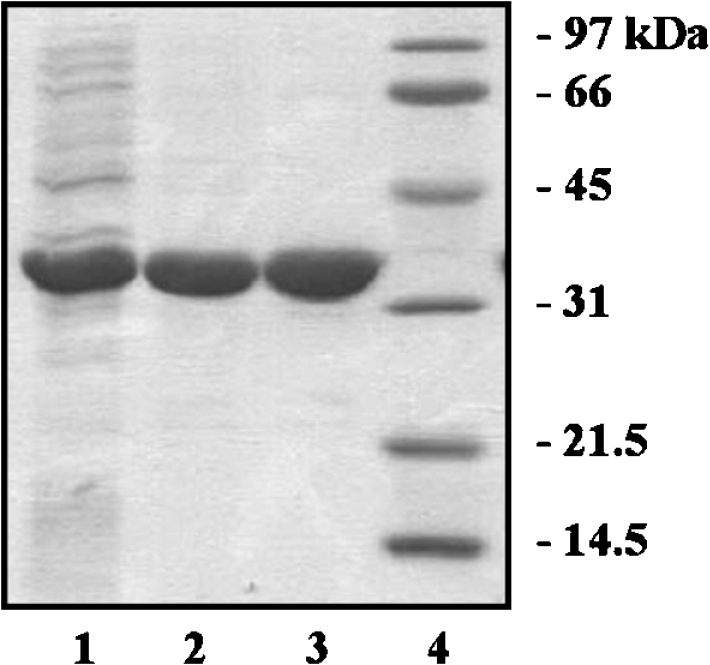
Approximately 10 μg of protein was loaded on to an SDS/12% polyacrylamide gel from each step of the purification. Lane 1, crude extract after sonication; lane 2, pooled fractions after Q-Sepharose anion-exchange chromatography; lane 3, post ammonium sulphate precipitation and dialysis; lane 4, molecular mass standards.
Molecular mass determinations
MALDI-MS indicated the presence of a single protein species (M+1 35447 and M+2 17767), agreeing well with the calculated mass (35486 Da), and suggesting that the internal methionine is indeed the correct native N-terminus. The native mass as determined by gel filtration was 145±7 kDa, indicating that the quaternary structure of KpsF is tetrameric (∼4.1 subunits).
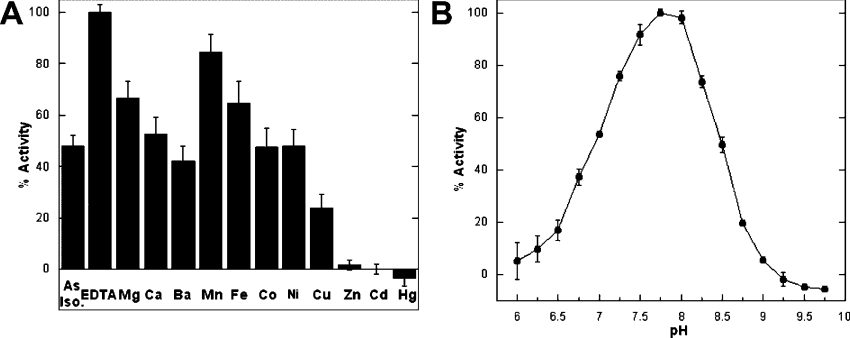
Effect of metals and pH on the activity of KpsF
The metal content of KpsF as isolated was 0.85±0.1 mol of bivalent metal cation per subunit, as determined by high-resolution inductively coupled plasma-MS, approx. 95% of which was Ca2+ and Zn2+ ions. Samples that had been treated with EDTA and subsequently isolated by gel filtration contained only trace amounts of metals, including both Ca2+ and Zn2+ (<0.01 mol per monomer), indicating that EDTA effectively chelates KpsF-bound metal cations.
As KpsF co-purified with nearly a stoichiometric amount of metal ions, the effect of bivalent metals on activity was investigated. The addition of EDTA to KpsF as isolated to remove the stoichiometric amount of bound cations, without removal of either the EDTA–metal complex or excess free EDTA, increased the observed activity approx. 2-fold (Figure 2A). The inclusion of surplus amounts of the late-transition metals (10 μM Zn2+, Cd2+ and Hg2+) significantly inhibited the activity of KpsF, though this inhibition could be reversed in situ by titration of the KpsF-bound metal with excess EDTA. In contrast, addition of certain other metals (Mg2+, Ca2+, Ba2+, Mn2+, Fe2+ and Co2+) either had no appreciable effect on activity or was slightly stimulating. In order to maintain KpsF metal-free under the variety of assay conditions used to determine the optimum pH, substrate specificity and kinetic parameters, 1 mM EDTA was included in all reactions to chelate any bivalent metals.
Figure 2. Effects of metals and pH on the activity of recombinant KpsF.
(A) Metal analysis of recombinant KpsF. Activity of KpsF as isolated was measured at 37 °C in the presence of various metals or EDTA (50 mM Tris/HCl, pH 7.8, 80 nM KpsF, 10 mM A5P and 10 μM metal or EDTA). ‘As Iso’, as isolated. (B) The pH versus rate profile of recombinant KpsF (100 mM BTP, 80 nM KpsF, 1 mM EDTA and 10 mM A5P). All pH values were measured at 37 °C.
A plot of apparent maximum velocity versus pH produced a bell-shaped activity profile (Figure 2B), consistent with a diprotic mechanism involving both an acidic and a basic active site residue. The pH optimum was narrow and determined to be close to physiological pH at 7.75.
Substrate specificity
A panel of naturally abundant phosphorylated pentoses and hexoses were tested as potential substrates for KpsF (Table 1). However, D-ribose 5-phosphate, D-glucose 6-phosphate, D-glucose 1-phosphate, D-glucosamine 6-phosphate and D-mannose 6-phosphate were not converted into their respective ketoses as monitored by an increase in the ratio of absorbance at 540 nm of the sample to that of the control. D-Arabinose was also not a substrate, indicating that there is a requirement for phosphorylation. Within the limits of detection of the assay, KpsF thus appears to be a specific phospho-sugar aldo/keto isomerase catalysing the interconversion of A5P and Ru5P.
Table 1. Substrate specificity of recombinant E. coli KpsF.
Substrates were assayed at 10 mM with 1 mM EDTA in 50 mM Tris/HCl (pH 7.8) for 10 min with 90 nM KpsF at 37 °C.
| Substrate | Absorbance ratio* |
|---|---|
| A5P | 2.5±0.1 |
| D-Ribose 5-phosphate | <1 |
| D-Arabinose | 0.96±0.06 |
| D-Glucose 6-phosphate | 1.01±0.05 |
| D-Mannose 6-phosphate | 0.94±0.06 |
| α-D-Glucose 1-phosphate | 1.00±0.09 |
| D-Glucosamine 6-phosphate | 1.01±0.01 |
* Ratio of absorbance at 540 nm of the sample to that of the control.
Kinetic parameters
Standard Michaelis–Menten kinetic analysis was applied to KpsF in order to determine the catalytic constants in both the forward (A5P to Ru5P) and the reverse (Ru5P to A5P) directions. The Km was 0.57±0.04 mM for A5P and 0.30±0.03 mM for Ru5P, while the kcat in the A5P to Ru5P direction was determined to be 15±1 and 19±2 s−1 in the Ru5P to A5P direction. The equilibrium constant was measured by integration of the 31P NMR peaks (results not shown). The equilibrium lies well in favour of A5P, with the Keq being equal to 0.48±0.02 ([Ru5P]/[A5P]). The measured Keq constant was in agreement, within error, with the calculated Keq using the kinetic constants as defined by the Haldane equation for reversible reactions (Kcalceq=0.42).
