Abstract
Furin catalyzes the proteolytic maturation of many proproteins within the trans-Golgi network (TGN)/endosomal system. Furin's cytosolic domain (cd) directs both the compartmentalization to and transit between its manifold processing compartments (i.e., TGN/biosynthetic pathway, cell surface, and endosomes). Here we report the identification of the first furin cd sorting protein, ABP-280 (nonmuscle filamin), an actin gelation protein. The furin cd was used as bait in a yeast two-hybrid screen to identify ABP-280 as a furin-binding protein. Binding analyses in vitro and coimmunoprecipitation studies in vivo showed that furin and ABP-280 interact directly and that ABP-280 tethers furin molecules to the cell surface. Quantitative analysis of both ABP-280-deficient and genetically replete cells showed that ABP-280 modulates the rate of internalization of furin but not of the transferrin receptor, a cycling receptor. However, although ABP-280 directs the rate of furin internalization, the efficiency of sorting of the endoprotease from the cell surface to early endosomes is independent of expression of ABP-280. By contrast, efficient sorting of furin from early endosomes to the TGN requires expression of ABP-280. In addition, ABP-280 is also required for the correct localization of late endosomes (dextran bead uptake) and lysosomes (LAMP-1 staining), demonstrating a pleiotropic role for this actin binding protein in the organization of cellular compartments and directing protein traffic. Finally, and consistent with the trafficking studies on furin, we showed that ABP-280 modulates the processing of furin substrates in the endocytic but not the biosynthetic pathways. The novel roles of ABP-280 and the cytoskeleton in the sorting of furin in the TGN/ endosomal system and the formation of proprotein processing compartments are discussed.
Genetic and biochemical studies have shown that the regulation of membrane and protein traffic between cellular compartments requires the orchestrated interactions of a large number of components including lipid, cytosolic, and membrane proteins and elements of the cytoskeleton (Cole and Lippincott-Schwartz, 1995; Drubin and Nelson, 1996; Mays et al., 1994; Stack et al., 1995). These interactions have been best described in studies on the organization of the secretory pathway and the localization and routing of proteins through its manifold compartments. The secretory pathway compartments can be subdivided into two central membrane populations, the endoplasmic reticulum/Golgi system and the trans-Golgi network (TGN)1/endosomal system (Roth et al., 1985). The TGN/ endosomal system is central to membrane-associated protein routing and recovery (Griffiths and Simons, 1986; Gruenberg and Howell, 1989). In addition to housing several biochemical reactions, including proprotein cleavage, the TGN orchestrates the sorting of proteins traversing the secretory pathway into lysosomes, regulated and constitutive exocytic vesicles, and, in polarized cells, to the basolateral and apical membranes (Pfeffer and Rothman, 1987; Tooze et al., 1990). Similarly, the plasma membrane is a site of dynamic regulation where proteins are internalized or recycled back to the same membrane domain. Thus, excluding proteins from nascently formed vesicles becomes an important determinant in the maintenance of the correct functional and structural organization of the membrane. Membrane proteins recovered from the cell surface into early endosomes can be recycled, transferred to lysosomal and/or late endosomal/prelysosomal compartments, transcytosed, or delivered to the TGN.
Despite the importance of the TGN/endosomal compartments to the organization and catalysis of cellular processes, relatively little is known about how these structures are formed and maintained. Clearly, however, the cytoskeleton is an important and contributing factor. Components of the cytoskeleton participate in many aspects of the localization, shape, and trafficking of organelles that comprise the membrane-limited compartments of the biosynthetic and endocytic pathways (Cole and Lippincott-Schwartz, 1995; Costa de Beauregard et al., 1995; Drubin and Nelson, 1996; Hu et al., 1995; Rodriguez-Boulan and Powell, 1992; van Deurs et al., 1995).
Microfilaments have a central role in directing vesicle and protein traffic. In yeast, genes encoding microfilament- associated proteins are important for receptor internalization, early steps of endocytosis, and vesicle transport (Liu and Bretscher, 1992; Munn et al., 1995). Studies with cytochalasin D show that in polarized animal cells microfilaments are necessary for the internalization of membrane proteins and fluid phase markers from the apical surface (Gottlieb et al., 1993). Furthermore, several actin binding proteins such as ABP-280, α-actinin, and ankyrin direct the organization of resident plasma membrane proteins (e.g., cell adhesion molecules, ion pumps, and receptors) by cross-linking them to microfilaments (Carpen et al., 1992; Lokeshwar et al., 1994).
Unlike the stable tethering of several resident cell-surface membrane proteins (by actin binding protein-mediated cross-linking to microfilaments), the routing of TGN/ endosomal itinerant membrane proteins is a dynamic process requiring transient interactions with as yet unidentified components of the cellular sorting machinery. Studies of the trafficking of a number of itinerant membrane proteins have demonstrated that their routing and compartmentalization requires sequences and motifs within their cytosolic domains (Trowbridge and Collawn, 1993). One example of such an itinerant membrane protein that is routed between compartments within the TGN/endosomal system is the proprotein convertase furin.
Furin is a membrane-associated, calcium-dependent serine endoprotease that proteolytically activates a large number of proprotein molecules in multiple cellular compartments by cleavage of these substrates at the COOH-terminal side of the consensus sequence -Arg-X-Lys/Arg-Arg↓ - (Molloy et al., 1992, for review see Steiner et al., 1992). In the TGN/biosynthetic pathway, furin cleaves a number of proprotein substrates at consensus furin sites including those for neurotrophic factors (Bresnahan et al., 1990), serum proteins (Misumi et al., 1991; Wasley et al., 1993; Wise et al., 1990), receptors (Bravo et al., 1994; Komada et al., 1993), and growth factors (Dubois et al., 1995). In addition, furin catalyzes the proteolytic maturation of several viral coat proteins including NDV-Fo (Gotoh et al., 1992), HIV-1 gp160 (Hallenberger et al., 1992), measles virus Fo (Watanabe et al., 1995), cytomegalovirus gB (Vey et al., 1995), and avian influenza HA (Steinecke-Grober et al., 1992; Walker et al., 1994). Similar to these viral pathogens, cleavage of many bacterial toxins by furin molecules present at the plasma membrane (e.g., anthrax toxin) and/or in endosomal compartments (e.g., Pseudomonas, Diphtheria, and Shiga toxins) is critical for their virulence (Chiron et al., 1994; Garred, et al., 1995; Gordon et al., 1995; Inocencio et al., 1994; Klimpel et al., 1992). The ability of furin to process endogenous secretory proteins and viral envelope glycoproteins in the TGN/biosynthetic pathway, as well as bacterial toxins at the cell surface and in endosomes, points to an important biological role for this protease in the activation of proproteins in multiple cellular compartments. It further suggests that factors that direct the trafficking of furin through these varied compartments will have a profound effect on the distribution and hence biological activity of the enzyme. Thus, a key to understanding the manifold roles of furin is the determination of the trafficking pathway of this enzyme and the cellular factors that govern its movement through the TGN/endosomal system.
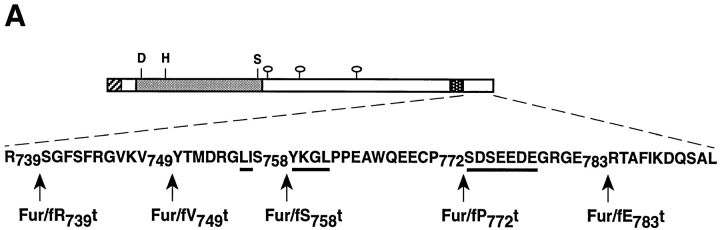
Furin is concentrated in the TGN and cycles between this compartment and two other furin processing compartments: the cell surface and endosomes (Chapman and Munro, 1994; Molloy et al., 1994). Both the TGN localization and the intracellular routing of furin require sorting signals in its 56-amino acid cytosolic domain (cd) (Chapman and Munro, 1994; Molloy et al., 1994). Sorting signals in the furin-cd include canonical Tyr-based and di-leucine-like internalization signals, as well as an acidic cluster that is required for localization of furin to the TGN (Jones et al., 1995; Schafer et al., 1995; Takahashi et al., 1995; Voorhees et al., 1995) (see Fig. 1 A).The routing of furin is thus predicted to be governed by the compartment-specific interactions of sequences within its cd with appropriate binding proteins.
Figure 1.
(A) Schematic of epitope-tagged furin (Fur/f) and COOH-terminal cytosolic domain truncated forms. The NH2-terminal cross-hatched box shows the FLAG epitope inserted COOH-terminal to the autoproteolytic maturation site (Molloy et al., 1994). The FLAG epitope cross-reacts with both mAbs M1 and M2. Binding of the mAb M1 requires the free amino terminus of the FLAG tag, whereas mAb M2 does not. Because autoproteolytic cleavage of the furin propeptide is localized to the RER (Molloy et al., 1994), all furin molecules trafficking in the TGN/endosomal compartments are capable of binding both mAbs. The lightly shaded area represents the catalytic domain with the Asp (D), His (H) and Ser (S) residues that form the catalytic triad, the hooked ovals represent NH2-linked carbohydrates and the dark-stippled box represents the membrane spanning domain. The sequence of the 56-amino acid cd is shown including the location of each of the COOH-terminal truncation sites. The Tyr-based and di-leucine-like internalization motifs, as well as the acidic cluster necessary for TGN localization of furin are underlined. (B) Two-hybrid analysis of furin cd truncations (bait), with library clone TP107, is identified in the initial screen. Yeast cells were cotransformed with pTP107 (Leu− medium) and bait plasmids (Trp− medium) expressing either furin cd or one of the furin cd truncation mutants (A) and selected for growth on Leu−/Trp− medium. Colonies from the Leu−/Trp− plates were streaked onto His- plates and scored for growth.
Although the cytoskeleton is important to the organization and trafficking through the TGN/endosomal system, a specific role for any individual protein has not been reported previously. Here, we report a new role for the actin filament cross-linking protein, ABP-280, in directing the trafficking of furin in the TGN/endosomal system and modulating the processing of furin substrates in the endocytic pathway.
Materials and Methods
Yeast Two-Hybrid Screen
The methods and plasmids used in the two hybrid screen are described elsewhere (Bartel and Fields, 1995). The mouse embryo cDNA library used in these studies (Vojtek et al., 1993) was generated from RNA isolated from 9.5- and 10.5-d embryos. The library contained both oligo dT and random primed inserts (size selected for 350–700 bp) subcloned into the plasmid pVP16, which directs the expression of protein chimeras containing the Herpes virus transcriptional activation domain fused to the subcloned insert.
pBTM116Fur Constructs.
DNA sequences encoding the furin-cd or the furin-cd truncation mutants depicted in Fig. 1 A were amplified by PCR with Taq polymerase (Boehringer Mannheim, Mannheim, Germany) using standard methods (Sambrook et al., 1989). pZVneo:furin constructs encoding each protein (Jones et al., 1995; Molloy et al., 1994) were used as templates and GTCTTCCGGATCCCGGGGCTGCGCTCTGGC (Oligo 1) and CGTTTGCCATACGCTCA (Oligo 2) as the primers. The PCR products encoding either the entire furin cd (digested with BamHI) or each of the furin cd truncation mutants (digested with BamHI and PstI) were subcloned into the two-hybrid bait plasmid pBTM116 (Bartel and Fields, 1995). pBTM116 directs the expression of a chimeric protein containing the LexA DNA-binding domain fused to sequences encoded by the inserted DNA.
Yeast Transformation.
Media recipes are described in Sherman (1991). Yeast strain L40 (MATa his3, trp, ade2, LYS::(lexAop)4-HIS3 URA::(lexAop)8-lacZ GAL4) containing pBTM116-Furcd were grown overnight in 100 ml yeast complete (YC) media with added leucine and histidine (100 μg/ml each). The overnight culture was used to inoculate 1 liter of yeast extract, peptone, adenine, dextrose (YPAD) and cultured for an additional 4 h. The cells were washed with 500 ml TE and resuspended in 20 ml 100 mM LiAc/0.5× TE (5 mM Tris, pH 7.5, 0.5 mM EDTA). 10 mg of denatured salmon sperm DNA and 500 μg of the pVP16 mouse embryo cDNA library were mixed with the cells before addition of 140 ml of 100 mM LiAC/40% PEG-3350/TE. After an incubation at 30°C for 30 min, DMSO (12% final) was added and the cells incubated at 42°C for 6 min. The cells were washed with YPA media, resuspended in one liter of YPAD medium, and incubated at 30°C for 1 h. The cells were pelleted, washed with YC media minus leucine and tryptophan, and then grown in the same media for 8 h. The yeast were applied onto YC minimal media plates minus leucine, tryptophan, and histidine. After 3 d, the His+ colonies were replica plated on nitrocellulose filters and tested for β galactosidase activity. The initial screen of 1.5 × 108 cotransformed library clones resulted in the identification of 200 positive yeast colonies (selected for growth on his− medium and positive for β galactosidase activity).
Plasmids and Bacterial Fusion Protein Production
Production of pGEX Plasmids and Fusion Proteins.
The pZVneo:furin plasmid containing the furin cd (Jones et al., 1995) was used as the PCR template. The 5′ primer was GCGCGCGGATCCTGCGCTCTGGCTTTAGT and the 3′ primer was Oligo 2. The PCR product containing the sequence encoding the furin-cd and was digested with BamHI. The insert was then subcloned into pGEX-3X (Pharmacia LKB Biotechnology, Inc., Piscataway, NJ). Methods for production and purification of GST-fusion proteins were performed as described by the manufacturer (Pharmacia LKB Biotechnology, Inc.).
Production of pET16b-ABP.
DNA sequences encoding mouse ABP-280 in pTP107 were amplified by PCR using the oligonucleotides ACTCTTCTCGAGGCGCCCAAAGGCCTGGTG (5 prime) and TAGCGGGAATTCAGCCACCGTGTATGT (3 prime) as primers. The amplification product was digested with XhoI, and then ligated into pET-16b that had been digested first with BamHI, made blunt-ended, and then digested with XhoI. His-tagged proteins were isolated as described by the manufacturer (Novagen, Inc., Madison, WI).
GST-binding Experiments
6 μg of HistagABP-280 (containing residues 1490–1607 of mouse ABP-280) were combined with 10 μg of either GST or a GST fusion protein containing the entire furin cd. The protein pairs were incubated in PBS, pH 7.0, containing 0.5% NP-40 and 0.1 mg/ml BSA for 2 h at room temperature. Glutathione agarose (Sigma Chemical Co., St. Louis, MO) was then added and the sample was incubated an additional hour. The beads were washed three times with binding buffer (without BSA). Bound proteins were eluted in SDS-sample buffer, heated, and separated by SDS-PAGE (10% acrylamide).
Coimmunoprecipitation
Replicate plates of BSC-40 cells were infected with wild-type vaccinia virus or vaccinia recombinants (multiplicity of infection, moi = 5) expressing either fur/f or the fur/f-cd truncation mutant fur/fR739t (Jones et al., 1995; Molloy et al., 1994). The cells were incubated for 16 h after infection in MEM with 10 mM hydroxurea (Sigma Chemical Co.), and then washed in PBS and incubated in PBS containing 1% NP-40, 1% pancreatic DNAase, 1 mM PMSF, 5 μg/ml pepstatin, and 5 μg/ml leupeptin on ice for 30 min. The DNase-treated detergent extracts were then scraped from the plates and clarified by centrifugation. The clarified supernatants were incubated overnight with the antifilamin(ABP-280) mAb M2-2A (Gorlin et al., 1990). Samples were then treated with protein G Sepharose and bound antibodies were isolated by centrifugation. The immunoprecipitates were washed three times in PBS (pH 7.0) containing 1% NP-40 and the bound proteins were dissolved in SDS-sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose. Coimmunoprecipitating FLAG epitope-tagged furin constructs were detected by incubating the blot with mAb M1, followed by color development with an alkaline phosphatase–conjugated goat anti–mouse antibody (Fisher Scientific Co., Santa Clara, CA). In some experiments, the protein G Sepharose-containing immunoprecipitates were boiled for 5 min in mRIPA (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, and 1% sodium deoxycholate) containing 1% SDS. The samples were diluted 10-fold with mRIPA and incubated overnight with an anti– furin cd antiserum (1:1,000 dilution; Affinity Bioreagents, Inc., Golden, CO). The samples were then treated with protein A Sepharose and the washed immunoprecipitates were dissolved in SDS-sample buffer and processed for Western blot analysis as described above. Biotinylated proteins (below) were detected by incubating the blots with avidin-HRP (NEN Research Products, Boston, MA) followed by chemiluminescence (NEN Research Products).
Cell Surface Biotinylation.
Replicate plates of BSC-40 cells were infected with either wild-type vaccinia virus or a vaccinia recombinant expressing fur/f (Molloy et al., 1994) (moi = 10). At 6 h after infection, the cells were washed in ice-cold PBS and the washed cells were incubated at 4°C in the presence of 0.3 mg/ml EZ-NHS-biotin (Pierce Chemical Co., Rockford, IL) in PBS. After a 1-h incubation, the cells were washed three times with ice-cold PBS containing 50 mM glycine. The cells were then harvested and processed for immunoprecipitation of ABP-280 as described above.
Cell Culture and Vaccinia Virus
BSC-40 cells are grown in minimal essential medium (MEM; BioWhittaker, Walkersville, MD) including 10% fetal bovine serum (HyClone, Logan, UT) and 25 μg/ml gentamycin (Gibco Laboratories, Grand Island, NY) as described previously (Thorne et al., 1989). M2 cells (Cunningham et al., 1992) are grown in MEM plus 10% fetal bovine serum and 25 μg/ml gentamycin. A7 cells (Cunningham et al., 1992) are grown in the same medium but with 500 μg/ml geneticin (G418) to maintain selection of the ABP-280 cDNA. Generation and expression of all other vaccinia virus constructs have been described previously (Jones et al., 1995; Molloy et al., 1994).
Quantitative Internalization Assays
Transferrin Receptor.
The rate of [125I]-transferrin (Tf) uptake was measured using a method described previously (Warren and Enns, 1997). Briefly, cells (1–2 × 106 per well) were washed twice with DMEM buffered with 20 mM Hepes, pH 7.4. At time zero, 1 ml of either 35 nM [125I]-Tf or 35 nM [125I]-Tf with 1 mg/ml unlabeled Tf were added. Cells were incubated at 37°C for 2, 4, 6, or 8 min, after which they were placed on ice and incubated with an acidic buffer at 4°C to remove any surface bound [125I]-Tf. Each well was washed four times at 4°C, solubilized in 0.1 N NaOH, 0.1% Triton X-100, and counted on a gamma counter. Each time point was done in quadruplicate. All results are corrected by subtracting nonspecific uptake.
Furin.
Protein A purified mAb M1 was mixed with [125I]-NaI (1 mCi) to which chloramine T (0.1 mg/ml final in 150 mM sodium phosphate, pH 7) was added. After 40 s at room temperature, the reaction was terminated with addition of an equal volume of sodium metabisulfite (1 mg/ml in 150 mM sodium phosphate, pH 7). The [125I]-mAb M1 was separated from free iodine with a Kwik-Sep column (Pierce Chemical Co.). Cells grown in 35-mm plates were infected with the vaccinia recombinant expressing fur/f (moi = 10). At 4 h after infection, 2 × 106 cpm [125I]-mAb M1 was added to the cultures. Control cultures were placed at 4°C, while the remaining plates were incubated at 37°C for the indicated times, and then placed at 4°C. The cells were washed twice with ice-cold DMEM without serum, and then incubated in 1.5 ml DMEM containing 1 mg/ml Proteinase K (Boehringer Mannheim) for 30 min on ice. Cells were washed and the amount of internalized [125I]-mAb M1 was quantified using a gamma counter. All data points were obtained in triplicate.
Immunofluorescence
Fur/f.
Cells grown on glass coverslips were infected with VV:hFur/f (moi = 10). At 4 h after infection, mAb M1 (6 μg/ml final concentration, IBI) was added to the culture medium for an additional hour. The cells were then fixed with paraformaldehyde, permeabilized with Triton X-100, and the samples were incubated with mAb M2 (IBI) to determine the steady state distribution of furin. mAb M1 was detected using a goat anti–mouse IgG2b-FITC secondary antibody and mAb M2 was visualized using a goat anti–mouse IgG1-TXR.
γ-Adaptin.
γ-Adaptin was detected using mAb 100/3 (E. Ungewickell, Washington University, St. Louis, MO), followed by incubation with goat anti–mouse IgG2b-FITC.
LAMP-1.
LAMP-1 was detected using mAb H4A3 (Developmental Studies Hybridoma Bank, Johns Hopkins University, Baltimore, MD), followed by incubation with goat anti–mouse IgG1-FITC.
Transferrin Uptake.
Parallel plates of A7 and M2 cells grown on coverslips were incubated with iron-loaded rhodamine-transferrin (r-Tf, 40 ng/ml; Molecular Probes, Inc., Eugene, OR) at 37°C for 1 h. The cultures were then washed and the cells were fixed with paraformaldehyde and processed for fluorescence microscopy.
Texas Red–Dextran Bead Uptake.
Cells attached to glass coverslips were placed cell face down on 50 μl of MEM-E containing 10 mg/ml Texas red (TXR)-dextran beads (10,000 mol wt; Molecular Probes, Inc.) and incubated in a humidified chamber for 30 min at 37°C. Cells were washed twice with PBS and prepared for immunofluorescence microscopy.
Proprotein Processing
Pro-βNGF.
Metabolic labeling and immunoprecipitation to assess pro-βNGF processing were performed as described previously (Bresnahan et al., 1990; Molloy et al., 1994), with the following exceptions. Cells were infected with VV:mNGF at moi = 1 and were metabolically labeled at 3 h after infection with 100 μCi [35S]-Met/Cys (Express label; NEN Research Products) for 2 h. The protein A-bound precipitates were washed three times with mRIPA and once with NET-N (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 0.1% SDS, and 0.01% NaN3). The samples were resolved on a 15% SDS-polyacrylamide gel and radiolabeled proteins were detected by phosphoimage analysis.
Pseudomonas Toxin.
Replicate cultures of M2 and A7 cells grown in 24-well plates were washed with PBS, and then placed in 450 μl of MCDB 202 (Thorne et al., 1989) containing 0.2% BSA and 50 μl Pseudomonas exotoxin A (10−8 M final, provided by R. Draper, University of Texas at Dallas) and incubated at 37°C for the times indicated. The cultures were washed twice with PBS, and then placed in 0.25 ml of DMEM (−Met) containing 22 μCi [35S]-Met/Cys for an additional 30 min at 37°C. The labeled cells were washed once with PBS and harvested by trituration in 0.25 ml of cold mRIPA containing benzonase(10−3 U/ml) (EM Science, Gibbtown, NJ). Cellular proteins were precipitated with TCA and the precipitates were passed over GF/C filters (Whatman Inc., Clifton, NJ). Incorporation of [35S]-Met/Cys into protein was determined by scintillation counting of the dried filters.
Results
Identification of ABP-280 as a Furin cd Binding Protein
The furin cd was employed as bait in a yeast two-hybrid screen to identify interacting clones from a mouse embryo cDNA library. To examine whether binding is dependent on previously identified trafficking motifs, plasmids expressing positive library clones were then examined for their ability to direct the interaction of their cloned inserts with a set of furin cd truncations (Fig. 1 A). The analysis for one of these clones, TP107, is shown in Fig. 1 B. Activation of reporter gene (HIS3/β-galactosidase) expression by TP107 required furin cd sequences membrane-proximal to Ser758. Thus, binding of TP107 to the furin cd does not require motifs previously shown to be important for furin trafficking, including the acidic cluster necessary for TGN localization and the tyrosine-based internalization signal. Furthermore, mutational analyses showed that interaction between these two proteins similarly does not require the putative di-leucine-like internalization signal (data not shown).
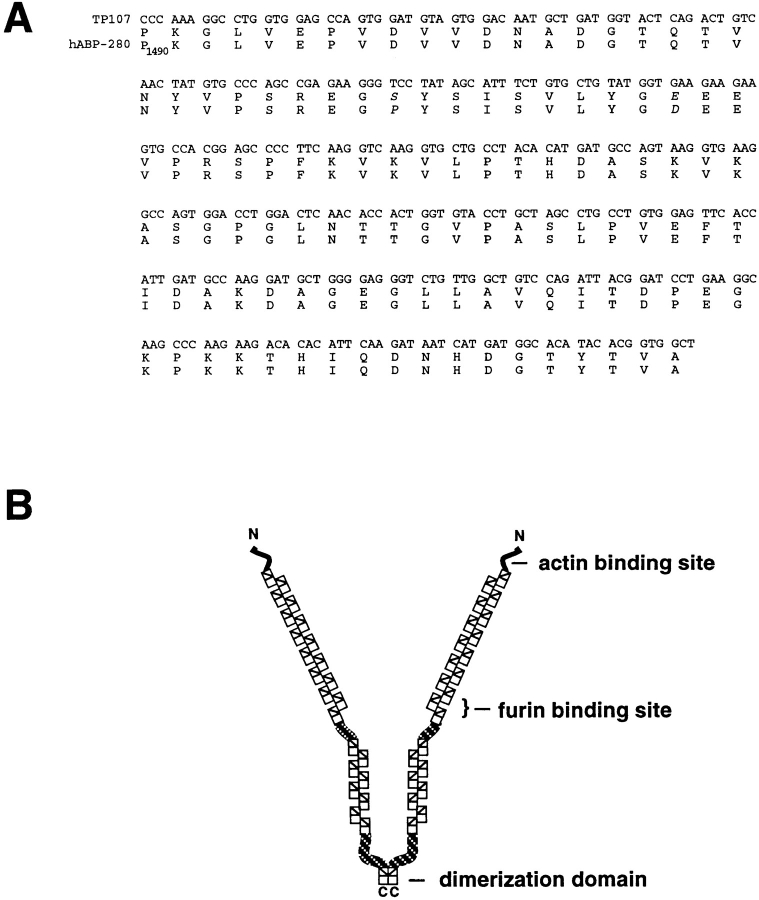
Sequence analysis of TP107 showed that it encodes a 118 amino acid segment of the actin binding protein ABP-280 (nonmuscle filamin) (Fig. 2). Alignment of TP107 with the sequence for human ABP-280 (Gorlin et al., 1990) shows that the isolated mouse clone represents a segment bridging the 13th and 14th repeats corresponding to residues 1490–1607 in human ABP-280. