Abstract
The suppressor of the dis2 mutant (sds22+) has been shown to be an essential regulator in cell division of fission and budding yeast where its deletion causes mitotic arrest. Its role seems to take place through the activation of PP1 (protein phosphatase type 1) in Schizosaccharomyces pombe. In the trematode Schistosoma mansoni, we have identified the Sds22 homologue (SmSds), and the PP1 (SmPP1). We showed by using a GST (glutathione S-transferase) pull-down assay that the SmSds gene product interacts with SmPP1 and that the SmSds–SmPP1 complex is present in parasite extracts. Furthermore, we observed that SmSds inhibited PP1 activity. Functional studies showed that the microinjection of SmSds into Xenopus oocytes interacted with the Xenopus PP1 and disrupted the G2/M cell-cycle checkpoint by promoting progression to GVBD (germinal vesicle breakdown). Similar results showing the appearance of GVBD were observed when oocytes were treated with anti-PP1 antibodies. Taken together, these observations suggest that SmSds can regulate the cell cycle by binding to PP1.
Keywords: cell cycle, leucine-rich repeat (LRR), protein phosphatase 1 (PP1), Schistosoma mansoni, schistosomiasis, suppressor of the dis2 mutant (Sds22)
Abbreviations: cRNA, capped RNA; DAPI, 4′,6-diamidino-2-phenylindole; EST, expressed sequence tag; GST, glutathione S-transferase; GVBD, germinal vesicle breakdown; HA, haemagglutinin; LRR, leucine-rich repeat; OA, okadaic acid; ORF, open reading frame; pNPP, p-nitrophenyl phosphate; PP1, protein phosphatase type 1; RACE, rapid amplification of cDNA ends; Sds22, suppressor of the dis2 mutant; Sm, Schistosoma mansoni; Sp, Schizosaccharomyces pombe
INTRODUCTION
Schistosomiasis is an important parasitic disease, infecting about 250 million people and causing morbidity and mortality [1]. Schistosoma mansoni, the most widespread causative species [2], has a complex life cycle involving a mammalian host, where adult worms of the two sexes mate and deposit eggs; a free-living aquatic stage (miracidium), derived from eggs excreted into the environment; a molluscan stage, with active asexual division; and a second free-living aquatic stage (cercaria). Beyond the fact that Schistosoma mansoni continuously maintains complex and dynamic molecular exchanges involved in communications with its host, it is clear that it must have developed appropriate and orchestrated mechanisms that are necessary for its own development and growth. It is well known that reversible phosphorylation of proteins is a ubiquitous mechanism crucial for regulation of most cellular functions [3,4]. Thus protein kinases and phosphatases are essential for normal functioning of signalling pathways. In Schistosoma mansoni, a phosphatase 2B (calcineurin) has been described as a heterodimer with a catalytic subunit and a regulatory subunit whose binding with Ca2+ increased the phosphatase activity [5,6]. No other Schistosoma mansoni phosphatases have so far been described, although they represent crucial molecules for the parasite and hence potential chemotherapeutic targets. It is of interest to note that PP1 (protein phosphatase type 1) is one of the major types of protein phosphatases. It comprises a catalytic subunit that interacts with different regulatory subunits, generating enzymes with unique substrate affinities [7–11]. In this context, while investigating the mitotic metaphase/anaphase transition in Schizosaccharomyces pombe, Ohkura and Yanagida [12] showed that a novel phosphatase regulator partner referred to as suppressor of the dis2 mutant (SpSds22) is essential in the progression from metaphase to anaphase by enhancing the PP1 activity [12]. Furthermore, the deletion of the sds22+ gene caused a mitotic arrest. These results, together with those independently obtained in Saccharomyces cerevisiae [13,14], suggest that the binding of regulatory proteins to phosphatases is a fundamental mechanism in the progression of cell division (for a review, see [15]). Subsequently, human Sds22 was identified and has also been shown to interact with PP1 [16]. Such gene-regulatory products are of considerable interest because they are key players in the activity of phosphatases.
In the present study, we identify in the trematode Schistosoma mansoni the Sds homologue of sds22+ of Schizosaccharomyces pombe, designated SmSds, and the PP1 (SmPP1) gene products, and find evidence that SmSds is capable of binding to PP1, to inhibit its activity and finally to disrupt a G2/M cell-cycle checkpoint of Xenopus oocytes, suggesting a possible role in the regulation of the cell cycle.
EXPERIMENTAL
Parasites and antigen extracts
A Puerto Rican strain of Schistosoma mansoni was maintained by passage through Biomphalaria glabrata snails and Mesocricetus auratus hamsters. Cercariae were prepared as described previously [17]. Adult schistosomes were collected by portal perfusion from infected hamsters. SWAP (soluble worm antigen preparation) was prepared for Western blot and ELISA experiments. Parasites were homogenized in PBS with a protease inhibitor cocktail containing AEBSF [4-(2-aminoethyl) benzenesulphonyl fluoride], pepstatin A, E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane], bestatin, leupeptin and aprotinin (Sigma–Aldrich). The homogenate was sonicated and centrifuged at 10000 g for 10 min at 4 °C. The supernatant fractions were recovered and used as the source of proteins. The integrity of protein extracts was checked on polyacrylamide gels with Coomassie Blue staining.
Identification of SmPP1 and SmSds homologue genes
To identify the Schistosoma mansoni PP1 gene, the TIGR database (http://www.tigr.org) was queried by tblastn with the ORFs (open reading frames) of human, rabbit and Schizosaccharomyces pombe PP1 (accession numbers P62136, P62139 and CAA22875). A partial PP1 sequence was identified (Schistosoma mansoni Gene Index EST accession number TC6011). The complete PP1 cDNA sequence was obtained by PCR from a mixed-sex adult reverse transcription prepared using RACE (rapid amplification of cDNA ends). RACE was carried out using a Clontech kit. The 5′ end was cloned using a reverse primer (R1 PP1, 5′-CGCGAGACTTCAGACACAGCCCGCGG-3′) and the adapter primer according to the manufacturer's instructions. The 3′ end was obtained using the forward primer (F1 PP1, 5′-TAATCATGAATGTGCCTCTATTAACCG-3′) in the presence of the adapter primer. Each PCR product was then cloned in TOPO 2.1 TA cloning vector (Invitrogen), and sequenced using the Dye Terminator Cycle Sequencing kit and analysed on an ABI Prism 377 DNA sequencer (PerkinElmer Biosystems). To obtain the complete sequence, a PCR was performed with the specific primers (F2 PP1, 5′-ATGGCAGGGGATGATAAGGTGAATA-3′, and R2 PP1, 5′-TTATAATTTCCCTTTCGCTTTAGCCCC-3′) covering the whole gene. The ORF was verified by nucleotide sequencing.
For SmSds, a partial sequence homologous with yeast sds22+ (GenBank® accession number AAA35342) gene product was identified in the GenBank® EST (expressed sequence tag) database (MEEG04305.M1R; accession number AI740260) and cloned by RACE PCR according to the method described above. The F1 Sds primer, 5′-ACGTATCGAAAATTTGGAAAATCTAAGT-3′, and the R1 Sds primer, 5′-GAGGTTTACTCTGGTCATCCATCAG-3′, were used to obtain the 5′ and 3′ ends respectively. The full-length gene was confirmed further by nucleotide sequencing of a PCR product obtained with the F2 Sds primer, 5′-ATGGATGACCAGAGTAAACCTC-3′, and the R2 Sds primer, 5′-TCAGATCCCAACTCCCGTAAGA-3′, that covers the entire sequence. Analysis of SmPP1 and SmSds proteins was performed using DNAStar and ClustalW, Pfam database (http://www.sanger.ac.uk) and NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Recombinant protein production and purification
The entire ORF of SmSds was amplified by PCR using Taq DNA polymerase (Promega) and the above primers modified to contain BamHI and HindIII restriction sites for subcloning afterward into the pQE30 expression plasmid (Qiagen) that contains a coding sequence for a His6 tag 5′ to the protein of interest. Primers used were F2 Sds, 5′-GGATCCATGGATGACCAGAGTAAACCTC-3′, and R2 Sds-like, 5′-AAGCTTTCAGATCCCAACTCCCGTAAGA-3′. The ORF in pQE30 was checked by nucleotide sequencing. To obtain recombinant soluble protein, SmSds was prepared as follows. An overnight culture of Escherichia coli M15 carrying the pQE30 SmSds was diluted in culture medium supplemented with 100 μg/ml ampicillin and 25 μg/ml kanamycin. At a D600 of 0.6, expression was induced by the addition of 0.5 mM IPTG (isopropyl β-D-thiogalactoside), and cells were grown overnight at 16 °C. Cells were then harvested and lysed by sonication on ice in lysis buffer (25 mM Tris/HCl, pH 8.0, 100 mM NaCl, 1 mM 2-mercaptoethanol, 0.1% Triton X-100 and 10% glycerol) containing 1 mM PMSF, EDTA-free inhibitor cocktail (Boehringer Mannheim) and 100 μg/ml lysozyme. After centrifugation at 2500 g for 10 min, the supernatant was incubated with Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) resin for 1 h at 4 °C to allow binding. The resin was washed ten times in sonication buffer containing 20 mM imidazole, and bound protein was eluted with elution buffer (25 mM Tris/HCl, pH 8.0, 100 mM NaCl, 10% glycerol and 250 mM imidazole). Supernatants were filtered and dialysed against PBS containing 5% glycerol, and protein purity was checked by SDS/PAGE (4–12% gels). The purified proteins were stored at −20 °C until use.
Antisera production
Rats were immunized by subcutaneous injection of 100 μg of recombinant SmSds in the presence of complete Freund's adjuvant and boosted in the presence of incomplete Freund's adjuvant 3 weeks later. For SmPP1, sera were raised in mice according to the above scheme. Sera were collected 2 weeks after the boost, and their reactivity and specificity were tested by ELISA and Western blotting against recombinant proteins.
GST (glutathione S-transferase) fusion proteins and pull-down assay
The expression vector pGEX-4T3 (Amersham Biosciences) containing the entire ORF of SmPP1 was transformed in E. coli, and GST-fused protein was expressed and purified according to the manufacturer's instructions. Protein of full-length SmSds was translated in vitro with [35S]methionine at 30 °C using rabbit reticulocyte lysates (Promega) and 1 μg of pCDNA6 plasmid (Invitrogen) containing the complete SmSds gene in a final volume of 50 μl. In vitro-translated SmSds protein (10 μl) was added to GST-fused PP1 protein, immobilized on glutathione–Sepharose 4B beads (Amersham Biosciences). After overnight incubation at 4 °C, the beads were washed extensively with pull-down buffer (20 mM Tris/HCl, pH 8, 1 mM EDTA, 100 mM NaCl, 0.002% BSA, 0.002% Tween 20 and 0.2 mM PMSF), and potential bound protein was eluted with SDS/PAGE loading buffer and separated on SDS/4–12% polyacrylamide gels. Following electrophoresis, gels were analysed by autoradiography. As a positive control, 20% of input was loaded.
Microcystin–agarose affinity column and co-immunoprecipitation experiments
A 1 ml volume of parasite extracts (2 mg/ml) was incubated with 50 μl of microcystin–agarose beads (Upstate Biotechnology) for 30 min at 4 °C. Beads were then washed four times with 50 mM Tris buffer, pH 7.4, containing 0.1 mM EGTA, 5% glycerol, 0.5 M NaCl, 0.1% 2-mercaptoethanol and protease inhibitors. For co-immunoprecipitation assays, 50 μl of SmSds antiserum or control serum was incubated for 4 h at 4 °C with 100 μl of Protein G–Sepharose in 0.5 ml of TNE (100 mM Tris/HCl, 0.1% Nonidet P40 and 2 mM EDTA). A 2 mg amount of Schistosoma mansoni extracts was then added to the mixture. After overnight incubation at 4 °C, Sepharose beads were washed extensively in TNTE (TNE supplemented with 0.1% Triton X-100). In all experiments, bound proteins were eluted, fractionated on 6–20% gradient SDS/polyacrylamide gels and transferred on to nitrocellulose membranes. Filters were probed with anti-SmSds or anti-SmPP1 antibodies (1:100) produced as described above. In some experiments, anti-SpPP1 (Schizosaccharomyces pombe PP1) antibodies described previously [21] were used. Detection with horseradish-peroxidase-conjugated anti-rat IgG [H+L (heavy and light chains)] or anti-mouse IgG (H+L) (Sigma) at 1:5000 dilution was carried out as described previously [18].
Yeast complementation assays
For complementation experiments in Schizosaccharomyces pombe, SmPP1 cDNA containing the entire coding region was subcloned in the plasmid Rep1 which carries the thiamine-inducible nmt1 promoter [19], the LEU2 gene as a selectable marker and a coding sequence for HA (haemagglutinin) and His6 3′ to the protein of interest.
The alkali cation method was performed to transform Schizosaccharomyces pombe cells by plasmids [20]. The expression of Schistosoma mansoni protein by Schizosaccharomyces pombe was checked by Western blot using anti-HA monoclonal antibody, and rabbit SpPP1 (Dis2) antisera were used [12,21]. The Schizosaccharomyces pombe wild-type and mutated strains used in the present study have been described previously [12,21,22]. The medium, growth conditions and standard genetic methods have been described in detail previously [12,21]. The restrictive temperature for the PP1 mutant strain (dis2-11) is 20 °C and the permissive temperature 33 °C. DAPI (4′,6-diamidino-2-phenylindole) was used for staining Schizosaccharomyces pombe chromosomal DNA [23].
Effect of recombinant SmSds on PP1 activity
The recombinant PP1 purchased from New England Biolabs was >95% homogeneous, with an activity of 7000 units/mg. The activity of PP1 with pNPP (p-nitrophenyl phosphate) at 5 mM as substrate was assayed as described previously [24]. To test the effect of SmSds on PP1 activity, different concentrations of rabbit PP1 in the range 0.5–5 units/assay were pre-incubated for 1 h at room temperature (20 °C) before adding substrate and reaction buffer with different amounts of SmSds before testing the phosphatase activity. The phosphatase activity was tested in a final volume of 200 μl. Experiments were repeated three times and each assay was performed in triplicate. As positive controls, the different concentrations of rabbit PP1 were pre-incubated with different concentrations of OA (okadaic acid) before the addition of reaction buffer and pNPP. As negative controls, the recombinant GST protein produced from the pGEX-4T3 plasmid was used at the same concentrations.
Preparation of SmSds cRNA (capped mRNA) and microinjection into Xenopus oocytes
cRNA was synthesized using the T7 or SP6 mMessage mMachine kit (Ambion). SmSds-pCDNA3 plasmid was linearized using XhoI. cRNA transcribed from 1.5 μg of linearized plasmid was precipitated with 2.5 M LiCl, washed in 70% ethanol, resuspended in 20 μl of diethyl pyrocarbonate-treated water, and was then quantified by spectrophotometry. A 1 μg sample of cRNA was analysed on a denaturating agarose gel. Gel staining with 10 μg/ml ethidium bromide allowed confirmation of the size of cRNA and the absence of abortive transcripts. cRNA control was obtained from pSP64T-Grb2 P49L that has been described previously [25].
Preparation of Xenopus oocytes and microinjection experiments were performed as described previously [26]. Briefly, after anaesthesia with 1 g/l MS 222 (Sandoz), Xenopus laevis ovarian fragments were surgically removed and placed in ND96 medium (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2 and 5 mM Hepes, adjusted to pH 7.4 with NaOH) supplemented with 50 μg/ml streptomycin/penicillin (Eurobio), 225 μg/ml sodium pyruvate (Sigma) and 30 μg/ml soybean trypsin inhibitor (Sigma). Stage VI oocytes were harvested after a 1 h treatment with 1 mg/ml collagenase A (Roche Applied Science). Complete defolliculation of oocytes was achieved by manual dissection. Oocytes were kept at 19 °C in ND96 medium before microinjection in the equatorial region. In each assay, 20 oocytes removed from at least two different animals were microinjected with different concentrations of OA prepared in PBS, or with anti-PP1 antibodies or SmSds cRNA. PBS, rat pre-bleed sera and Grb2 P49L cRNA were used as controls. GVBD (germinal vesicle breakdown) was detected by the appearance of a white spot at the centre of the animal pole. The expression of the SmSds gene product was checked by immunoblot analysis of whole oocyte extracts using anti-SmSds antibodies. For microcystin affinity chromatography, extracts corresponding to 20 oocytes were incubated with 50 μl of beads and were processed as described above for immunoblot analysis, except that antibody complexes were detected by the advanced enhanced chemiluminescence Western blotting detection system (Amersham Biosciences).
RESULTS
Isolation of the Schistosoma mansoni gene homologue of Schizosaccharomyces pombe sds22+
A 489 bp EST (MEEG04305.M1R AI740260) fragment with similarity to sds22+ of Schizosaccharomyces pombe was identified in the publicly available Schistosoma mansoni raw database (NCBI-dbest). In order to obtain the full-length sequence, 5′-RACE and 3′-RACE were performed on cDNA from adult worms with reverse and forward primers as detailed in the Experimental section. Sequencing of the amplified products allowed the design of primers for amplification of the full-length cDNA. The amplified 1343 bp cDNA contained an ORF of 981 bp with an initiation codon at position 186 and a stop codon at position 1169 (GenBank® accession number AY886887). The encoded protein contains 327 amino acids, and has a predicted molecular mass of 38 kDa and a calculated pI of 4.79.
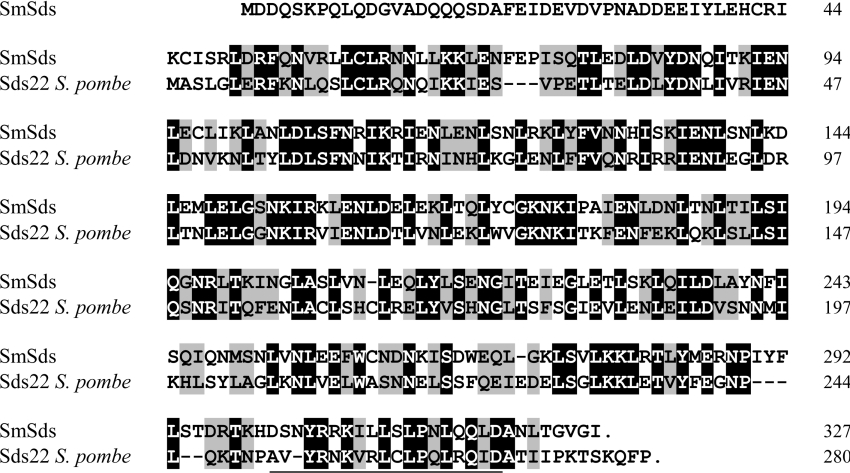
The SmSds deduced amino acid sequence shares 48% overall identity with the PP1 regulatory subunit sds22+ of Schizosaccharomyces pombe, and 62% similarity when taking into account the conserved substitution of residues (Figure 1). Regions conserved in the Sds22 families were found in SmSds which encompass several LRRs (leucine-rich repeats) that are known to be involved in protein–protein interactions. A close examination of the sequence using the Pfam database revealed that SmSds is related to LRR proteins, with the presence of 11 consensus sequences LXXLXLXXNXXLXLXXL [in which X can be any amino acid, and leucine can be replaced by valine, isoleucine or phenylalanine] (Figure 2A) [27]. The LRRs comprise the central 248 residues of the protein sequence and are surrounded by 55 and 25 amino acids at the N-terminal and C-terminal extremities respectively. The sequence at the C-terminal end contains a 19-amino-acid (301–319) sequence similar to a consensus sequence (YPXXΦXXXΦPXΦXXLD, where X is any amino acid and Φ represents any hydrophobic residue) considered to be an LRR cap.
Figure 1. Protein sequence alignment of SmSds with Schizosaccharomyces pombe Sds22+ (GenBank® accession number AAA35342).
Identical amino acid residues are in white-on-black reverse font, and similar residues are shaded in grey. The LRR cap sequence is underlined.
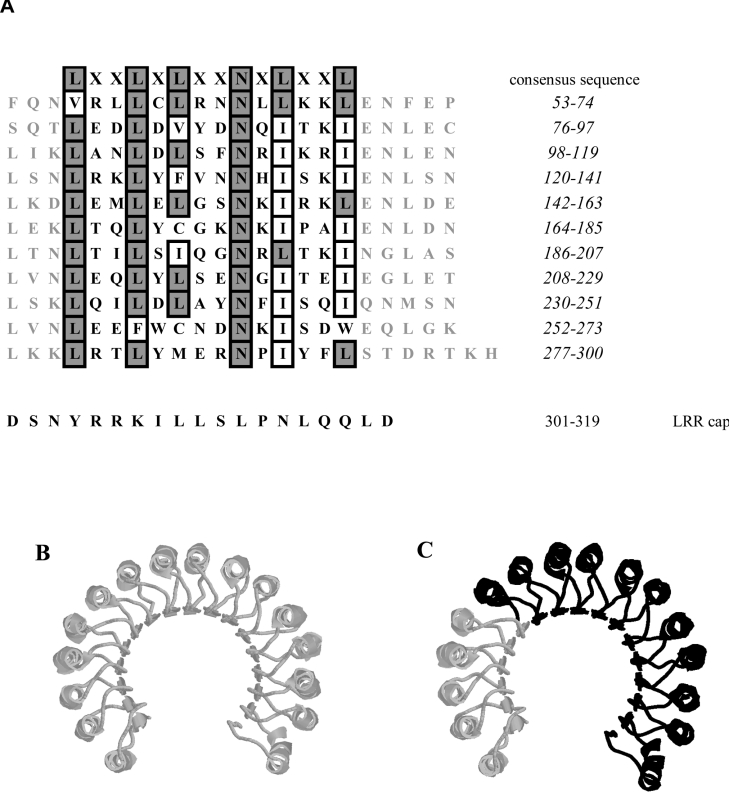
Figure 2. LRR motifs of SmSds.
(A) The 11 LRRs of SmSds are aligned to indicate the conserved positions. Numbers at the right side indicate the amino acid positions of each LRR motif. The leucine/isoleucine (position 4 in the consensus sequence), asparagine (position 9) and leucine/isoleucine (position 11) residues are absolutely conserved and only seven repeats out of 11 exactly fit the consensus. (B) Structural model of RNasin. α-Helices, β-sheets and coils are shown in monochrome shading. (C) Superposition of SmSds and RNasin proteins. The overlapping domains are shown in black.
Further analysis of the amino acid sequence using the Conserved Domain Database showed that the three-dimensional structure of the SmSds protein superposes a part of the architecture of the LRRs of the RNase inhibitor RNasin resolved by crystallization of the corresponding recombinant protein. RNasin contains several tandem copies of a 20–22-residue LRR, giving a curved horseshoe structure with 15 parallel β-sheets on the concave side and helical elements on the convex side (Figure 2B). The overlapping three-dimensional structure of SmSds fits with the 11 tandem repeats from the C-terminal side of a crystallized LRR protein, the RNasin (Figure 2C).
Characterization of SmPP1
We investigated further whether Schistosoma mansoni expresses the possible partner of SmSds, SmPP1. Public databases analysis allowed us to find in the TIGR Schistosoma mansoni database a 636 bp EST (TC6011) presenting a high degree of identity with PP1. To obtain the full-length cDNA, we performed 5′- and 3′-RACE as described in the Experimental section. The full sequence length of SmPP1 (1865 bp) revealed an ORF of 981 bp (GenBank® accession number AY886888). Comparative analysis of SmPP1 with known PP1 sequences showed 90% and 83% identity respectively with rabbit PP1 (GenBank® accession number P62139) and SpPP1 (GenBank® accession number CAA22875) (Figure 3). The deduced amino acid sequence presents the signature of serine/threonine phosphatases, LRGNHE (positions 121–126).
Figure 3. Protein sequence alignment of SmPP1 with rabbit PP1 and SpPP1 (GenBank® accession numbers P62139 and CAA22875).
Identical amino acid residues are in white-on-black reverse font. The six-amino-acid sequence specific to serine/threonine phosphatases is boxed.
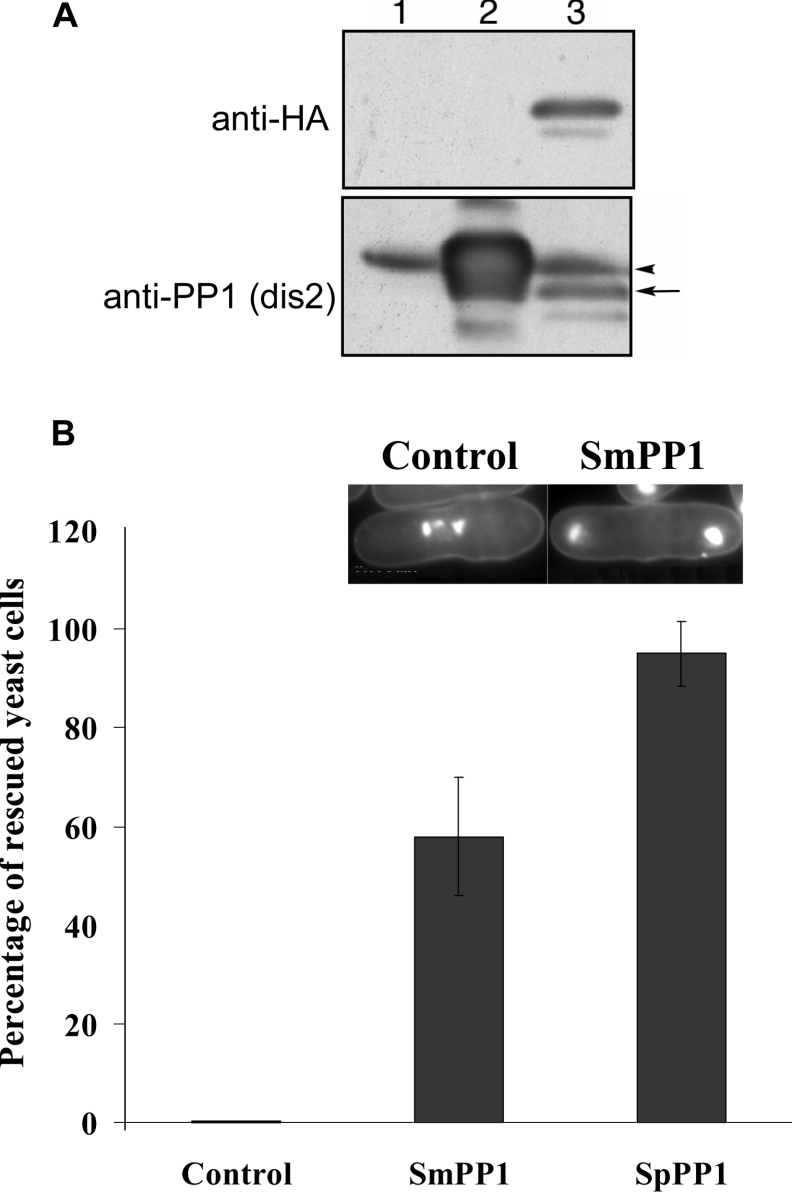
To assess the functional role of SmPP1, we examined its effect in a yeast model using a Schizosaccharomyces pombe strain mutated in the PP1 gene, with a conditional point mutation in the dis2+ gene. This strain was transformed with Rep1–SmPP1 or Rep1–SpPP1. First, it was essential to check whether SmPP1 could be expressed by Schizosaccharomyces pombe. Western blot experiments using anti-HA monoclonal antibody showed efficient expression (Figure 4A). In addition, SmPP1 was recognized by antibodies directed against SpPP1 (Figure 4A). At the restrictive temperature, the mutant yeast transformed with empty Rep1 arrested in mitosis with condensed chromosomes (results not shown), confirming previous results [22]. In contrast, transformants containing Rep1–SpPP1 or Rep1–SmPP1 were able to grow at the restrictive temperature, although we observed slower growth in the latter case (results not shown). However, all of the transformants isolated and tested independently showed complete recovery of the growth of the dis2 mutant complemented with SmPP1. To analyse further the effects of SmPP1 complementation on the abnormal mitotic phenotype of the mutated Schizosaccharomyces pombe strain, the chromosome segregations were investigated. In Figure 4(B), the frequencies of rescued cells with SmPP1 vector or SpPP1 vector (Dis-2) are presented, showing that approx. 60 and 95% of mutated cells exhibited the normal mitotic phenotype respectively. However, the transformation with control vector did not show any effect. Examination of chromosomes by DAPI staining revealed a correct segregation (Figure 4B, right inset) of chromosomes of mutated yeast complemented with SmPP1 when compared with control (Figure 4B, left inset).
Figure 4. Complementation of PP1-mutated yeast by SmPP1.
(A) SmPP1 expression in yeast. Western blot analysis of SmPP1 from wild-type Schizosaccharomyces pombe expressing empty vector (lane 1), Rep1–SpPP1 Dis2 (lane 2) and HA-tagged Rep1–SmPP1 (lane 3). The extracts of each transformant were immunoblotted with anti-HA (upper panel) or anti-SpPP1 (lower panel) antibodies. The arrow indicates the SmPP1 and the arrowhead indicates the SpPP1. (B) Frequencies of rescued yeast cells. The SpPP1-mutated strain was transformed with empty vector (Rep1) or Rep1–SmPP1 or Rep1–SpPP1 vector. Results are means±S.D. for two independent experiments performed with two different clones. In this yeast model, transformation with empty vector yielded up to 44.4% of abnormal mitosis. In the case of Dis2 and SmPP1 transformations, the percentage of cells with abnormal mitosis was 16.66% and 2.5% respectively. For clarity, we considered that the percentage of abnormal cells obtained with the empty vector alone is 100%. The percentages of abnormal cells obtained after transformation with Dis2 or SmPP1 vectors were calculated using the following formula: X=(percentage of abnormal cells obtained with either Dis2 or SmPP1 vector/percentage of abnormal cells obtained with empty vector alone)×100, thus allowing us to present data as frequencies of rescued cells (100−X%). The inset shows DAPI staining microscopy of Schizosaccharomyces pombe. Left: SpPP1-null single yeast showing condensed chromosomes typical of mitosis deficiency; right: yeast rescued with SmPP1, where the chromosomes are found at both ends of the cell.
Interaction of SmSds with SmPP1 by GST pull-down and in Schistosoma mansoni extracts
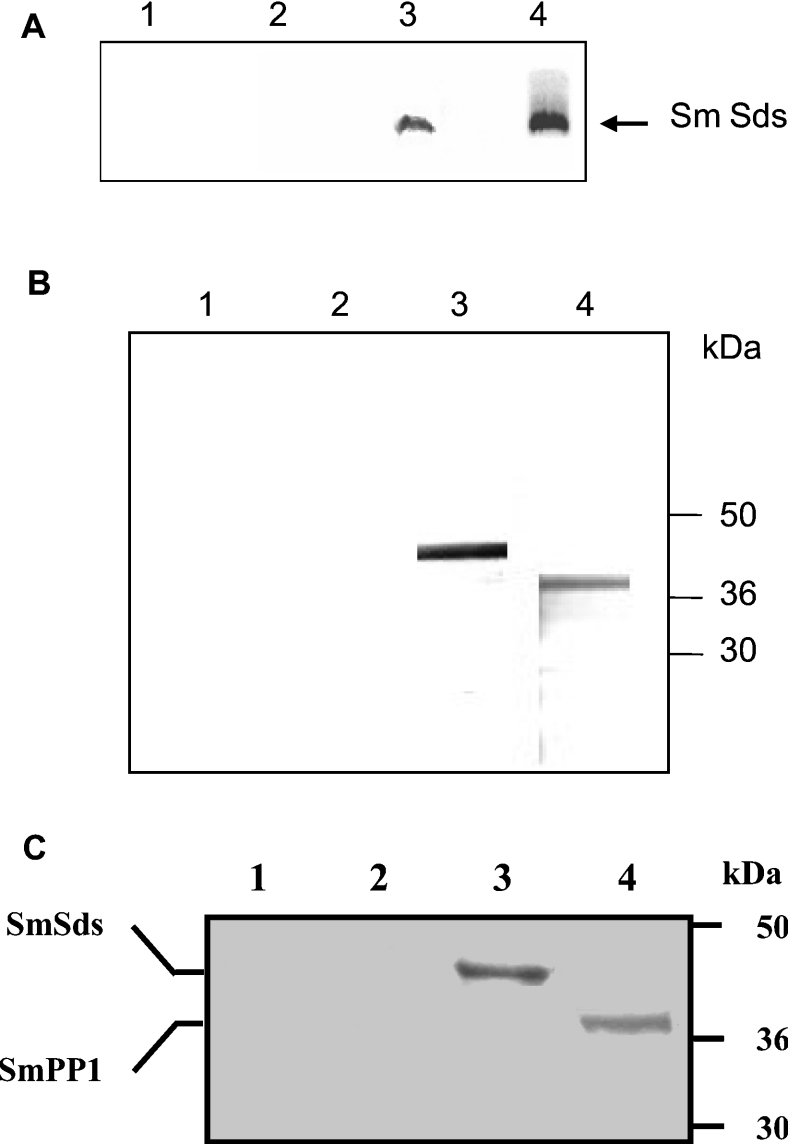
Analysis of SmSds and SmPP1 protein expression indicated that both proteins are present at the expected size in Schistosoma mansoni extracts (results not shown). Next, we searched for a possible interaction of these proteins as has been observed for their homologues in other species. To this end, we performed a GST pull-down assay in which the fusion protein GST–PP1 was incubated with in vitro-translated [35S]Met-labelled SmSds and then bound to glutathione–Sepharose beads. The results presented in Figure 5(A) indicate that GST–PP1 (lane 3), but not GST alone, was able to efficiently bind to recombinant SmSds protein.
Figure 5. Interaction of SmSds with SmPP1 in vitro and in Schistosoma mansoni.
(A) GST pull-down assays were performed with 35S-labelled SmSds incubated with beads alone (lane 1), GST bound to beads (lane 2) and GST–PP1-bound beads (lane 3). Lane 4 represents 20% of the input. (B) Immunoblot analysis of proteins eluted from microcystin–agarose. Eluates were separated by SDS/PAGE (4–12% gels) and transferred on to nitrocellulose. Lanes 1 and 2 represent the negative controls (pre-immune rat and mouse sera respectively). Lanes 3 and 4 represent the detection of SmSds and the detection of SmPP1 respectively. (C) Co-immunoprecipitation of the SmSds–SmPP1 complex with anti-SmSds antibodies from Schistosoma mansoni extracts. Immunoprecipitates from control pre-bleed sera or from SmSds antisera were eluted, separated by SDS/PAGE (4–12% gels) and transferred on to nitrocellulose. Immunoblot analysis was performed with anti-SmSds (lanes 1 and 3) and anti-SmPP1 (lanes 2 and 4) antibodies. Molecular-mass sizes are given in kDa.
The interaction of the two proteins was then investigated in the parasite by the isolation of complexes of SmSds–SmPP1 using microcystin, a cyclic heptapeptide that binds to PP1 [28]. Microcystin is routinely coupled to agarose beads for purification of PP1 along with associated regulatory subunits [29,30]. As can be seen in Figure 5(B), immunoblot analysis of proteins eluted from microcystin–agarose revealed the presence of both SmPP1 and SmSds, suggesting an interaction between these two proteins in the parasite. It is interesting to note that recombinant SmSds did not bind directly to microcystin–agarose (results not shown). To ascertain further the interaction between SmSds and SmPP1, immunoprecipitation experiments with anti-SmSds antibodies were performed. Immunoblot analysis of SmSds immunoprecipitates showed that PP1 had been co-immunoprecipitated with the SmSds from the parasite extracts (Figure 5C). Taken together, our data demonstrate that SmSds interacts physically with SmPP1 within Schistosoma mansoni.
Inhibitory effect of SmSds on PP1
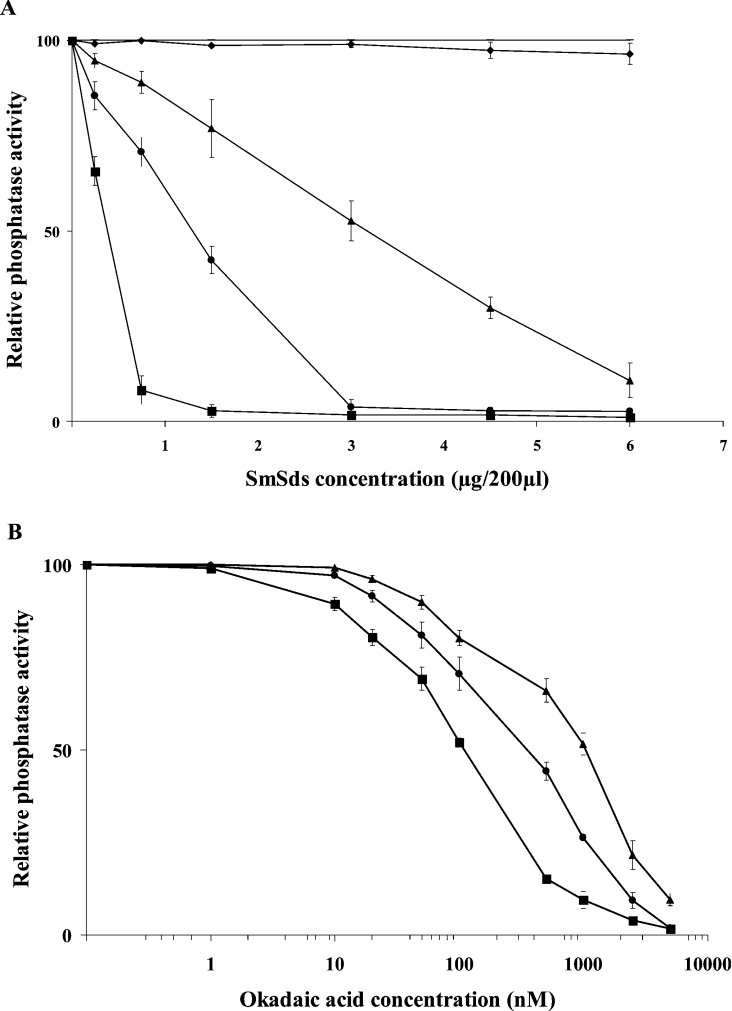
Next, the effect of SmSds on PP1 activity was assessed. To this end, we incubated different concentrations of an active PP1 with different concentrations of recombinant SmSds and measured the phosphatase activity against pNPP. The results presented in Figure 6(A) reveal that the addition of SmSds decreased the phosphatase activity up to 90% according to the concentration of PP1 used. The IC50 values obtained based on the predicted molecular mass of SmSds ranged between 500 and 4100 nM. The pre-incubation of PP1 with GST control protein did not decrease PP1 activity whatever the concentration used, indicating the specificity of inhibition by SmSds. For comparison, OA, a potent inhibitor of PP1, was used. As shown in Figure 6(B), OA decreased the phosphatase activity with IC50 values in the range 100–1000 nM, according to the concentration of PP1 used. These results, unlike those obtained with yeast Sds promoting an activation of PP1, support the idea that SmSds can be considered as one of the inhibitors of PP1.
Figure 6. Effects of recombinant SmSds on PP1 activity.
(A) Different amounts of recombinant PP1 (7000 units/mg) ranging from 0.5 to 5 units per assay were pre-incubated for 1 h at room temperature with different concentrations of SmSds before testing for phosphatase activity in a final volume of 200 μl. −, Activity in medium alone (100%); ♦, relative activity of PP1 in the presence of GST protein control (no inhibition was observed whatever the concentration used). Only the effect of GST on 0.5 unit of PP1 is represented; no significant effects were observed on 2 and 5 units of PP1 (results not shown). ▲, ● and ■ represent the relative activity of 5, 2 and 0.5 units respectively of PP1 in the presence of different concentrations of recombinant SmSds. (B) Effects of OA on PP1 activity. ▲, ● and ■ represent the relative activity of 5, 2 and 0.5 units respectively of PP1 in the presence of different concentrations of OA. Results are means±S.D. for three representative experiments performed in triplicate.