DISCUSSION
The role of KpsF, and Kdo in general, within group 2 capsule biogenesis has not been fully elucidated. In order to begin to understand better the function of Kdo, the kpsF gene from the uropathogenic strain E. coli CFT073 (O6:K2:H1) was cloned and characterized. KpsF was highly overexpressed as a soluble cytoplasmic protein (Figure 1). Previous attempts to overexpress the now identified KpsF reading frame cloned from E. coli K1 resulted in the nearly complete partitioning of recombinant protein into the membrane pellet, and thus required urea solubilization for purification [19]. In contrast, the vast majority of recombinant KpsF was readily soluble using the expression conditions reported here. KpsF certainly may be membrane-associated as was suggested [19], either through non-specific hydrophobic interactions with the membrane surface or perhaps through specific protein–protein interactions within a complex involving other integral membrane Kps proteins. As no other kps genes were co-overexpressed, KpsF may remain soluble without the presence of these Kps proteins to recruit it to the membrane, or simply once all the non-specific membrane association sites are filled, KpsF remains cytoplasmic. The difference may also stem from the fact that we did not use a cloning vector construct with a His tag because of the marked propensity of bivalent cations to exacerbate aggregation of KpsF.
Recombinant KpsF consistently co-purified with substantial quantities of bivalent cations, which could be removed in situ with excess EDTA chelating reagent. As the enzymatic activity actually increased upon sequestration of these cations, there does not appear to be a metal cofactor requirement for KpsF. The observed activity of the untreated enzyme preparation did slightly increase with certain other metals, particularly Mn2+, though we attribute this to displacement of more inhibitory Zn2+ cations that had fortuitously bound during purification. A protein-stabilizing, auxiliary, or perhaps even regulatory role for the metal-binding site cannot be ruled out at this point, however. Reversible zinc inhibition at low micromolar concentrations has been observed in many other enzymes, and it has been proposed that metal ion inhibition may represent an under-appreciated post-transcriptional control mechanism in vivo [20]. It is unclear if the observed zinc inhibition in KpsF is biologically relevant, particularly when the cytoplasmic concentration of free unsequestered zinc in E. coli has been estimated to be in the femtomolar range [21].
The biochemical properties of KpsF are consistent with its identification as an API, and would suggest no other catalytic function. KpsF displayed reasonable affinity for both A5P (Km=0.57±0.04 mM) and Ru5P (Km=0.30±0.03 mM), and was specific for only these two substrates among a panel of other phosphorylated monosaccharides (Table 1). E. coli strains expressing group 2 capsules thus have three distinct API genes; kpsF from the K-antigen cluster (K-API), in addition to the two conserved API genes present in all E. coli strains {kdsD from the LPS (L-API) biosynthetic pathway [15] and gutQ from D-glucitol phosphotransferase operon (G-API) [22]}. While all three API paralogues catalyse the same reaction, a survey of their biochemical properties indicates that L-API and G-API are more similar to each other than to K-API (Table 2). Of particular difference are the values for the catalytic constant, with kcat in each direction being more than 10-fold lower for K-API. The measured kcat values were consistently lower across multiple independent protein preparations, regardless of the purification method. Further, measured activities in the crude KpsF extracts immediately after cell disruption were significantly lower than with the other APIs, suggesting that the decrease in catalytic efficiency is indeed real and not an artifact caused by differences in protein stability or purity. All three API proteins share greater than 40% sequence identity, and yet the kcat for K-API is an order of magnitude lower. A similar decrease in catalytic efficiency was noted for the only other Kdo pathway enzyme on the kps operon, KpsU (CMP Kdo synthetase) from E. coli K5 [23]. When KpsU was compared with its LPS biosynthetic counterpart (KdsB), Km values were 10-fold higher and the Vmax 100-fold lower.
Table 2. Biochemical properties of API paralogues from E. coli.
| E. coli API paralogue | |||
|---|---|---|---|
| Property | L-API* | G-API† | K-API |
| Km (A5P) | 0.61±0.06 mM | 1.2±0.1 mM | 0.57±0.04 mM |
| Km (Ru5P) | 0.35±0.08 mM | 0.64±0.08 mM | 0.30±0.03 mM |
| kcat (A5P to Ru5P) | 157±4 s−1 | 218±4 s−1 | 15±1 s−1 |
| kcat (Ru5P to A5P) | 255±16 s−1 | 242±11 s−1 | 19±2 s−1 |
| Keq (calc.)‡ | 0.47 (0.35) | 0.47 (0.48) | 0.48 (0.42) |
| Optimum pH | 8.4 | 8.25 | 7.75 |
| Specific for A5P/Ru5P§ | Yes | Yes | Yes |
| Zn2+/subunit∥ | 1.0±0.1 mol | 1.4±0.2 mol | 0.85±0.1 mol |
| Inhibition by 10 μM Zn2+¶ | Yes | Yes | Yes |
| Activation by 10 μM EDTA | Yes | Yes | Yes |
| Subunit mass** (calc.) | 35104 Da (35196 Da) | 33909 Da (34031 Da) | 35447 Da (35487 Da) |
| Native mass†† | 122±5 kDa (tetramer) | 133±4 kDa (tetramer) | 145±7 kDa (tetramer) |
* Data from [15].
† Data from [21].
‡ Measured by 31P NMR (calculated from Haldane relationship [Ru5P]/[A5P]).
§ See the Experimental section for tested substrates.
∥ Amount of Zn2+ per monomer as determined by high-resolution inductively coupled plasma-MS.
¶ Less than 5% activity remaining.
** Determined by electrospray ionization or MALDI-MS; calculated from protein sequence.
†† Determined by gel filtration.
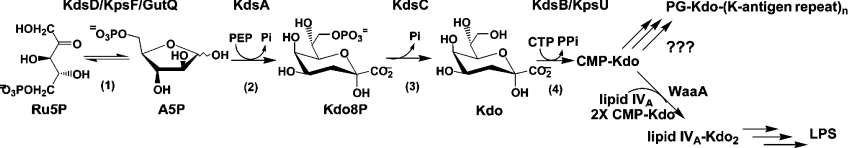
Kdo is a carbohydrate constituent of numerous group 2 capsule serotypes [24]. A single Kdo molecule can be found as the reducing sugar linking the K-antigen polysaccharide to the lipid α-glycerophosphate membrane anchor, as a repeating component of the capsular polysaccharide, or curiously, not at all in certain strains (K1 and K92) [3]. Although Kdo is not in the exported K1 polysaccharide, it may still play a role as an intermediate carrier for the polysaccharide chain, which is then subsequently displaced during the ligation of the lipid, as was proposed for the related capsule system from Neisseria [13]. Despite the prominent role of Kdo in capsule biogenesis, only two Kdo-related genes (kpsF and kpsU) are located on the kps operon (Scheme 1). Neither Kdo synthetase [25] nor Kdo phosphatase [26] is duplicated. Genetic studies in E. coli K-12 K5 have shown that KpsF and KpsU are not essential for capsule synthesis [27,28]. Paradoxically, a ΔkpsF mutant in an E. coli K-12 K1 hybrid resulted in an intermediate phenotype, where there was a 10-fold decrease in capsule polymer being transported to the surface [19]. Failure to clearly observe the ΔkpsF/ΔkpsU phenotypes was reasonably attributed to partial complementation by their homologues. Further, we would expect that a clear phenotype will only be manifested when the capsule biosynthetic demand for Kdo exceeds the capacity of the LPS pathway enzymes to supply it. These apparent discrepancies may therefore stem from differences in the kinetics of polysaccharide assembly apparatus which are unique to each individual K-antigen, i.e. intracellular pools of Kdo in a ΔkpsF construct become limiting for one but not for the other. It would be interesting to construct these same mutants in E. coli serotype strains that have Kdo in the repeat unit, where 40–60% of the capsular polysaccharide mass consists of Kdo [29]. One would expect that the availability of Kdo would become rate-limiting, and therefore be able to directly observe the ΔkpsF phenotype. It is possible that all group 2 capsules arose from a prototype group 2 strain with Kdo in the repeat unit. Thus post acquisition via horizontal transfer of region 2 genes that do not direct the synthesis of a Kdo-containing polysaccharide, the functions of KpsF and KpsU have become redundant.
Scheme 1. The Kdo biosynthetic pathway.
Enzymes involved are (1) API (KdsD, KpsF and GutQ), (2) Kdo8P synthase (KdsA), (3) Kdo8P phosphatase (KdsC) and (4) CMP-Kdo synthetase (KdsB and KpsU). The LPS branch is initiated with the transfer of CMP-Kdo to lipid IVA acceptor by Kdo transferase (WaaA). Enzymes responsible for incorporating activated CMP-Kdo into the capsule are unknown. PG, phosphatidylglycerol.
Probing the role of Kdo in capsule biogenesis in E. coli is complicated by the presence of complementing genes for both KpsU and KpsF. KpsU was reported to be membrane-associated [23], and perhaps may be a component of a Kps protein complex. An argument could be made that KpsU is present in order to generate the activated sugar nucleotide CMP-Kdo ‘on-site’ within the immediate proximity of the Kps protein complex in order to accommodate the remarkably short half-life of CMP-Kdo [30]. Further, the decreased catalytic competency of KpsU may ensure that if Kdo pools ever do become limited, there is a hierarchal distribution of Kdo in favour of KdsB, and thus to the essential LPS biosynthesis pathway. The presence of K-API may reflect the need to supplement the basal level of A5P produced during the capsule production phase, suggesting that the total intracellular API level dictates the net flux through the Kdo pathway. The existence of an alternative specific role for K-API in protein–protein interactions, as either a stabilizing component or signalling mechanism, cannot be ignored. However, the high sequence identity over the entire length of the reading frame between the API paralogues would suggest that if such a role exists, they would be able to complement each other in this hypothetical function along with the established catalytic one. We recently constructed a viable ΔKdo strain in E. coli K-12 [31]. The construction of group 2 capsule hybrids in this host will hopefully enable further probing of the role of KpsF and Kdo in capsule biogenesis.
Acknowledgments
This research was supported by the Pfizer Fellowship in Medicinal Chemistry (T. C. M.) and the Chemical Biology Interface (CBI) training grant (T. C. M.). We thank Professor Harry L. T. Mobley for providing E. coli CFT073 and Professor Ted Houston (Department of Geology, University of Michigan) at the W. M. Keck Elemental Geochemistry Laboratory for measuring metal content. We also thank other members of the Woodard group for helpful discussions.
References
- 1.Orskov F., Orskov I. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 1992;38:699–704. [PubMed] [Google Scholar]
- 2.Moxon E. R., Kroll J. S. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield C., Roberts I. S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 4.Roberts I. S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 5.Bliss J. M., Silver R. P. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol. Microbiol. 1996;21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 6.Rowe S., Hodson N., Griffiths G., Roberts I. S. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E coli. J. Bacteriol. 2000;182:2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson D. A., Hammarton T. C., Roberts I. S. Transcriptional organization and regulation of expression of region 1 of the Escherichia coli K5 capsule gene cluster. J. Bacteriol. 1996;178:6466–6474. doi: 10.1128/jb.178.22.6466-6474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens M. P., Clarke B. R., Roberts I. S. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol. Microbiol. 1997;24:1001–1012. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- 9.Cieslewicz M., Vimr E. Thermoregulation of kpsF, the first region 1 gene in the kps locus for polysialic acid biosynthesis in Escherichia coli K1. J. Bacteriol. 1996;178:3212–3220. doi: 10.1128/jb.178.11.3212-3220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt M. A., Jann K. Phospholipid substitution of capsular (K) polysaccharide antigens from Escherichia coli causing extra-intestinal infections. FEMS Microbiol. Lett. 1982;14:69–74. [Google Scholar]
- 11.Raetz C. R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finke A., Jann B., Jann K. CMP-KDO-synthetase activity in Escherichia coli expressing capsular polysaccharides. FEMS Microbiol. Lett. 1990;57:129–133. doi: 10.1016/0378-1097(90)90426-q. [DOI] [PubMed] [Google Scholar]
- 13.Tzeng Y. L., Datta A., Strole C., Kolli V. S., Birck M. R., Taylor W. P., Carlson R. W., Woodard R. W., Stephens D. S. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-D-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J. Biol. Chem. 2002;277:24103–24113. doi: 10.1074/jbc.M200931200. [DOI] [PubMed] [Google Scholar]
- 14.Tan L., Darby C. Yersinia pestis is viable with endotoxin composed of only lipid A. J. Bacteriol. 2005;187:6599–6600. doi: 10.1128/JB.187.18.6599-6600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meredith T. C., Woodard R. W. Escherichia coli YrbH is a D-arabinose 5-phosphate isomerase. J. Biol. Chem. 2003;278:32771–32777. doi: 10.1074/jbc.M303661200. [DOI] [PubMed] [Google Scholar]
- 16.Welch R. A., Burland V., Plunkett G., III, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dische Z., Borenfreund E. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J. Biol. Chem. 1951;192:583–587. [PubMed] [Google Scholar]
- 18.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 19.Cieslewicz M., Vimr E. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol. Microbiol. 1997;26:237–249. doi: 10.1046/j.1365-2958.1997.5651942.x. [DOI] [PubMed] [Google Scholar]
- 20.Maret W., Jacob C., Vallee B. L., Fischer E. H. Inhibitory sites in enzymes: zinc removal and reactivation by thionine. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1936–1940. doi: 10.1073/pnas.96.5.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Outten C. E., O'Halloran T. V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 22.Meredith T. C., Woodard R. W. Identification of GutQ from Escherichia coli as a D-arabinose 5-phosphate isomerase. J. Bacteriol. 2005;187:6936–6942. doi: 10.1128/JB.187.20.6936-6942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenow C., Roberts I. S., Jann K. Isolation from recombinant Escherichia coli and characterization of CMP-Kdo synthetase, involved in the expression of the capsular K5 polysaccharide (K-CKS) FEMS Microbiol. Lett. 1995;125:159–164. doi: 10.1111/j.1574-6968.1995.tb07352.x. [DOI] [PubMed] [Google Scholar]
- 24.Jann B., Jann K. Structure and biosynthesis of the capsular antigens of Escherichia coli. Curr. Top. Microbiol. Immunol. 1990;150:19–42. doi: 10.1007/978-3-642-74694-9_2. [DOI] [PubMed] [Google Scholar]
- 25.Dotson G. D., Nanjappan P., Reily M. D., Woodard R. W. Stereochemistry of 3-deoxyoctulosonate 8-phosphate synthase. Biochemistry. 1993;32:12392–12397. doi: 10.1021/bi00097a017. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Woodard R. W. Escherichia coli yrbI is 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase. J. Biol. Chem. 2003;278:18117–18123. doi: 10.1074/jbc.M301983200. [DOI] [PubMed] [Google Scholar]
- 27.Bronner D., Sieberth V., Pazzani C., Smith A., Boulnois G., Roberts I., Jann B., Jann K. Synthesis of the K5 (group II) capsular polysaccharide in transport-deficient recombinant Escherichia coli. FEMS Microbiol. Lett. 1993;113:279–284. doi: 10.1111/j.1574-6968.1993.tb06527.x. [DOI] [PubMed] [Google Scholar]
- 28.Bronner D., Sieberth V., Pazzani C., Roberts I. S., Boulnois G. J., Jann B., Jann K. Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the K5 gene cluster. J. Bacteriol. 1993;175:5984–5992. doi: 10.1128/jb.175.18.5984-5992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jann K., Jann B. Capsules of Escherichia coli. In: Sussman M., editor. Escherichia coli: Mechanisms of Virulence. Cambridge, U.K.: Cambridge University Press; 1997. pp. 113–143. [Google Scholar]
- 30.Lin C. H., Murray B. W., Ollmann I. R., Wong C. H. Why is CMP-ketodeoxyoctonate highly unstable? Biochemistry. 1997;36:780–785. doi: 10.1021/bi962055c. [DOI] [PubMed] [Google Scholar]
- 31.Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 2006 doi: 10.1021/cb0500015. in the press. [DOI] [PubMed] [Google Scholar]