Abstract
Myosin heavy-chain kinase A (MHCK A) catalyses the disassembly of myosin II filaments in Dictyostelium cells via myosin II heavy-chain phosphorylation. MHCK A possesses a ‘coiled-coil’-enriched domain that mediates the oligomerization, cellular localization and actin-binding activities of the kinase. F-actin (filamentous actin) binding by the coiled-coil domain leads to a 40-fold increase in MHCK A activity. In the present study we examined the actin-binding characteristics of the coiled-coil domain as a means of identifying mechanisms by which MHCK A-mediated disassembly of myosin II filaments can be regulated in the cell. Co-sedimentation assays revealed that the coiled-coil domain of MHCK A binds co-operatively to F-actin with an apparent KD of approx. 0.5 μM and a stoichiometry of approx. 5:1 [actin/C(1–498)]. Further analyses indicate that the coiled-coil domain binds along the length of the actin filament and possesses at least two actin-binding regions. Quite surprisingly, we found that the coiled-coil domain cross-links actin filaments into bundles, indicating that MHCK A can affect the cytoskeleton in two important ways: (1) by driving myosin II-filament disassembly via myosin II heavy-chain phosphorylation, and (2) by cross-linking/bundling actin filaments. This discovery, along with other supporting data, suggests a model in which MHCK A-mediated bundling of actin filaments plays a central role in the recruitment and activation of the kinase at specific sites in the cell. Ultimately this provides a means for achieving the robust and highly localized disruption of myosin II filaments that facilitates polarized changes in cell shape during processes such as chemotaxis, cytokinesis and multicellular development.
Keywords: actin-binding domain, cross-linking, Dictyostelium, filamentous actin (F-actin), myosin II, myosin heavy-chain kinase (MHCK)
Abbreviations: DSS, disuccinyl suberate; DTT, dithiothreitol; F-actin, filamentous actin; GFP, green fluorescent protein; GST, glutathione S-transferase; MHCK, myosin heavy-chain kinase
INTRODUCTION
Myosin II is a molecular motor that plays a central role in driving the highly localized and tightly co-ordinated contraction of actin filaments that is associated with critical cellular processes such as cytokinesis, directed cell locomotion, intracellular trafficking and maintenance of cell shape [1–5]. The role of myosin II in these cellular processes is often reflected in its specific localization in the cell. For example, myosin II is enriched at the rear of migrating cells, where it drives contraction and movement of the cell body forward [1], and at the cleavage furrow of a dividing cell, where it facilitates division and separation of the resulting daughter cells [6]. Regardless of the cellular process or its localization, myosin II must first assemble into bipolar filaments before it can mediate the contraction of actin filaments [7]. This requirement highlights the importance of understanding not only the basic processes controlling myosin II-filament assembly, but also how variations in such regulatory mechanisms can ultimately specify which of the diverse roles myosin II plays in the highly dynamic environment of a non-muscle cell.
In the amoeba Dictyostelium, myosin II heavy-chain phosphorylation regulates the dynamic equilibrium between a cytoplasmic pool of myosin II monomers and a cytoskeleton-associated assembly of myosin II bipolar filaments [8]. More specifically, the phosphorylation of three specific threonine residues (amino acids 1823, 1833 and 2029) at the C-terminal end of the myosin II heavy chain drives the disassembly of myosin II filaments into soluble monomers [9]. These regulatory sites were identified, in part, as a consequence of mapping the myosin II heavy-chain phosphorylation sites that are targeted by the enzyme, MHCK A (myosin heavy-chain kinase A) [10]. MHCK A was the first kinase shown to catalyse myosin II-filament disassembly both in vitro [11] and in vivo [12] and is the most extensively studied of the MHCKs that have been identified in Dictyostelium. It is particularly noteworthy that the cloning and analysis of the MHCK A gene led to the discovery of a new and functionally diverse family of protein kinases (α-kinases) that share homologous catalytic domains and are found in a broad range of eukaryotic organisms and cell types [13,14]. While there are at least six members of the α-kinase family in Dictyostelium, recent studies by Yumura et al. [15] indicate that only three (MHCK A, MHCK B [16] and MHCK C [17,18]) appear to have physiologically significant functions in regulating myosin II-filament disassembly in the cell.
Upon initial examination, MHCK A, B and C appear to represent functionally redundant activities because they share a common ability to drive myosin II-filament disassembly in the cell; however, several studies indicate that these enzymes may actually play distinct roles in regulating different aspects of myosin II-dependent activities of the cell [15,17,18]. The functional differences among the MHCKs are reflected, in part, by their distinct localization patterns during dynamic cellular processes such as cell migration and cytokinesis. In a migrating cell, for example, MHCK A is preferentially enriched at sites of pseudopod formation at the cell anterior, where the kinase is expected to catalyse the disassembly of myosin II-based cross-linking of actin filaments which, in turn, may facilitate the reorganization of the actin cytoskeleton necessary for the formation of cellular extensions [19]. In contrast, MHCK C co-localizes with myosin II at the trailing edge of locomoting cells, suggesting a role for this enzyme in the co-ordinated turnover of myosin II filaments mediating contraction of the cell rear [17,18]. At a structural level, the three kinases share similar catalytic and C-terminal WD-repeat-motif domains. Studies of recombinant MHCK A and MHCK B have revealed that their WD-repeat domains facilitate myosin II heavy-chain phosphorylation by physically targeting the kinases to myosin II bipolar filaments [20]; it is not known if the WD (Trp/Asp)-repeat domain of MHCK C serves a similar function. Of the three kinases, only MHCK A possesses an extensive region of predicted coiled-coil structure [13]; our studies of the novel actin-binding characteristics of this domain are the primary focus of the present paper.
The loosely termed ‘coiled-coil’ domain of MHCK A is located at the N-terminal end of the kinase and is defined by a stretch of approx. 500 amino acids that possess a high propensity to form coiled-coil structures interspersed with short, relatively unstructured regions of the protein (see Figure 1). It is important to note that the first 130 amino acids of the coiled-coil domain are actually not predicted to fold into coiled-coil structure; thus a central goal of the work described in the present paper is to better characterize the functions of the structurally distinct regions of the coiled-coil domain of MHCK A. In general terms, the coiled-coil is a widespread structural motif formed when two or more complementary coiled-coil regions wrap around each other to form a higher-ordered ‘supercoiled’ structure [21]. The broad range of protein activities requiring coiled-coil domains can be attributed mainly to the well-characterized ability of these domains to form multimeric assemblies [22]. Early structure–function studies of MHCK A demonstrated a fairly predictable role for the coiled-coil domain, in the formation of MHCK A homo-oligomers containing three to five MHCK A molecules [23]. Subsequent biochemical and cellular studies have revealed that this domain is a functionally dynamic region of the kinase that plays direct roles in regulating the cellular and biochemical activities of the MHCK A enzyme [24,25]. More specifically, we found that the coiled-coil domain of MHCK A is both necessary and sufficient to mediate translocation of the kinase to the actin-rich cortex of Dictyostelium cells [24]. We have also shown that the coiled-coil domain binds directly to actin filaments and that this interaction leads to a 40-fold increase in MHCK A catalytic activity [25]. These results are particularly notable because they represent the identification of a novel actin-binding domain exhibiting no detectable sequence identity with any known actin-binding proteins. In a broader context, these studies support a model in which the coiled-coil domain of MHCK A mediates the recruitment of the kinase to actin-rich sites in the cell where there is an increased possibility of a direct interaction between the coiled-coil domain and actin filaments. The resulting activation of MHCK A is proposed to drive the highly localized disassembly of myosin II filaments that is required for specific changes in cell shape.
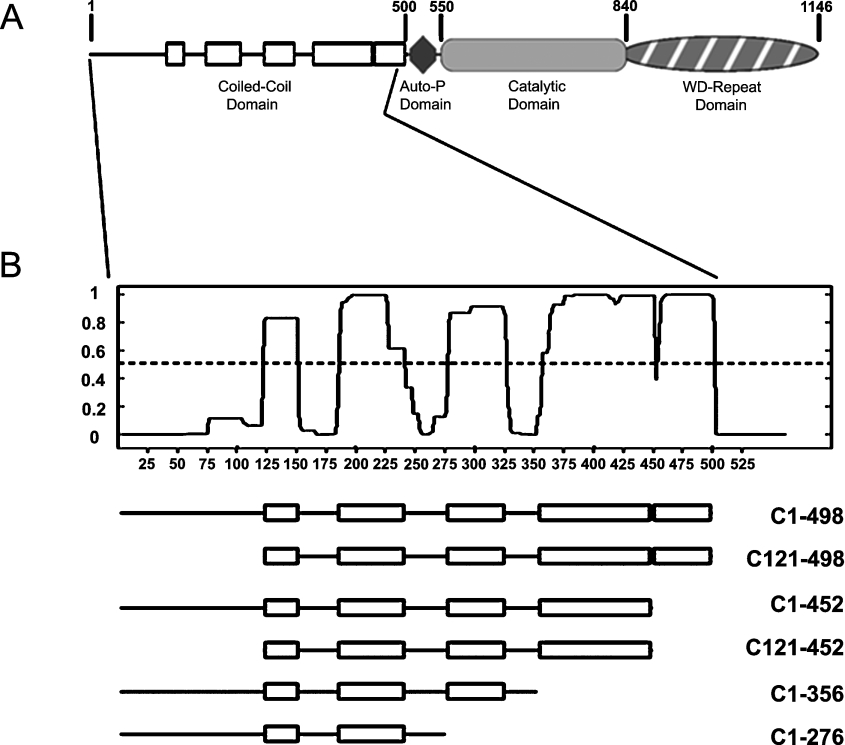
Figure 1. Structural domains of MHCK A.
(A) Unshaded rectangles represent regions in the N-terminal coiled-coil domain (amino acids 1–500) exhibiting a high probability (>0.5) to fold into left-handed α-helical coiled-coil structures based upon analysis of the sequence with the COILS algorithm [35]. The diamond represents the putative autophosphorylation (Auto-P) domain of the kinase. The central catalytic domain is the prototype for the α-kinase family of novel serine/threonine kinases [13]. The C-terminal WD-repeat domain contains a 7-fold WD-repeat motif that is characteristic of the β-subunit of heterotrimeric G-proteins; this domain physically targets MHCK A to phosphorylate myosin II filaments [20]. The amino acid numbering indicated corresponds to GenBank™ accession number P42527 for MHCK A. (B) The graph shows the probability (y-axis) of each residue contributing to coiled-coil formation as a function of its position in a 21-residue scanning window and with respect to its overall sequence (x-axis values). Below the graph are schematic representations of MHCK A coiled-coil-domain truncations used in the studies described in the present paper. The positions of the coiled-coil regions (rectangles) and non-coil regions (lines) are based on the coiled-coil prediction analysis represented in the plot.
The experiments described in the present paper were designed to explore the actin-binding properties of the coiled-coil domain as a means of gaining insight into the mechanisms by which the catalytic and cellular translocation activities of MHCK A might be regulated in the cell. Our analyses revealed that the coiled-coil domain of MHCK A exhibits co-operative binding to F-actin and may bind lengthways along the actin filament because it competes with tropomyosin for actin binding. Further studies led to the highly interesting discovery that the coiled-coil domain of MHCK A not only binds to F-actin but also cross-links actin filaments into bundle-like structures. This unexpected result indicates that MHCK A, when recruited to actin-rich sites of the cell cortex, has the potential to alter the actin–myosin cytoskeleton in two important ways: (1) by driving myosin II-filament disassembly via myosin II heavy-chain phosphorylation, and (2) by cross-linking actin filaments into bundles.
EXPERIMENTAL
Dictyostelium cell growth
Dictyostelium MHCK A-null cells [12] expressing GFP (green fluorescent protein)-tagged fusion proteins were grown on plastic Petri dishes (150 mm diameter) containing liquid HL5 medium [26] supplemented with 10 μg/ml Geneticin (G418; Gibco), at 20 °C for 3 days or until the cell density was approx. 1×106 cells/ml. Cells were prepared for chemoattractant-response studies by inducing development to the aggregation stage, as has been described previously [19]. Briefly, growth-phase Dictyostelium cells were collected by centrifugation (at 1200 g for 7 min at 20 °C), washed twice in starvation buffer (20 mM Mes, pH 6.8, 0.2 mM CaCl2, 2 mM MgCl2), and then re-suspended to a density of 4×107 cells/ml. The cell suspension was shaken (150 rev./min) for 2 h at 20 °C and then pulsed every 6 min with 100 nM cAMP for 6–8 h using a Branson syringe pump to apply the concentrated cAMP drip. The cells were then brought to a ‘primed’ stimulation state by adding caffeine to a final concentration of 2.5 mM and then shaking (150 rev./min at 20 °C) for an additional 30 min before stimulation with cAMP for translocation studies.
Construction of recombinant plasmids
All amino acid numbering in this text refers to the GenBank™ accession number P42527 for MHCK A [13]. The recombinant plasmid for bacterial expression of the entire coiled-coil domain [C(1–498)] with an N-terminal GST (glutathione S-transferase) tag (pGEX-2T vector; Amersham Biosciences) has been reported previously [25]. This same method was used for cloning all of the coiled-coil truncations into the pGEX-2T vector to be used in the present study. Briefly, the truncations described in Figure 1 were amplified by standard PCR methodology using full-length MHCK A gene as template, and in-frame primers containing BamHI sites for ligation into expression plasmids. Subsequent manipulations followed the same methods described in detail previously [25], and involved the initial cloning of PCR product into pDrive vector (Qiagen) and subsequent subcloning of truncations from pDrive into the pGEX-2T expression vector. Plasmids were maintained in NovaBlue bacterial cells (Novagen). All GFP-tagged fusion constructs were based on the expression vector pTX–GFP [27], and involved the same cloning and subcloning manipulations of coiled-coil-domain truncations mentioned above for the production of recombinant GST-based plasmids. All the plasmid inserts were sequenced to ensure that they did not contain errors.
Protein expression and purification
The bacterial expression and affinity purification of all of the GST-tagged fusion proteins used in these studies were performed as described previously for the GST-tagged coiled-coil domain [GST–C(1–498)] [25]. Purified proteins were dialysed in buffer [20 mM Tris/HCl, pH 7.5, 20% (v/v) glycerol, 70 mM NaCl, 1 mM EDTA, and 1 mM DTT (dithiothreitol)] and stored for up to 3 months at −80 °C, without apparent breakdown or loss of activity. Removal of the GST tag from GST–C(1–498) involved binding bacterially expressed fusion protein to glutathione–agarose beads (Novagen; diameter ranged from approx. 40–165 μm) and then removing unbound cellular materials by washing the beads thoroughly with 20 column vol. [100 mm height×10 mm internal diameter (Pierce Biotechnology, Rockford, IL, U.S.A.)] of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). The bead-bound GST–C(1–498) was washed with 2 column vol. of thrombin cleavage (20 mM Tris/HCl, pH 8.4, 150 mM NaCl, 2.5 mM CaCl2) and then resuspended to a 50% slurry with the same buffer. Biotinylated thrombin (6 units; Thrombin Cleavage Capture Kit, Novagen) was added to the bead slurry and the mixture was incubated for 12 h at 4 °C with rotation (12 rev./min). After incubation, the unbound material containing C(1–498) protein (minus GST tag) was separated from the beads via filtration through a standard chromatography column (100 mm×10 mm) and the collected material was incubated (1 h at 4 °C with rotation at 12 rev./min) with streptavidin–agarose beads to bind the biotinylated thrombin. The bead-bound thrombin was removed by filtration and the soluble fraction [containing C(1–498) protein] was dialysed and then stored as described above for the purified GST-tagged fusion proteins.
Purified proteins were quantified by subjecting the protein preparations, along with a series of known amounts of BSA, to SDS/PAGE (10% gels). After Coomassie-Blue (Sigma–Aldrich) staining, the protein bands were quantified by scanning densitometry and the concentration of each purified fusion protein was determined by comparing the densitometry values for the purified fusion proteins with those of the BSA standards. The level of purity for each of the GST-tagged fusion proteins was greater than 95% as determined from Coomassie-Blue-stained SDS/PAGE gels of the purified samples. In addition, immunoblot analyses using anti-GST (Novagen) and anti-(MHCK A) antibodies [20] revealed that the fusion proteins did not show significant breakdown upon purification (results not shown). Full-length MHCK A was over-expressed in Dictyostelium cells and purified to homogeneity as described previously [23].
For translocation studies, recombinant plasmids for the expression of GFP-tagged fusion proteins were electroporated into MHCK A null cells [12] as described previously [19]. Cells containing the expression plasmid of interest were selected by growth in liquid HL5 medium containing G418 (10 μg/ml). The expression of each GFP-tagged truncation was confirmed visually by GFP fluorescence of transformed cells and by immunoblot analyses of cell lysates using anti-GFP polyclonal antibody (Molecular Probes™).
Actin-binding assays
Actin co-sedimentation assays were performed as described previously [25]. Briefly, rabbit skeletal-muscle actin (Cytoskeleton, Inc.) was induced to polymerize by the addition of 100 mM KCl, 2 mM MgCl2, and 1 mM ATP from a concentrated stock. Purified coiled-coil-domain proteins were incubated (15 min at 20 °C) with F-actin (3 μM) in a 50 μl reaction mixture containing 5 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.5 mM DTT, 100 mM KCl, 2 mM MgCl2, 1 mM ATP, and 1×Complete (Mini) Protease Inhibitor Cocktail (Roche Diagnostics). Negative-control reactions contained all the components of the assay mixture except F-actin. After incubation, reaction mixtures were centrifuged at 100000 g for 20 min at 4 °C to sediment actin filaments. Equal volumes of the resulting pellet and supernatant fractions were resolved by SDS/PAGE (10% gels) followed by Coomassie-Blue staining, to detect the actin and GST-tagged fusion proteins present in each fraction. The level of fusion protein co-sedimentation with F-actin was quantified via scanning densitometry of stained gels.
Co-sedimentation experiments for assessing competition between GST–C(1–498) and tropomyosin were performed essentially as described by Pacholsky et al. [28] for studies of the muscle protein Xin. Purified tropomyosin isolated from porcine skeletal muscle was obtained as a lyophilized powder (Sigma–Aldrich) and contained tropomyosin isoforms 1 and 2 [29,30]. The competition-reaction mixture contained 3 μM F-actin, 3 μM GST–C(1–498) and 1 μM tropomyosin. Positive-control reactions involved incubating 3 μM F-actin with either 3 μM GST–C(1–498) alone or 1 μM tropomyosin alone. Negative-control reactions did not contain F-actin. All reactions were incubated (at 22 °C for 30 min) and centrifuged (at 100000 g for 20 min at 4 °C) and fractions were analysed via SDS/PAGE (10% gels) followed by Coomassie-Blue staining to quantify tropomyosin co-sedimentation with actin filaments.
Chemical cross-linking assay
Untagged C(1–498) protein (2 μM) was incubated (60 min at 25 °C) with 50 μM of the chemical cross-linker DSS (disuccinyl suberate) dissolved in DMSO in a 50 μl reaction mixture containing 1×PBS. In preparation for the cross-linking experiment, the C(1–498) protein was dialysed against PBS to remove any Tris buffer reagent that might interfere with DSS-cross-linking of primary amine groups on adjacent coiled-coil-domain polypeptides. Negative-control reactions contained the same components, except that DMSO vehicle was added in place of DMSO-dissolved DSS. After incubation, the cross-linking reaction was quenched by adding 10 mM Tris, pH 7.2, to the mixture. The reactions were then analysed by SDS/PAGE and Coomassie-Blue staining to visualize a shift of C(1–498) to a slower migrating form upon cross-linking with DSS.
Actin cross-linking assays
The cross-linking activity of the coiled-coil domain was assessed via low-speed centrifugation assays [31] and imaging of fluorescently labelled actin filaments [32–34] incubated with coiled-coil-domain proteins. Low-speed centrifugation assays were prepared, processed and quantified exactly as described for actin co-sedimentation assays except that actin-cross-linking-reaction mixtures were centrifuged at only 8500 g for 20 min at 4 °C. Under these conditions, only F-actin aggregates will sediment [31]. All reaction mixtures contained 3 μM F-actin incubated with GST–C(1–498) (0, 0.3, 1 or 3 μM), 1 μM C(1–498), 1 μM GST–C(121–498), or 1 μM GST–C(1–452). To minimize background sedimentation of coiled-coil-domain proteins, a ‘pre-clearing’ step (centrifugation at 8500 g for 20 min at 4 °C) was performed before adding the coiled-coil-domain proteins to the reaction mixture.
Assays of F-actin organization using fluorescence microscopy involved incubating 3 μM of the indicated GST-tagged fusion protein for 30 min at 20 °C with 3 μM F-actin that contained 10% of the actin filaments saturated with Texas Red® phalloidin (Molecular Probes). Bundles of F-actin labelled in this way can be visualized using fluorescence microscopy with minimal background fluorescence from non-cross-linked actin filaments. After incubation, the reaction mixtures were applied to coverslips and imaged by epifluorescence microscopy using an Olympus IX81 motorized microscope system with one of the following objective lenses: UplanApo, ×20 [NA (numerical aperture) 0.7]; UplanFL, ×40, oil immersion (NA 1.3); or PlanApo, ×60 (NA 1.4).
Cell stimulation and fluorescence microscopy
Approx. 2×105 aggregation-competent, caffeine-treated Dictyostelium cells were transferred to a glass coverslip in a 100 μl vol. of starvation buffer as described previously [19,24]. The cells were stimulated with 100 μM cAMP and epifluorescent images were collected at zero time (before addition of cAMP) and after 30 s and 60 s, with an Olympus IX81 motorized microscope/confocal laser system using an UPlanFL ×40 oil immersion objective lens (NA 1.3).
RESULTS
Previous studies have revealed that the structural determinants for the actin-binding and cellular-translocation activities of MHCK A reside in the coiled-coil domain of the kinase (Figure 1A) [24,25]. This region of MHCK A (amino acids 1–498) comprises five distinct ‘subdomains’ that are each predicted to fold into a coiled-coil conformation; the N-terminal end of this domain contains an ‘unstructured’ stretch of 120 amino acids possessing no predicted structural motifs or patterns [35]. As a means of identifying the minimal region of the coiled-coil domain responsible for MHCK A binding to actin filaments, we constructed a set of truncations designed around the predicted coiled-coil subdomains of the kinase (Figure 1B). These truncations were cloned for expression in bacteria as GST-tagged fusion proteins. The expressed proteins were affinity-purified and then assayed for co-sedimentation with actin filaments.
As a point of reference for mapping the actin-binding domain(s) of MHCK A, we performed initial actin co-sedimentation assays with the full-length coiled-coil domain [C(1–498)], which has been studied previously for its ability to bind F-actin [25]. In the present study, the GST–C(1–498) fusion protein exhibited concentration-dependent binding to actin filaments (Figures 2A and 2B) with an apparent KD of approx. 0.5 μM F-actin, and saturation binding was reached at an actin/GST–C(1–498) stoichiometry of approx. 5:1. These values are within the range of those reported previously [25]. It is noteworthy that the saturation-binding curve of GST–C(1–498) is sigmoidal, signifying that the interaction between the full-length coiled-coil domain and F-actin occurs in a co-operative fashion (Figure 2B, see inset). We also found that full-length coiled-coil-domain protein [C(1–498)] lacking a GST tag still exhibited sigmoidal actin-binding characteristics (Figure 2B). This is an important result because it supports the notion that coiled-coil-domain binding to F-actin is indeed co-operative and that it is not a consequence of altered binding characteristics introduced upon GST tag dimerization [36]. Negative-control assays revealed that GST alone does not co-sediment with F-actin under our experimental conditions (results not shown).
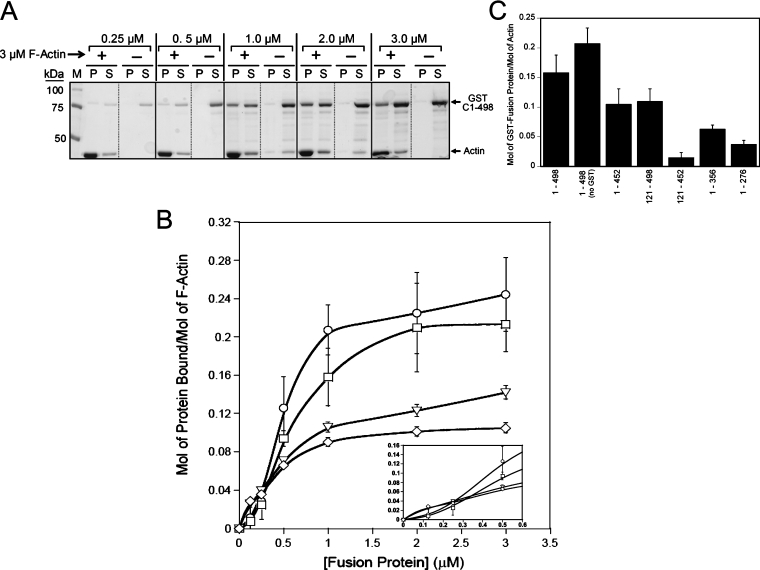
Figure 2. Mapping of the actin-binding region of the coiled-coil domain.
(A) Coomassie Blue-stained SDS-polyacrylamide gel showing the distribution of GST–C(1–498) (at the concentrations indicated above the lanes) in pellet (P) and supernatant (S) fractions in the absence or presence of 3 μM F-actin (compared with marker lane, M). GST–C(1–498) was incubated (at 25 °C for 15 min) with 3 μM rabbit skeletal-muscle F-actin and then subjected to ultracentrifugation as described in the Experimental section, and the resulting P and S fractions were subjected to SDS/PAGE and Coomassie-Blue staining to visualize co-sedimentation with F-actin in the P fraction. (B) Saturation binding curves for full-length and truncated coiled-coil-domain proteins. F-actin binding by different coiled-coil-domain proteins was assessed (as described in the Experimental section) over a range of recombinant protein concentrations (0.5–3.0 μM) and at a constant concentration (3 μM) of rabbit skeletal-muscle F-actin. Co-sedimentation with actin filaments was quantified by scanning densitometry of stained gels and the amount of each recombinant protein co-sedimenting with F-actin was plotted against the concentration of the protein: GST-tagged full-length coiled-coil domain [GST–C(1–498), □], untagged full-length coiled-coil domain [C(1–498), ○] and GST-tagged coiled-coil domain truncations [GST–C(121–498), ○, and GST-C(1–452), ◇]. Values are means±S.E.M. from at least three separate experiments. (C) The actin-binding activity of the coiled-coil domain and its truncations. Each bar represents the level of binding to F-actin (3 μM) for the GST-tagged fusion proteins (1 μM) as indicated. Values are means±S.E.M. from at least three separate experiments.
Mapping of the actin-binding region of the coiled-coil domain involved assessing the actin-binding activities of the truncations listed in Figure 1(B). Analysis of the C(121–498) truncation, which contained all five of the predicted coiled-coil subdomains of MHCK A, revealed that removal of the first 120 amino acids of the coiled-coil domain resulted in an approx. 50% reduction in the capacity to bind to actin filaments (Bmax approx. 0.12 mol/mol of F-actin) (Figure 2B). By contrast, there was no apparent change in the affinity of this truncation for F-actin (apparent KD approx. 0.5 μM). Likewise, a truncation missing the coiled-coil subdomain nearest to the C-terminal [C(1–452)] exhibited a similar reduction in its capacity to bind to F-actin (Bmax approx. 0.10 mol/mol of F-actin), also with no significant change in its affinity for actin (Figure 2B). The hyperbolic saturation binding curves for both the C(121–498) and C(1–452) truncations indicate that their respective interactions with F-actin do not occur in a co-operative manner (Figure 2B, see inset).
Although both the C(121–498) and C(1–452) truncations demonstrate clear (but reduced) binding to actin filaments, a truncation lacking both the N- and C-terminal ends of the coiled-coil domain (C121-452) did not exhibit concentration-dependent co-sedimentation with actin filaments (Figure 2C). In fact, interaction of C121-452 with 3 μM F-actin was not detectable even when the concentration of C121-452 was raised to 4 μM (results not shown). Taken together, these results indicate that the capacity of MHCK A to bind to F-actin is related directly to the presence (or accessibility) of structural determinants residing at the N- and C-termini of the coiled-coil domain. Further analyses of coiled-coil-domain truncations also revealed a general trend in which the sequential removal of coiled-coil subdomains from the C-terminal end of the coiled-coil domain led to progressive decreases in actin-binding capacity (Figure 2C), suggesting that the coiled-coil domain may contain multiple actin-binding sites that correspond to individual coiled-coil subdomains, or that the coiled-coil structure of this domain is important for the proper folding and presentation of the actin-binding region(s). Attempts to generate either shorter C- or N-terminal truncations were not successful in producing soluble and/or intact fusion protein that could be used in actin-binding assays.
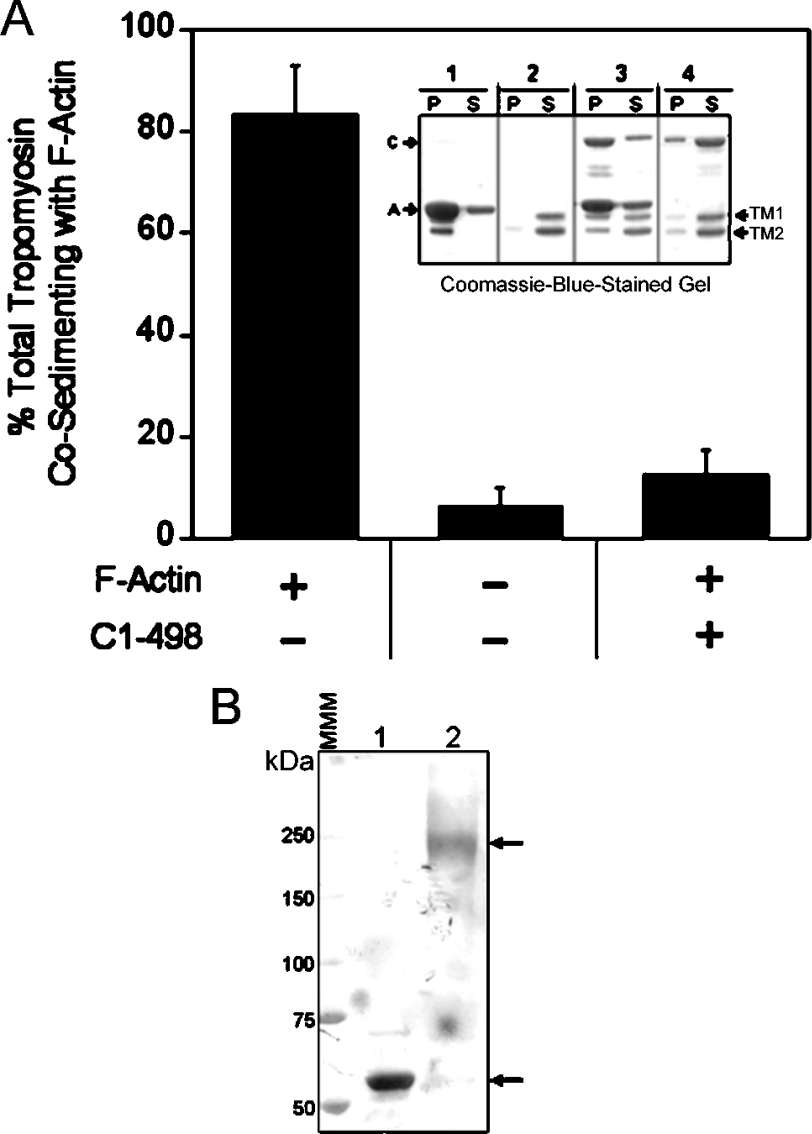
The studies presented above have identified specific regions of the coiled-coil domain (i.e. amino acids 1–120 and 453–498) required for maximal binding to actin filaments. These regions, as well as the entire MHCK A protein, exhibit no significant sequence identity with any known actin-binding proteins; thus the results described here highlight the fact that the coiled-coil domain of MHCK A is indeed a unique actin-binding domain. Despite the lack of identifiable homologues, the fact that a large portion of this domain possesses a high probability of forming α-helical coiled-coil structures (Figure 1; [13,35]) suggests that it may interact with actin filaments in a manner analogous to that of tropomyosin, a well-characterized actin-binding protein that almost entirely comprises coiled-coil structure [37]. To investigate this possibility, we tested the coiled-coil domain of MHCK A (C1–498) for its ability to compete with porcine skeletal-muscle tropomyosin for binding to F-actin (Figure 3A); Dictyostelium does not appear to possess a gene for tropomyosin [38]. For these experiments, tropomyosin (1 μM; TM1 and TM2 isoforms from porcine skeletal muscle; [29,30]) was assayed for co-sedimentation with actin filaments (3 μM) in the presence or absence of C(1–498) protein (3 μM). In the absence of C(1–498), the vast majority (83±9.8%, n=4) of tropomyosin bound to actin filaments. However, inclusion of saturating concentrations of C(1–498) in the assay mixture resulted in a striking decrease in tropomyosin co-sedimentation with F-actin (12.4±4.9%, n=4). This inhibition suggests that the coiled-coil domain of MHCK A may bind lengthways along the actin filament in a manner similar to tropomyosin, and that these two proteins may compete for interaction with the same or overlapping regions on F-actin. Complementary experiments revealed that treatment of purified C(1–498) protein (no GST tag) with the chemical cross-linking agent DSS led to a shift in the electrophoretic migration pattern of C(1–498) from approx. 56 kDa to approx. 240 kDa (Figure 3B). This result indicates that the coiled-coil domain, like tropomyosin, exhibits higher-level structural organization by assembling into oligomers. Unlike tropomyosin, which forms two-stranded coiled-coil structures, the coiled-coil domain of MHCK A appears to form assemblies containing four to five coiled-coil monomers; this is consistent with previous analyses of the oligomerization state of full-length MHCK A, which is proposed to form oligomers of three to six MHCK A molecules [23]. It is noteworthy that other binding assays performed in our laboratory indicate that C(1–498) and tropomyosin do not bind to each other and thus are not likely to form heteromeric complexes via interactions between their coiled-coil regions (results not shown).
Figure 3. The coiled-coil domain of MHCK A competes with tropomyosin for binding to F-actin and forms oligomers with itself.
(A) Tropomyosin (1 μM) was incubated (25 °C for 15 min) with or without 3 μM F-actin before the addition of GST-tagged C(1–498) (3 μM) or an equal volume of fusion protein storage buffer [20 mM Tris/HCl, pH 7.5, 20% (v/v) glycerol, 70 mM NaCl, 1 mM EDTA and 1 mM DTT] to the assay mixture. The amount of tropomyosin present in the high-speed pellet was quantified as described in the Experimental section. Results are means±S.E.M from four separate experiments. Inset: a representative gel from one competition experiment. Lane 1 shows the co-sedimentation of tropomyosin with F-actin. The amount of tropomyosin present in the P and S fractions in the absence of F-actin and GST–C(1–498) (lane 2), in the presence of both F-actin and GST–C(1–498) (lane 3) and in the presence of GST–C(1–498) only (lane 4) is shown. A, F-actin; C, GST–C(1–498); TM1 and TM2, tropomyosin isoform 1 and 2 respectively. (B) Untagged C(1–498) protein (2 μM) was incubated (60 min at 25 °C) with 50 μM of the chemical cross-linker DSS (lane 2) or an equivalent volume of DMSO vehicle (lane 1) and the reactions were analysed for cross-linking by SDS/PAGE and Coomassie-Blue staining. The upper arrow indicates the position of the major C(1–498) band (approx. 240 kDa) present after cross-linking with DSS. The lower arrow indicates the position of the C(1–498) (approx. 56 kDa) in the absence of DSS. MMM, molecular mass marker.
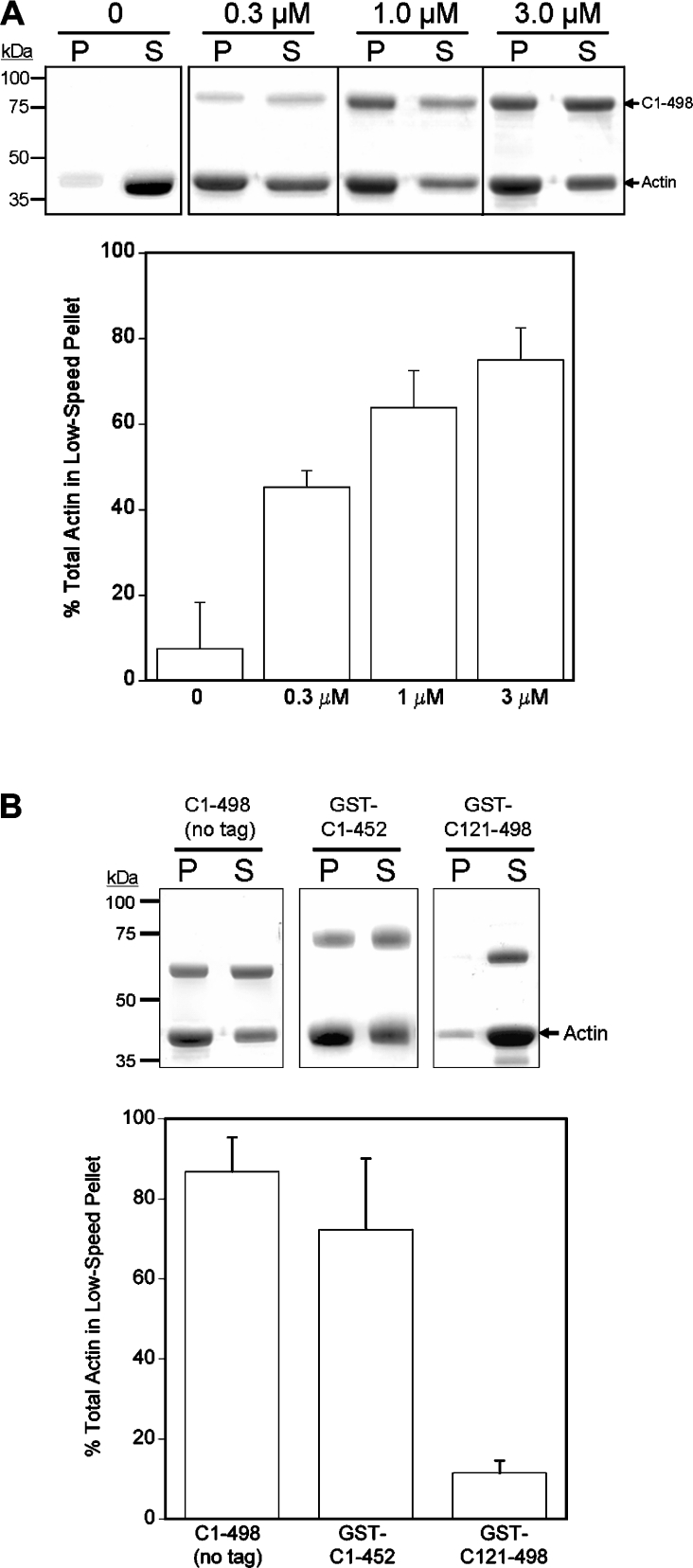
Although the experiments described above indicate that the coiled-coil domain of MHCK A may bind along the length of the actin filament, the possibility exists that this domain also interacts with actin in a manner that leads to the formation of a cross-linked meshwork of actin filaments. A similar ability both to cross-link and to bind laterally along actin filaments has been reported recently for the repeat region of the cardiac and skeletal-muscle protein Xin [28]. To explore this possibility, we analysed the effect of C(1–498) protein on the formation of cross-linked F-actin aggregates that sediment at low centrifugal forces (8500 g) [31]. Results from these experiments (n=7) revealed that the amount of F-actin detected in low-speed pellets increased with increasing concentrations of C(1–498) protein in the assay mixture (Figure 4A). By contrast, F-actin remained in the soluble fraction in the absence of fusion protein. Removal of the GST tag did not alter C(1–498)-induced aggregation of actin filaments (Figure 4B). Taken together, these results indicate that the coiled-coil domain has the potential not only to bind along the length of an actin filament, but also to cross-link adjacent filaments into aggregates. We next analysed whether the C(121–498) and C(1–452) truncations, both of which possess reduced actin-binding activity, were able to induce the formation of cross-linked aggregates of F-actin. Our results showed that F-actin aggregates were detectable in the presence of C(1–452), but not C(121–498) (Figure 4B), thus indicating that the structural determinants for F-actin cross-linking were lost upon removal of the N-terminal unstructured portion of the coiled-coil domain.
Figure 4. The coiled-coil domain cross-links F-actin into aggregates that sediment at low speeds.
(A) Coomassie-Blue-stained SDS/polyacrylamide gels showing the distribution of F-actin (3 μM) in pellet (P) and supernatant (S) fractions resulting from low-speed centrifugation (8000 g for 20 min at 4 °C) in the absence or presence of 0.3, 1.0 or 3.0 μM GST–C(1–498). The corresponding graph shows the amount of F-actin (3 μM) quantified in low-speed pellets in the presence or absence of GST–C(1–498) (means±S.E.M. from at least three separate experiments). (B) Coomassie-Blue-stained gels showing the actin present in low-speed pellets (P) and supernatants (S) in the presence of 1 μM C(1–498), 1 μM GST–C(1–452) or 1 μM GST–(121–498). The corresponding graph shows the amount of F-actin quantified in low-speed pellets in the presence of each fusion protein (means±S.E.M. from at least three separate experiments).
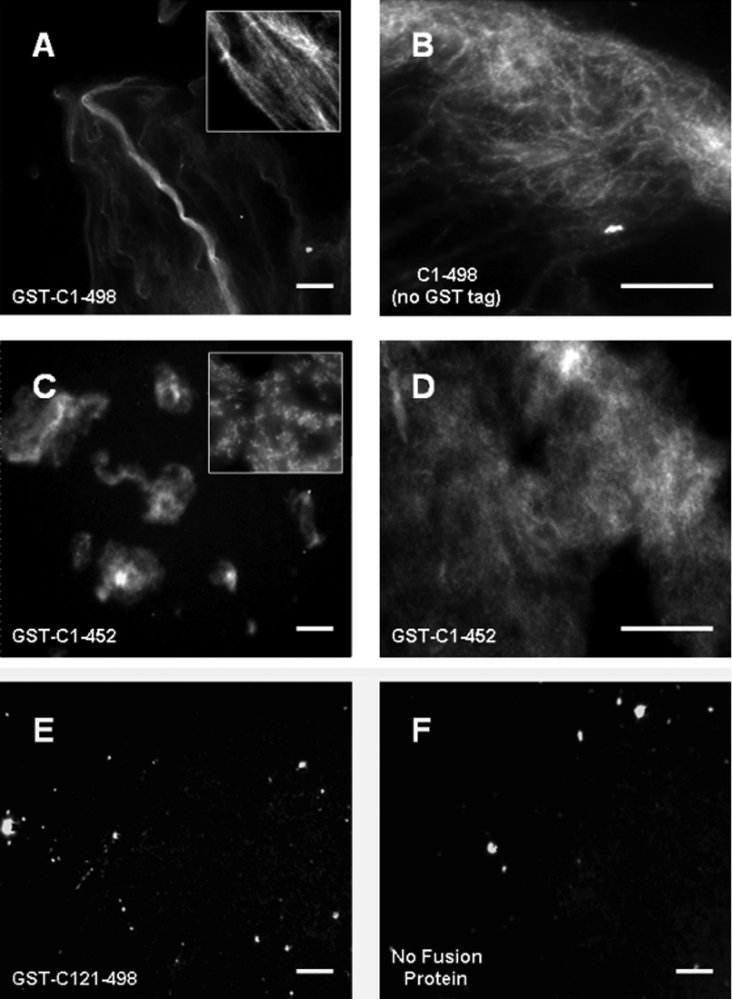
To explore the cross-linking activity of the coiled-coil domain further, we analysed the structural arrangement of fluorescently labelled actin filaments [32] that had been incubated in the presence of saturating concentrations of C(1–498) protein. The addition of GST–C(1–498) to F-actin led to the formation of extended actin ‘cables’ (Figure 5A), suggesting that the full-length coiled-coil domain cross-links F-actin into bundle-like structures. The ability of C(1–498) to induce the reorganization of actin filaments into bundles was not affected by the removal of the GST tag (Figure 5B). In contrast, there was no detectable formation of actinfilament aggregates or bundles in the absence of fusion protein or in the presence of the GST–C(121–498) fusion protein (Figures 5E and 5F). However, robust, but unorganized, aggregates of F-actin were detectable in the presence of GST–(C1–452), consistent with its ability to induce the formation of F-actin aggregates detectable in low-speed pellets (Figures 5C and 5D). Collectively, these results suggest that the full-length coiled-coil domain of MHCK A possesses a previously unrecognized ability to cross-link and bundle actin filaments. However, the possibility exists that the bundling activity of the coiled-coil domain is not exhibited by the full-length MHCK A protein. To address this issue we assessed the F-actin cross-linking/bundling activity of full-length MHCK A purified from Dictyostelium cells [23] and found that the full-length kinase possesses the same cross-linking (Figure 6A) and bundling (Figures 6B and 6C) activities exhibited by the coiled-coil domain alone. Taken together, our data suggest that cortically localized MHCK A has the dual capacity to alter the actin–myosin cytoskeleton not only by catalysing the inactivation of myosin II via the disassembly of myosin II bipolar filaments but also by directly organizing actin filaments into bundles.
Figure 5. The coiled-coil domain of MHCK A cross-links actin filaments into bundles.
F-actin (3 μM) was labelled with Texas Red® phalloidin as described in the Experimental section and incubated with the indicated fusion protein (3 μM). The organization of actin filaments in the presence of (A) GST–C(1–498) (×20 magnification), (B) untagged C(1–498) (×40), (C) GST–C(1–452) (×20), (D) GST–C(1–452) (×40), (E) GST–C(121–498) (×20), or (F) no fusion protein (×20), was visualized directly using fluorescence microscopy. Insets in panels (A) and (C) are high-magnification images (×60) of F-actin incubated with GST–C(1–498) and GST–C(1–452) respectively. Scale bars, 10 μm.
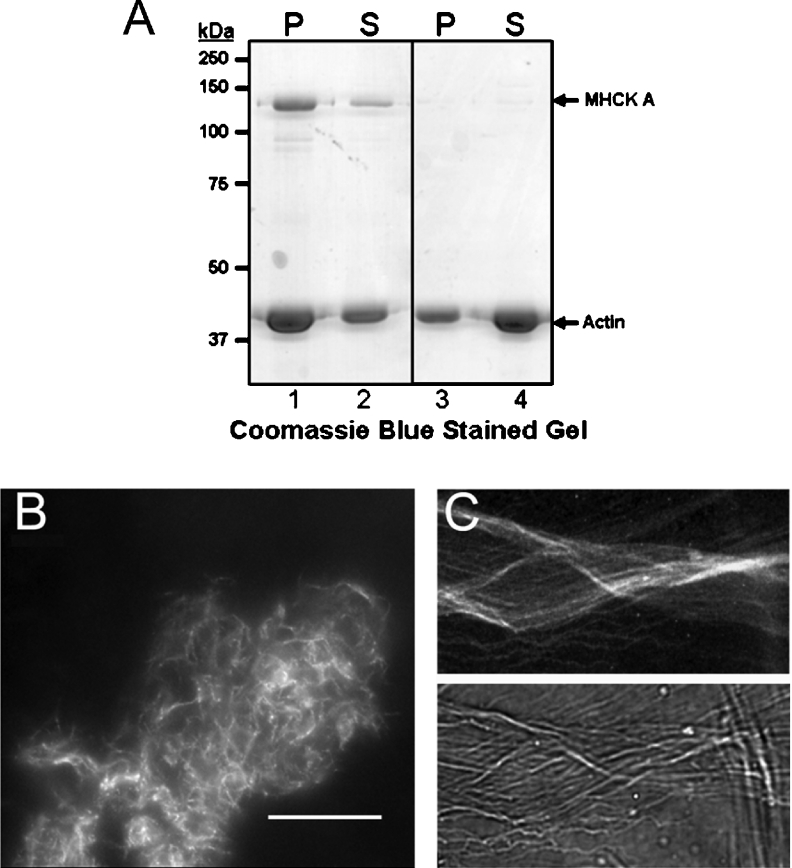
Figure 6. Full-length MHCK A cross-links and bundles actin filaments.
(A) Coomassie-Blue-stained SDS-polyacrylamide gel showing the distribution of F-actin (3 μM) in pellet (P) and supernatant (S) fractions resulting from low-speed centrifugation (at 8000 g for 20 min at 6 °C) in the presence (lanes 1 and 2) or absence (lanes 3 and 4) of 1 μM MHCK A. (B) F-actin (3 μM) labelled with Texas Red® phalloidin was incubated with 1 μM MHCK A and the organization of actin filaments was visualized directly using fluorescence microscopy (×40 magnification). Scale bar, 10 μm. (C) High-magnification (×60) images of phalloidin-labelled actin-filament bundles formed in the presence of 1 μM MHCK A. Upper and lower panels represent the fluorescent and phase-contrast images of the actin bundles respectively.
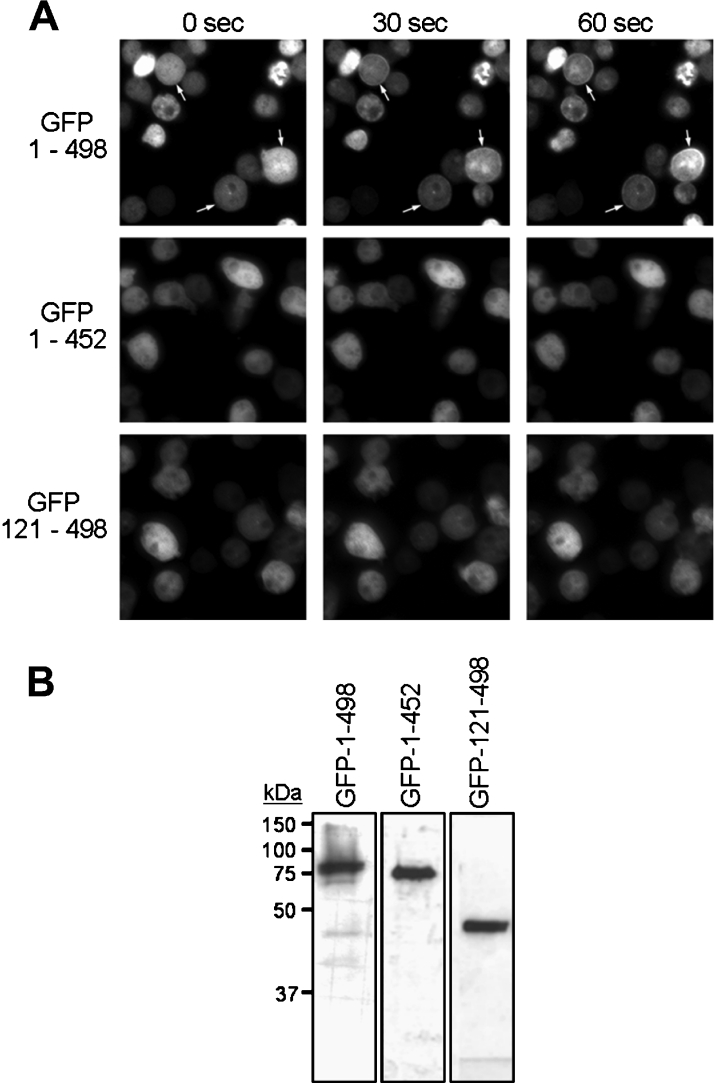
We have reported previously that the coiled-coil domain possesses all of the determinants required for chemoattractant-induced translocation of MHCK A to the cell cortex in Dictyostelium cells [25]. We have also shown that the translocation activity of the coiled-coil domain is dependent on the presence of an intact actin cytoskeleton in the cell [24]. To explore the relationship between the actin-binding and translocation activities of the coiled-coil domain further, we monitored the chemoattractant-induced translocation of GFP-tagged C(1–498), C(121–498) and C(1–452) expressed in live Dictyostelium cells lacking expression of endogenous MHCK A (Figure 7A). As reported previously, the full-length coiled-coil domain of MHCK A [GFP–C(1–498)] translocates rapidly to the cell cortex upon uniform stimulation of aggregation-phase Dictyostelium cells with cAMP (Figure 7A) [24]. By contrast, the cellular localization of the GFP–C(121–498) and GFP–C(1–452) truncations, both of which display only partial actin-binding activity (Figure 2B), remains diffuse and does not change upon stimulation of the cells with chemoattractant. It is important to note that the GFP-C(121–498) and GFP–C(1–452) fusion proteins were expressed at the same overall levels as GFP–C(1–498) and without significant degradation (Figure 7B); however, there is still some heterogeneity in expression levels among individual cells within a given cell line. Nevertheless, this heterogeneity does not affect the translocation activity, indicating that the absence of cAMP-induced enrichment in the cell cortex is probably not a consequence of instability or altered levels of GFP–C(121–498) and GFP–C(1–452) fusion proteins when expressed from plasmids in Dictyostelium cells. Based on these results, we propose that coiled-coil-domain recruitment to the actin-rich cortex depends on the ability of this domain to bind and assemble actin filaments into bundles, and that regulated changes in the accessibility of actin-binding determinants in the coiled-coil domain may be a mechanism by which MHCK A translocation, and the resulting localized disassembly of myosin II filaments, is targeted to specific sites in the cell.
Figure 7. The cellular translocation activity of MHCK A requires the intact coiled-coil domain.
(A) The in vivo translocation activities of GFP–C(1–498), GFP–C(121–498), and GFP–C(1–452) were analysed in chemoattractant-responsive cells. Chemoattractant (cAMP, 100 μM)-induced changes in the localization of the GFP-tagged fusion proteins were monitored using epifluorescence microscopy. Images were collected before the addition of cAMP (zero time) and at 30 s and 60 s after the addition of a uniform and saturating concentration of cAMP to the cells [24]. Arrows indicate regions of cortical recruitment of GFP–C(1–498) in response to stimulation with chemoattractant; translocation was not observed for cells expressing GFP–C(1–452) and GFP–C(121–498). (B) The level and stability of GFP-tagged fusion-protein expression was determined using immunoblot analysis (using anti-GFP antibody) of lysates from cells expressing the fusion protein indicated above each lane. Each lane was loaded with the entire lysate from 1×106 Dictyostelium cells. Molecular mass markers are indicated in kDa.
DISCUSSION
MHCK A is an atypical serine/threonine protein kinase that plays a prominent role in regulating myosin II activity and localization in Dictyostelium cells [8,13]. More specifically, MHCK A drives the disassembly of myosin II bipolar filaments into contraction-incompetent monomers by phosphorylating specific regulatory sites at the C-terminal end of the myosin II heavy chain. MHCK A contains a 500-amino-acid region at its N-terminus that is predicted to fold into two- or three-stranded α-helical coiled-coil structures (Figure 1) [13,35]. We have reported previously that this coiled-coil domain is a functionally dynamic region of MHCK A that participates in the oligomerization, cellular localization, and actin-binding activities of the kinase [23–25]. In the present study we explored in more detail the actin-binding properties of the coiled-coil domain with the goal of understanding the mechanisms by which MHCK A, and thus myosin II filament assembly, might be regulated in the cell.
While we refer to this region of MHCK A as the ‘coiled-coil’ domain, it is important to acknowledge that nearly one-third of this domain is not predicted to be coiled-coil in structure and does not possess any apparent structural motifs or folds. Nevertheless, results from chemical cross-linking experiments described here revealed for the first time that the coiled-coil domain of MHCK A indeed possesses the ability to form oligomers, each containing about containing four to six monomers (Figure 3B). This result is consistent with previous gel filtration and ultrastructural studies of MHCK A demonstrating that the native kinase forms oligomers, and that the coiled-coil domain is required for the assembly of the kinase into oligomers [23]. Taken together, our results demonstrate that the coiled-coil domain of MHCK A indeed possesses the assembly properties that are invariably associated with the coiled-coil protein motif [22]. Continuing studies of coiled-coil-domain oligomer organization, stability and regulation of formation are being pursued.
Initial characterization of the actin-binding activity of the coiled-coil domain revealed a sigmoidal saturation-binding curve (Figure 2B), which is diagnostic of co-operative binding to F-actin. In contrast, the C(121–498) and C(1–452) truncations exhibited non-co-operative interaction with F-actin. Together, these results suggest that functional determinants located at the C- and N-terminal ends of the coiled-coil domain may be involved in the initial interactions with F-actin that lead to higher efficiency (i.e. co-operative) recruitment of other MHCK A coiled-coil domains to F-actin. The idea that sites for interaction with F-actin are localized to the ends of the coiled-coil domain is supported by the finding that a truncation [C(121–452)] lacking both ends of the domain is unable to bind F-actin (Figure 2C). Yet it is still not clear that these ends actually represent independent actin-binding sites within the coiled-coil domain, because the C(1–276) truncation, which contains the entire N-terminal unstructured region and very little coiled-coil structure, does not bind F-actin well (Figure 2C). Thus it may be that only in the context of the intact coiled-coil domain are the two ends able to form proper contacts with the actin filament. Alternatively, the regions for binding actin may actually reside in the central, highly coiled-coil region of the domain, and structural determinants at the ends of the coiled-coil domain facilitate interaction of the central region with F-actin. We are currently exploring studies involving targeted mutagenesis to disrupt potentially important regions of the coiled-coil domain as a means of gaining insight into the specific interactions mediating interaction of MHCK A with F-actin.
The coiled-coil domain shares several properties with tropomyosin, including high coiled-coil content, assembly into oligomers (Figure 3B), similar predicted length, co-operative binding to F-actin (Figure 2B), and similar affinity and stoichiometry of binding to actin filaments (see [25] for a more detailed comparison). In the present study we tested the hypothesis that if the coiled-coil domain of MHCK A and tropomyosin share common mechanisms for interaction with F-actin, then the coiled-coil domain will compete with tropomyosin for binding to the actin filament. Numerous studies have established that tropomyosin binds in the long pitch groove along the actin filament, where it plays an important role in modulating the proper interaction of myosin II with F-actin that is required for activation of the ATPase activity of the myosin II head domain that drives cellular contraction [30]. Despite the large number of studies on this topic, details of the interaction between tropomyosin and F-actin are still unclear. At this time, it appears that the interaction between these two proteins is electrostatic in nature, with no specific site (or sites) on the actin molecule that mediates interaction with tropomyosin [30]. Detailed analysis of chicken striated-muscle tropomyosin sequence, as well as corresponding site-specific mutational studies, revealed that in addition to the heptad repeats related to its coiled-coil structure, tropomyosin also possesses a 7-fold repeat of equivalent, but non-identical, amino acid sequences. Each repeat is approx. 40 amino acids in length and may represent semi-independent sites for F-actin binding [39].
The present study revealed that the coiled-coil domain of MHCK A inhibits tropomyosin co-sedimentation with F-actin, suggesting that the coiled-coil domain, like tropomyosin, binds along the side of the actin filament (Figure 3). Thus it is tempting to speculate that MHCK A binding to F-actin has the same potential as tropomyosin to modulate myosin II-mediated contraction of actin filaments in Dictyostelium. While this model is appealing, recent studies in our laboratory indicate that over-expression (approx. 60-fold) of GFP–C(1–498) in Dictyostelium cells does not lead to any apparent defects in myosin II-dependent cellular activities such as growth in suspension culture or multicellular development (results not shown). Thus there may be subtle differences between F-actin binding by the coiled-coil domain of MHCK A and tropomyosin that could arise from the lack of similarity in oligomer size and sequence periodicity between the two proteins. It is important to note that sequence database searches indicate that a tropomyosin gene is not present in the Dictyostelium genome [38]; thus tropomyosin will not be involved in regulating myosin II activity or affecting MHCK A binding to F-actin. Nevertheless, the possibility exists that other Dictyostelium proteins that bind along the sides of actin filaments may regulate MHCK A binding to F-actin in the cell.
Quite unexpectedly, we found that coiled-coil-domain binding to F-actin led to the concentration-dependent cross-linking of actin filaments into aggregates that formed pellets when centrifuged at low speeds (Figure 4). Fluorescence microscopy of these aggregates revealed that the actin filaments were organized into bundled cables (Figure 5A and 5B) that were similar to those formed in the presence of well-characterized actin-bundling proteins such as fascin [32], filamin [33], WICH {WIP [WASP (Wiskott–Aldrich syndrome protein)-interacting protein]- and CR16-homologous protein} [34], AFAP (actin-filament-associated protein)-110 [40] and scruin [41]. In addition, we demonstrated that the full-length MHCK A protein retains the ability to induce the formation of actin-filament bundles (Figure 6), thus supporting the idea that this can be a physiologically relevant activity within the cell. These discoveries are particularly exciting because they indicate that MHCK A possesses the dual capacity to bring about changes in the actin–myosin II cytoskeleton directly via its actin-bundling activity and through its well-documented ability to drive myosin II bipolar-filament disassembly via myosin II heavy-chain phosphorylation. At this time, it is unclear if the bundling activity of MHCK A is involved in large-scale rearrangements of the cytoskeleton, because the estimated concentration of the kinase in a Dictyostelium cell is much lower (0.3 μM; [23]) than the concentration of actin present in actin-rich cortical structures such as forming pseudopods (>100 μM; [42]). On the other hand, recruitment of MHCK A to specific actin-rich regions of the cell [19] may result in localized increases in MHCK A concentration that are sufficient to drive in vivo bundling of actin filaments. A similar argument has been made for the 34 kDa protein, a physiologically prominent actin-bundling protein present at similar concentrations (approx. 1 μM) in Dictyostelium cells [43].
Our studies of the C(121–498) and C(1–452) truncations indicate that the bundling of actin filaments may also play a central role in driving co-operative binding of the kinase to F-actin. We found that C(1–452), which lacks the C-terminal coiled-coil subdomain (Figure 1B), induced the formation of unorganized F-actin aggregates lacking higher-order structures such as bundles (Figure 5C and 5D). In contrast, the C(121–498) truncation did not cause the formation of actin aggregates (Figure 5E) despite the fact that it binds actin filaments as well as C(1–452) (Figure 2). Neither the C(121–498) nor the C(1–452) truncations exhibited co-operative binding to F-actin (Figure 2A). Collectively, these results imply a direct relationship between co-operative binding to F-actin and the ability of the coiled-coil domain to bundle actin filaments. Thus we propose a model in which initial binding of the coiled-coil domain to F-actin leads to the formation and stabilization of actin-filament bundles that, in turn, serve as higher-affinity targets for more efficient recruitment of MHCK A coiled-coil domains to actin. This model is supported by our results showing that cross-linking alone (without bundling) by the C(1–452) truncation was not sufficient to drive co-operative binding to F-actin. A similar mechanism for co-operative recruitment to F-actin has been proposed for the Abl-related gene, Arg [44].
It is noteworthy that the stoichiometry of the F-actin–C(1–498) interaction appears to be higher [3 mol of F-actin/1 mol of C-498, at 3 μM C(1–498)] under low-speed centrifugation conditions compared with that observed under the conditions of the standard actin co-sedimentation assay (5 mol of F-actin/1 mol of C-498) (compare Figure 2A with the graph of Figure 4A). We suspect that this may reflect alterations in the organization of F-actin that may occur upon ‘compaction’ into a high-speed pellet. Such subtle alterations in actin-filament conformation or environmental conditions can often have a profound effect on the ability of a protein to interact with F-actin [45]. Nevertheless, our results demonstrate that the binding of the coiled-coil domain to F-actin induces a robust and concentration-dependent cross-linking of actin filaments into bundles.
Our previous studies have established that chemoattractant-induced translocation of MHCK A to the cell cortex relies on the ability of the kinase to bind F-actin [19,24]. In the present paper we show that even though the C(121–498) and C(1–452) truncations bind to F-actin (Figure 2), they do not exhibit cAMP-induced translocation to the actin-rich cortex (Figure 6A). These results suggest that simple binding to actin filaments is not sufficient to drive MHCK A recruitment to actin-rich structures and that the more complex processes (i.e. actin bundling) driving co-operative binding to F-actin are probably important for achieving rapid and robust recruitment of MHCK A to specific actin-rich sites in the cell. Thus an initially weak and transient signal could activate a small population of MHCK A molecules to associate with F-actin at a specific location in the cell. Signal-independent accumulation of MHCK A molecules could then occur via a feed-forward mechanism involving localized organization of actin filaments into bundles that serve as high-affinity targets for further recruitment of kinase molecules.
In summary, the studies presented in the present paper demonstrate that MHCK A possesses two methods by which it can alter the actin–myosin II cytoskeleton in the cell: (1) by driving myosin II-filament disassembly via the phosphorylation of myosin II heavy chain, and (2) by cross-linking actin filaments into bundles. The ability to facilitate cytoskeletal rearrangements via both kinase catalytic activity and F-actin bundling activity is uncommon among protein kinases, with only a few examples reported in the literature [44,46–47]. Thus the studies of MHCK A reported in the present paper are particularly exciting because they support the notion that this type of functional duality may be present in other cytoskeleton-associated protein kinases, but has simply not been explored. In addition, our studies have led to the identification of another member of the growing number of actin-bundling proteins present in Dictyostelium [48]. We suggest that the bundling activity of the coiled-coil domain plays a more focused role in modulating the actin-binding and translocation activities of MHCK A and thus represents a potent target (i.e. actin-binding) for the regulation of MHCK A by components of signal-transduction pathways in Dictyostelium cells [49]. Furthermore, the results reported in the present paper shed further light on the mechanisms by which the cell achieves the targeted disruption of myosin II filaments that is critical for the proper execution of localized changes in cell shape that drive dynamic cellular events such as cytokinesis, particle and fluid-phase uptake, and chemotaxis.
Acknowledgments
This work was supported by an National Institutes of Health grant to P. A. S. (1R15GM066789). Microscopy studies were supported in part by grants from the National Science Foundation (DBI-0319021) and the North Carolina Biotechnology Center (003-IDG-1011). We thank Phillip Brake, Orna Fisher and Jacinta Watkins for technical assistance.
References
- 1.De la Roche M. A., Côté G. P. Regulation of Dictyostelium myosin I and II. Biochim. Biophys. Acta. 2001;1525:245–261. doi: 10.1016/s0304-4165(01)00110-6. [DOI] [PubMed] [Google Scholar]
- 2.Laevsky G., Knecht D. A. Cross-linking of actin filaments by myosin II is a major contributor to cortical integrity and cell motility in restrictive environments. J. Cell Sci. 2003;116:3761–3770. doi: 10.1242/jcs.00684. [DOI] [PubMed] [Google Scholar]
- 3.Lo C. M., Buxton D. B., Chua G. C., Dembo M., Adelstein R. S., Wang Y. L. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Steimle P. A., Fulcher F. K., Patel Y. M. A novel role for myosin II in insulin-stimulated glucose uptake in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2005;331:1560–1565. doi: 10.1016/j.bbrc.2005.04.082. [DOI] [PubMed] [Google Scholar]
- 6.Robinson D. N., Girard K. D., Octtaviani E., Reichl E. M. Dictyostelium cytokinesis: from molecules to mechanics. J. Muscle Res. Cell Motil. 2002;23:719–727. doi: 10.1023/a:1024419510314. [DOI] [PubMed] [Google Scholar]
- 7.Lee R. J., Egelhoff T. T., Spudich J. A. Molecular genetic truncation analysis of filament assembly and phosphorylation domains of Dictyostelium myosin heavy chain. J. Cell Sci. 1994;107:2875–2886. doi: 10.1242/jcs.107.10.2875. [DOI] [PubMed] [Google Scholar]
- 8.De la Roche M. A., Smith J. L., Betapudi V., Egelhoff T. T., Côté G. P. Signaling pathways regulating Dictyostelium myosin II. J. Muscle Res. Cell Motil. 2002;23:703–718. doi: 10.1023/a:1024467426244. [DOI] [PubMed] [Google Scholar]
- 9.Egelhoff T. T., Lee R. J., Spudich J. A. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 10.Vaillancourt J. P., Lyons C., Côté G. P. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J. Biol. Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- 11.Côté G. P., Bukiejko U. Purification and characterization of a myosin heavy chain kinase from Dictyostelium discoideum. J. Biol. Chem. 1987;262:1065–1072. [PubMed] [Google Scholar]
- 12.Kolman M. F., Futey L. M., Egelhoff T. T. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J. Cell Biol. 1996;132:101–109. doi: 10.1083/jcb.132.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futey L. M., Medley Q. G., Côté G. P., Egelhoff T. T. Structural analysis of myosin heavy chain kinase A from Dictyostelium. Evidence for a highly divergent protein kinase domain, an amino-terminal coiled-coil domain, and a domain homologous to the β-subunit of heterotrimeric G-proteins. J. Biol. Chem. 1995;270:523–529. doi: 10.1074/jbc.270.2.523. [DOI] [PubMed] [Google Scholar]
- 14.Ryazanov A. G. Elongation factor-2 kinase and its newly discovered relatives. FEBS Lett. 2002;514:26–29. doi: 10.1016/s0014-5793(02)02299-8. [DOI] [PubMed] [Google Scholar]
- 15.Yumura S., Yoshida M., Betapudi V., Licate L. S., Iwadate Y., Nagasaki A., Uyeda T. Q., Egelhoff T. T. Multiple myosin II heavy chain kinases: roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol. Biol. Cell. 2005;16:4256–4266. doi: 10.1091/mbc.E05-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rico M., Egelhoff T. T. Myosin heavy chain kinase B participates in the regulation of myosin assembly into the cytoskeleton. J. Cell. Biochem. 2003;88:521–532. doi: 10.1002/jcb.10361. [DOI] [PubMed] [Google Scholar]
- 17.Nagasaki A., Itoh G., Yumura S., Uyeda T. Q. Novel myosin heavy chain kinase involved in disassembly of myosin II filaments and efficient cleavage in mitotic Dictyostelium cells. Mol. Biol. Cell. 2002;13:4333–4342. doi: 10.1091/mbc.E02-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang W., Licate L., Warrick H., Spudich J., Egelhoff T. Differential localization in cells of myosin II heavy chain kinases during cytokinesis and polarized migration. BMC Cell Biol. 2002;3:19. doi: 10.1186/1471-2121-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steimle P. A., Yumura S., Côté G. P., Medley Q. G., Polyakov M. V., Leppert B., Egelhoff T. T. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr. Biol. 2001;11:708–713. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 20.Steimle P. A., Naismith T., Licate L., Egelhoff T. T. WD repeat domains target Dictyostelium myosin heavy chain kinases by binding directly to myosin filaments. J. Biol. Chem. 2001;276:6853–6860. doi: 10.1074/jbc.M008992200. [DOI] [PubMed] [Google Scholar]
- 21.Lupas A. N., Gruber M. The structure of α-helical coiled coils. Adv. Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 22.Burkhard P., Stetefeld J., Strelkov S. V. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 23.Kolman M. F., Egelhoff T. T. Dictyostelium myosin heavy chain kinase A subdomains. Coiled-coil and WD repeat roles in oligomerization and substrate targeting. J. Biol. Chem. 1997;272:16904–16910. doi: 10.1074/jbc.272.27.16904. [DOI] [PubMed] [Google Scholar]
- 24.Steimle P. A., Licate L., Côté G. P., Egelhoff T. T. Lamellipodial localization of Dictyostelium myosin heavy chain kinase A is mediated via F-actin binding by the coiled-coil domain. FEBS Lett. 2002;516:58–62. doi: 10.1016/s0014-5793(02)02494-8. [DOI] [PubMed] [Google Scholar]
- 25.Egelhoff T. T., Croft D., Steimle P. A. Actin activation of myosin heavy chain kinase A in Dictyostelium: a biochemical mechanism for the spatial regulation of myosin II filament disassembly. J. Biol. Chem. 2005;280:2879–2887. doi: 10.1074/jbc.M410803200. [DOI] [PubMed] [Google Scholar]
- 26.Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 27.Levi S., Polyakov M., Egelhoff T. T. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 28.Pacholsky D., Vakeel P., Himmel M., Lowe T., Stradal T., Rottner K., Furst D. O., van der Ven P. F. M. Xin repeats define a novel actin-binding motif. J. Cell Sci. 2004;117:5257–5268. doi: 10.1242/jcs.01406. [DOI] [PubMed] [Google Scholar]
- 29.Greaser M. L., Gergely J. Reconstitution of troponin activity from three protein components. J. Biol. Chem. 1971;246:4226–4233. [PubMed] [Google Scholar]
- 30.Perry S. V. Vertebrate tropomyosin: distribution, properties and function. J. Muscle Res. Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 31.Pollard T. D., Cooper J. A. Methods to characterize actin filament networks. Methods Enzymol. 1982;85(Part B):211–233. doi: 10.1016/0076-6879(82)85022-2. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa R., Yamashiro S., Kohama K., Matsumura F. Regulation of actin binding and actin bundling activities of fascin by caldesmon coupled with tropomyosin. J. Biol. Chem. 1998;273:26991–26997. doi: 10.1074/jbc.273.41.26991. [DOI] [PubMed] [Google Scholar]
- 33.Prassler J., Stocker S., Marriott G., Heidecker M., Kellermann J., Gerisch G. Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin–myosin complexes. Mol. Biol. Cell. 1997;8:83–95. doi: 10.1091/mbc.8.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M., Takenawa T. WICH, a member of WASP-interacting protein family, cross-links actin filaments. Biochem. Biophys. Res. Commun. 2005;328:1058–1066. doi: 10.1016/j.bbrc.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 35.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 36.Parker M. W., Lo Bello M., Federici G. Crystallization of glutathione S-transferase from human placenta. J. Mol. Biol. 1990;213:221–222. doi: 10.1016/s0022-2836(05)80183-4. [DOI] [PubMed] [Google Scholar]
- 37.Pollard T. D., Almo S., Quirk S., Vinson V., Lattman E. E. Structure of actin binding proteins: insights about function at atomic resolution. Annu. Rev. Cell Biol. 1994;10:207–249. doi: 10.1146/annurev.cb.10.110194.001231. [DOI] [PubMed] [Google Scholar]
- 38.Kreppel L., Fey P., Gaudet P., Just E., Kibbe W. A., Chisholm R. L., Kimmel A. R. dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res. 2004;32:D332–D333. doi: 10.1093/nar/gkh138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hitchcock-DeGregori S. E., Varnell T. A. Tropomyosin has discrete actin-binding sites with sevenfold and fourteenfold periodicities. J. Mol. Biol. 1990;214:885–896. doi: 10.1016/0022-2836(90)90343-K. [DOI] [PubMed] [Google Scholar]
- 40.Qian Y., Baisden J. M., Cherezova L., Summy J. M., Guappone-Koay A., Shi X., Mast T., Pustula J., Zot H. G., Mazloum N., et al. PC phosphorylation increases the ability of AFAP-110 to cross-link actin filaments. Mol. Biol. Cell. 2002;13:2311–2322. doi: 10.1091/mbc.E01-12-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin J. H., Gardel M. L., Mahadevan L., Matsudaira P., Weitz D. A. Relating microstructure to rheology of a bundled and cross-linked F-actin network in vitro. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9636–9641. doi: 10.1073/pnas.0308733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janmey P. A. Creating a niche in the cytoskeleton: actin reorganization by a protein kinase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14745–14747. doi: 10.1073/pnas.011601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fechheimer M. The Dictyostelium discoideum 30,000-dalton protein is an actin filament-bundling protein that is selectively present in filopodia. J. Cell Biol. 1987;104:1539–1551. doi: 10.1083/jcb.104.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Miller A. L., Mooseker M. S., Koleske A. J. The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14865–14870. doi: 10.1073/pnas.251249298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlova A., Rybakova I. N., Prochniewicz E., Thomas D. D., Ervasti J. M., Egelman E. H. Binding of dystrophin's tandem calponin homology domain to F-actin is modulated by actin's structure. Biophys. J. 2001;80:1926–1931. doi: 10.1016/S0006-3495(01)76162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayakawa K., Okagaki T., Ye L. H., Samizo K., Higashi-Fujime S., Takagi T., Kohama K. Characterization of the myosin light chain kinase from smooth muscle as an actin-binding protein that assembles actin filaments in vitro. Biochim. Biophys. Acta. 1999;1450:12–24. doi: 10.1016/s0167-4889(99)00034-8. [DOI] [PubMed] [Google Scholar]
- 47.Vitavska O., Merzendorfer H., Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J. Biol. Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- 48.Noegel A. A., Schleicher M. The actin cytoskeleton of Dictyostelium: a story told by mutants. J. Cell Sci. 2000;113:759–766. doi: 10.1242/jcs.113.5.759. [DOI] [PubMed] [Google Scholar]
- 49.Affolter M., Weijer C. J. Signaling to cytoskeletal dynamics during chemotaxis. Dev. Cell. 2005;9:19–34. doi: 10.1016/j.devcel.2005.06.003. [DOI] [PubMed] [Google Scholar]