Induction of oocyte GVBD by SmSds
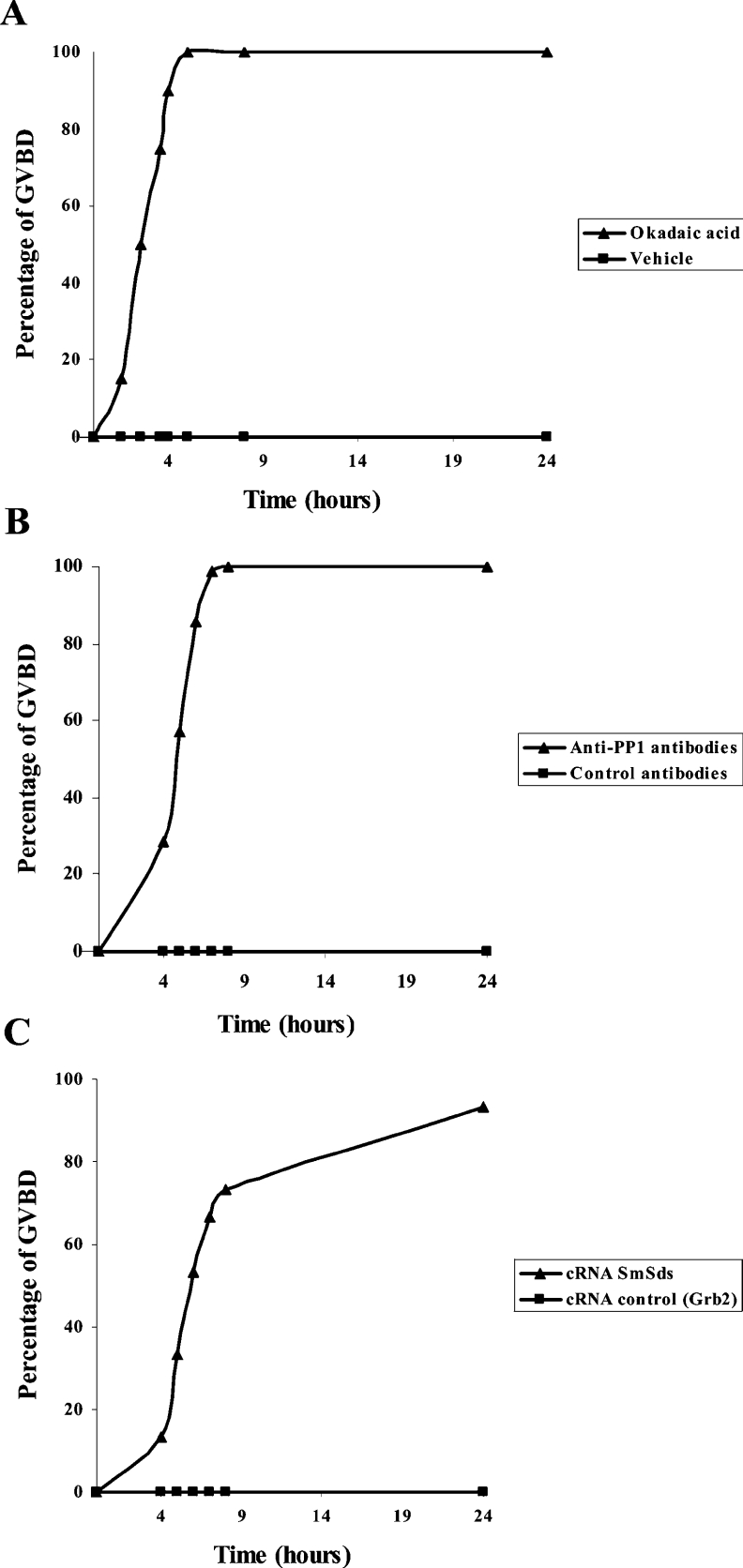
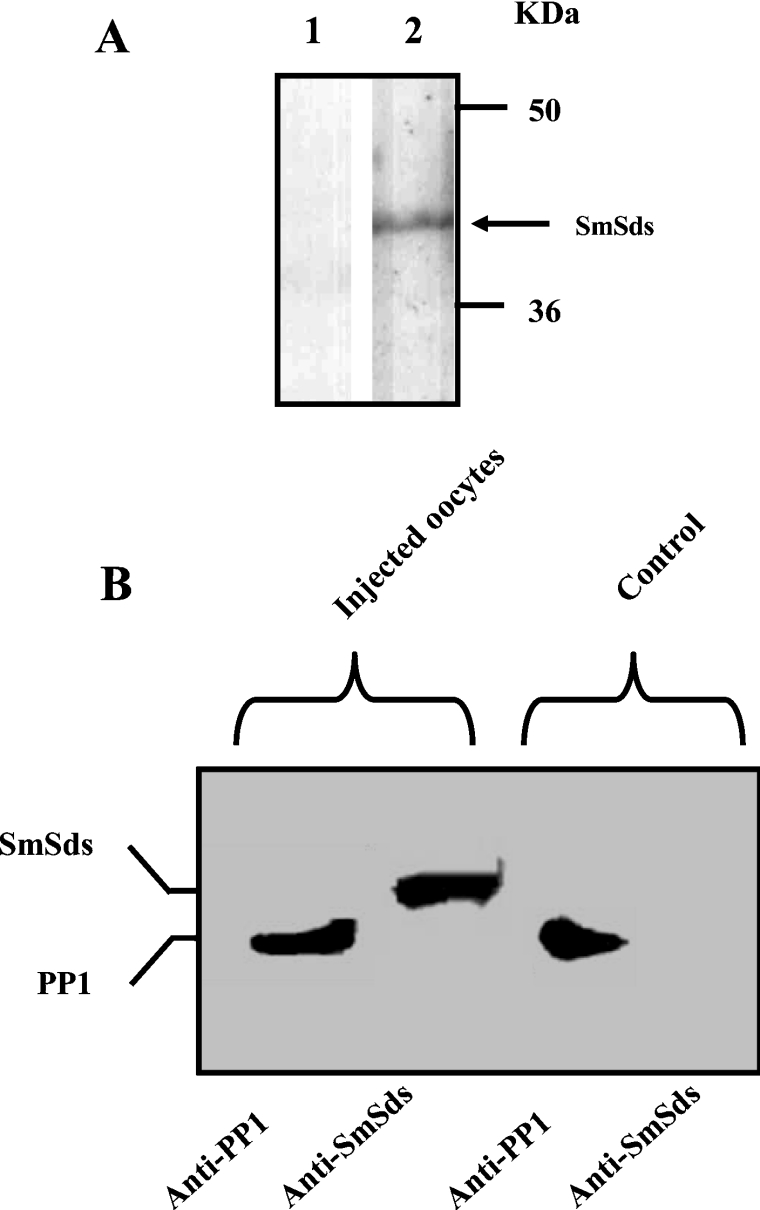
Xenopus oocytes, physiologically arrested at G2/M meiotic prophase I, have been investigated largely to analyse the phosphorylation/dephosphorylation process in the regulation of cell cycle (for a review, see [31]). Previously, it has been shown that meiotic progression can be achieved either by the microinjection of OA, a well-known phosphatase inhibitor, or anti-PP1 antibodies, leading to oocyte GVBD [32]. If SmSds inhibited PP1, then expression of SmSds in oocytes should lead to GVBD. We therefore microinjected SmSds RNA into oocytes and followed up the appearance of GVBD. First, we determined the different doses of OA, serum anti-PP1 and SmSds cRNA that were required for the initiation of GVBD. This allowed us to choose 10 μM for OA, 100 nl for serum anti-PP1 and 100 ng of SmSds cRNA for subsequent microinjections. As expected, OA induced GVBD very rapidly (90% within 4 h) (Figure 7A). Similarly to OA, anti-PP1 antibodies provoked GVBD, but reaching 85% at 6 h after microinjection (Figure 7B). With respect to SmSds, the microinjection of its cRNA resulted in 53% GVBD at 6 h and attained approx. 90% 24 h later (Figure 7C). If we compare the formation of 50% GVBD, we notice that OA acted more rapidly (2.5 h after injection) than anti-PP1 antibodies (5 h) and SmSds cRNA (6 h). This can be explained, at least in part, by the slow binding of anti-PP1 antibodies to PP1 or the translation step of SmSds cRNA that is required for protein expression. All negative controls including vehicle, mice pre-bleed serum and cRNA corresponding to a mutated Grb2 previously described [25] did not show any GVBD formation. The expression of SmSds protein in microinjected oocytes verified by Western blot on whole extracts revealed the presence of a band at the expected size (Figure 8A). Next, it was important to investigate whether SmSds may bind to Xenopus PP1. As can be seen in Figure 8(B), immunoblot analysis of eluted proteins from microcystin–agarose revealed the presence of both Xenopus PP1 and SmSds, suggesting an interaction between these two proteins in oocytes. As expected, the eluate of control oocytes that did not receive SmSds cRNA revealed only the presence of PP1.
Figure 7. Induction of GVBD in Xenopus oocytes.
Appearance of GVBD was monitored for 24 h after microinjection, and values are presented as percentages. Each experiment was performed using a set of 20 oocytes and was repeated two or three times. The kinetic study shown is representative of one experiment. (A) Percentage GVBD induced with 10 μM OA (▲) or vehicle (■). (B) Percentage GVBD induced with 100 nl of mouse anti-PP1 serum (▲) or control serum (■). (C) Percentage GVBD induced by the microinjection of 100 ng of SmSds cRNA (▲) or control cRNA (■).
Figure 8. SmSds expression in Xenopus oocytes and its interaction with endogenous PP1.
(A) Expression of SmSds protein by Xenopus oocytes microinjected with SmSds cRNA. Immunoblot analysis was performed on oocytes injected with vehicle (lane 1) or SmSds cRNA (lane 2). The arrow indicates SmSds. Molecular-mass sizes are given in kDa. (B) Interaction of SmSds with Xenopus PP1. Immunoblot analysis of eluates from oocytes extracts after binding to microcystin–agarose beads, revealed using either anti-PP1 antibodies or anti-SmSds antibodies. The two left-hand lanes correspond to SmSds cRNA-injected oocyte eluates, and the two right-hand lanes correspond to eluates of vehicle-injected oocytes.
DISCUSSION
In the process of searching for regulatory mechanisms of protein dephosphorylation, the Sds22 protein has been shown to regulate PP1 activity by a direct interaction. The identification of this regulatory protein and others has been invaluable in the analysis of molecular events that control important cell functions [33]. Thus the investigation of phosphatase-regulatory proteins in pathogens and parasites may constitute an additional step towards a better understanding of parasite biology and the discovery of potential therapeutic targets. In the helminth parasite Schistosoma mansoni, the presence of serine/threonine phosphatase activities (PP1, PP2A, PP2B and PP2C) has been shown, leading subsequently to the molecular characterization of PP2B [5,6]. The importance of Sds22 in the regulation of yeast and mammalian PP1 has prompted us to analyse the function of its homologue in Schistosoma mansoni. The comparative analysis of yeast Sds22 with the Schistosoma mansoni EST, in combination with reverse transcription–PCR and RACE, allowed us to characterize the Schistosoma mansoni Sds homologue gene. The predicted schistosome Sds is 48% identical with Schizosaccharomyces pombe Sds22, with an overall similarity of 62%. This degree of identity/similarity is similar to that observed between the Sds22 proteins of Schizosaccharomyces pombe and Saccharomyces cerevisiae. From sequence analysis, SmSds emerges as a prime candidate for regulation of PP1 function. First, it showed the presence of several LRRs that are well known to participate in protein–protein interactions. It is interesting to note that SmSds contains 11 LRRs, covering 80% of the whole protein. Each LRR of SmSds is composed of a 22-residue motif that is typical to the Sds subfamily of proteins [34]. From a structural point of view, the crystal structures of two LRR proteins, together with the predicted three-dimensional structures of many others [34–36], indicated that the LRR unit comprises a short β-strand (the first 11 amino acids), followed by a fragment folded into a helicoidal conformation. The LRRs are arranged so that the protein adopts a horseshoe shape with curved parallel β-strands lining on the concave side, while the α-helices flank its convex side. In the case of human Sds22, interaction studies showed that the concave surface formed by the LRRs is essential for binding to PP1 [16]. The SmSds sequence indicated the presence of such putative structures that are essential for its function. In addition, the SmSds gene product contains the motif RNPIYF (positions 287–292), similar to the consensus sequence RXXV(I)XF that is found in many PP1-binding subunits [37,38]. Binding studies with different peptides showed that the loss of Val-Xaa-Phe abolished the ability of the peptide to bind to PP1 [39], but, unexpectedly, yeast and human Sds22 proteins lack this motif, but retain their ability to bind to PP1 [40]. Finally, SmSds includes an LRR cap at the C-terminal end (positions 301–319) that matches perfectly with the consensus sequence described by Ceulemans et al. [41]. This conserved structure was proposed to play a role in the protection of the hydrophobic core of the LRRs that is required for the protein binding activity. It is also important to point out that mutational studies indicate that the LRR cap domain present in the yeast Sds22 seems to be involved in nuclear targeting, while the central repetitive domain interacts with PP1 [21].
It is well known that reversible protein phosphorylation plays a key role in a variety of cellular functions. PP1 has been shown to be involved in mitotic progression in several organisms from yeast to mammals [12,42,43]. The presence of PP1 in a wide range of organisms together with the presence of SmSds, a potential partner of PP1, has led us to investigate its expression in Schistosoma mansoni. Cloning experiments allowed us to isolate the PP1 gene of Schistosoma mansoni, designated SmPP1. Comparative analysis showed a high degree of identity (>85%) with several known PP1s (rabbit, human, rat and yeast). Furthermore, the deduced amino acid sequence of SmPP1 revealed a near perfect alignment of the catalytic domain (positions 10–303) and the presence of the signature sequence of serine/threonine phosphatases, LRGNHE. In order to evaluate the functional role of the SmPP1 gene product, complementation assays of a PP1 mutant Schizosaccharomyces pombe strain were conducted. Using this approach, SmPP1 was shown to be expressed and was able to rescue the loss-of-function mutation related to SpPP1. These results demonstrate that SmPP1 is not only similar to Schizosaccharomyces pombe PP1 at the sequence level, but also functionally complements the activity of PP1 in yeast.
PP1 is one of the most highly conserved eukaryotic proteins. In the cell, it seems to be often associated with regulatory proteins [44]. Some regulatory binding sites of mammalian PP1 have been mapped, including that for Sds22 [16]. It has been shown that Sds22 interacts with PP1 through a triangular region composed of three consecutive helix structures [16]. Such structures were also present in SmPP1. These observations, together with the expression of SmSds gene product by Schistosoma mansoni, a potential partner to SmPP1, led us to investigate whether they may interact. First, we observed using GST pull-down assays that SmSds bound directly with SmPP1. More importantly, we were able to isolate the complex SmSds–SmPP1 from Schistosoma mansoni extracts. This was achieved by using microcystin, a well-known interactor with PP1, and was confirmed further by the co-immunoprecipitation experiments with anti-SmSds antibodies. Our results are in agreement with those obtained from studies performed in yeast or mammalian cells [12,16,45]. Taken together, these data clearly demonstrate the interaction of SmSds and SmPP1, raising the question of a possible regulatory effect on PP1 activity. This hypothesis was validated by the fact that the addition of recombinant SmSds reduced up to 90% the phosphatase activity related to recombinant rabbit PP1, which shows 85% identity with SmPP1. Although these results are different from those observed in yeast, they are in agreement with data showing an inhibitory effect of rat Sds on PP1, but not PP2A, activity [45]. Finally, as PP1s are highly conserved among different species, it was pertinent to determine whether the inhibitory effect of SmSds on PP1 could be operative in vitro in cells. To address this point, we used Xenopus oocytes that were physiologically arrested in G2/M prophase I and in which OA, a specific protein phosphatase inhibitor, induced GVBD [46]. In the present study, we observed that the microinjection of SmSds into oocytes, similar to anti-PP1 antibodies and OA, promoted the progression of oocytes to M-phase by inducing GVBD. Our results regarding the effect of anti-PP1 antibodies are in agreement with those obtained in roscovitine-arrested mouse oocytes, indicating that the inhibition of PP1 is a crucial event in the regulation of nuclear envelope dissolution [32] and consequently of GVBD. The observation that microcystin beads isolated the complex SmSds–PP1 from oocyte extracts, together with the above results, supports the idea that driving the phosphorylation/dephosphorylation process in favour of the latter initiates oocyte GVBD. With respect to the effect of SmSds on active SmPP1, we have tried to express the latter in various expression systems, but we have invariably obtained aggregated recombinant protein without any activity after refolding.
In conclusion, we show that SmSds interacts with PP1 both in vitro as observed by GST pull-down experiments and in parasites by co-isolating the SmPP1–SmSds complex. This interaction seems to decrease the PP1-related activity. More importantly, our results provide the first direct evidence that SmSds can provoke the appearance of oocyte GVBD. Although the mode of action of SmSds in the parasite remains an open question, it is likely that it participates in the regulation of cell cycle in Schistosoma mansoni. In the future, three-dimensional structure analyses of SmSds would be helpful in order to design specific inhibitors that may prevent its binding with PP1 and that may affect parasite growth and/or survival.
Acknowledgments
This work was supported by Unité INSERM 547 and Institut Pasteur de Lille. We thank Dr Ray Pierce for a critical review of the manuscript.
References
- 1.King C. L. Initiation and regulation of disease in schistosomiasis. In: Mahmoud A. A. F., editor. Schistosomiasis. London: Imperial College Press; 2001. pp. 213–264. [Google Scholar]
- 2.WHO. Geneva: WHO; 2002. Schistosomiasis: Strategic Direction for Research ( http://www.who.int/tdr/diseases/schisto/files/direction.pdf) [Google Scholar]
- 3.Luan S. Protein phosphatases in plants. Annu. Rev. Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 5.Khattab A., Pica-Mattoccia L., Wenger R., Cioli D., Klinkert M. Q. Assay of Schistosoma mansoni calcineurin phosphatase activity and assessment of its role in parasite survival. Mol. Biochem. Parasitol. 1999;99:269–273. doi: 10.1016/s0166-6851(99)00014-6. [DOI] [PubMed] [Google Scholar]
- 6.Mecozzi B., Rossi A., Lazzaretti P., Kady M., Kaiser S., Valle C., Cioli D., Klinkert M. Q. Molecular cloning of Schistosoma mansoni calcineurin subunits and immunolocalization to the excretory system. Mol. Biochem. Parasitol. 2000;110:333–343. doi: 10.1016/s0166-6851(00)00287-5. [DOI] [PubMed] [Google Scholar]
- 7.Shenolikar S. Protein serine/threonine phosphatases – new avenues for cell regulation. Annu. Rev. Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 8.Wera S., Hemmings B. A. Serine/threonine protein phosphatases. Biochem. J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggen J. B., Nairn A. C., Chamberlin R. Regulation of protein phosphatase-1. Chem. Biol. 2000;7:R13–R23. doi: 10.1016/s1074-5521(00)00069-7. [DOI] [PubMed] [Google Scholar]
- 10.Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem. Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 11.Cohen P. T. Protein phosphatase 1 – targeted in many directions. J. Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 12.Ohkura H., Yanagida M. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- 13.Hisamoto N., Frederick D. L., Sugimoto K., Tatchell K., Matsumoto K. The EGP1 gene may be a positive regulator of protein phosphatase type 1 in the growth control of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:3767–3776. doi: 10.1128/mcb.15.7.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacKelvie S. H., Andrews P. D., Stark M. J. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 1995;15:3777–3785. doi: 10.1128/mcb.15.7.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagida M., Kinoshita N., Stone E. M., Yamano H. Protein phosphatases and cell division cycle control. Ciba Found. Symp. 1992;170:130–140. doi: 10.1002/9780470514320.ch9. [DOI] [PubMed] [Google Scholar]
- 16.Ceulemans H., Vulsteke V., De Maeyer M., Tatchell K., Stalmans W., Bollen M. Binding of the concave surface of the Sds22 superhelix to the α4/α5/α6-triangle of protein phosphatase-1. J. Biol. Chem. 2002;277:47331–47337. doi: 10.1074/jbc.M206838200. [DOI] [PubMed] [Google Scholar]
- 17.Dissous C., Grzych J. M., Capron A. Schistosoma mansoni shares a protective oligosaccharide epitope with freshwater and marine snails. Nature (London) 1986;323:443–445. doi: 10.1038/323443a0. [DOI] [PubMed] [Google Scholar]
- 18.Khalife J., Grzych J. M., Pierce R., Ameisen J. C., Schacht A. M., Gras-Masse H., Tartar A., Lecocq J. P., Capron A. Immunological crossreactivity between the human immunodeficiency virus type 1 virion infectivity factor and a 170-kD surface antigen of Schistosoma mansoni. J. Exp. Med. 1990;172:1001–1004. doi: 10.1084/jem.172.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maundrell K. nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 20.Okazaki K., Okazaki N., Kume K., Jinno S., Tanaka K., Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone E. M., Yamano H., Kinoshita N., Yanagida M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 1993;3:13–26. doi: 10.1016/0960-9822(93)90140-j. [DOI] [PubMed] [Google Scholar]
- 22.Ohkura H., Kinoshita N., Miyatani S., Toda T., Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- 23.Adachi Y., Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J. Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takai A., Mieskes G. Inhibitory effect of okadaic acid on the p-nitrophenyl phosphate phosphatase activity of protein phosphatases. Biochem. J. 1991;275:233–239. doi: 10.1042/bj2750233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cailliau K., Browaeys-Poly E., Broutin-L'Hermite I., Nioche P., Garbay C., Ducruix A., Vilain J. P. Grb2 promotes reinitiation of meiosis in Xenopus oocytes. Cell. Signalling. 2001;13:51–55. doi: 10.1016/s0898-6568(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 26.Vicogne J., Cailliau K., Tulasne D., Browaeys E., Yan Y. T., Fafeur V., Vilain J. P., Legrand D., Trolet J., Dissous C. Conservation of epidermal growth factor receptor function in the human parasitic helminth Schistosoma mansoni. J. Biol. Chem. 2004;279:37407–37414. doi: 10.1074/jbc.M313738200. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B., Kajava A. V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 28.Honkanen R. E., Zwiller J., Moore R. E., Daily S. L., Khatra B. S., Dukelow M., Boynton A. L. Characterization of microcystin-LR, a potent inhibitor of type 1 and type 2A protein phosphatases. J. Biol. Chem. 1990;265:19401–19404. [PubMed] [Google Scholar]
- 29.Moorhead G., MacKintosh R. W., Morrice N., Gallagher T., MacKintosh C. Purification of type 1 protein (serine/threonine) phosphatases by microcystin–Sepharose affinity chromatography. FEBS Lett. 1994;356:46–50. doi: 10.1016/0014-5793(94)01232-6. [DOI] [PubMed] [Google Scholar]
- 30.MacKintosh R. W., Dalby K. N., Campbell D. G., Cohen P. T., Cohen P., MacKintosh C. The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1. FEBS Lett. 1995;371:236–240. doi: 10.1016/0014-5793(95)00888-g. [DOI] [PubMed] [Google Scholar]
- 31.Philpott A., Yew P. R. The Xenopus cell cycle: an overview. Methods Mol. Biol. 2005;296:95–112. doi: 10.1385/1-59259-857-9:095. [DOI] [PubMed] [Google Scholar]
- 32.Swain J. E., Wang X., Saunders T. L., Dunn R., Smith G. D. Specific inhibition of mouse oocyte nuclear protein phosphatase-1 stimulates germinal vesicle breakdown. Mol. Reprod. Dev. 2003;65:96–103. doi: 10.1002/mrd.10258. [DOI] [PubMed] [Google Scholar]
- 33.Printen J. A., Brady M. J., Saltiel A. R. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- 34.Kajava A. V. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 35.Price S. R., Evans P. R., Nagai K. Crystal structure of the spliceosomal U2B″–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature (London) 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 36.Kobe B., Deisenhofer J. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhao S., Lee E. Y. A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J. Biol. Chem. 1997;272:28368–28372. doi: 10.1074/jbc.272.45.28368. [DOI] [PubMed] [Google Scholar]
- 38.Wakula P., Beullens M., Ceulemans H., Stalmans W., Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 39.Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X., Tatchell K. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry. 2001;40:7410–7420. doi: 10.1021/bi002796k. [DOI] [PubMed] [Google Scholar]
- 41.Ceulemans H., De Maeyer M., Stalmans W., Bollen M. A capping domain for LRR protein interaction modules. FEBS Lett. 1999;456:349–351. doi: 10.1016/s0014-5793(99)00965-5. [DOI] [PubMed] [Google Scholar]
- 42.Bloecher A., Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez A., Brautigan D. L., Lamb N. J. Protein phosphatase type 1 in mammalian cell mitosis: chromosomal localization and involvement in mitotic exit. J. Cell Biol. 1992;116:1421–1430. doi: 10.1083/jcb.116.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceulemans H., Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 45.Dinischiotu A., Beullens M., Stalmans W., Bollen M. Identification of sds22 as an inhibitory subunit of protein phosphatase-1 in rat liver nuclei. FEBS Lett. 1997;402:141–144. doi: 10.1016/s0014-5793(96)01514-1. [DOI] [PubMed] [Google Scholar]
- 46.Goris J., Hermann J., Hendrix P., Ozon R., Merlevede W. Okadaic acid, a specific protein phosphatase inhibitor, induces maturation and MPF formation in Xenopus laevis oocytes. FEBS Lett. 1989;245:91–94. doi: 10.1016/0014-5793(89)80198-x. [DOI] [PubMed] [Google Scholar]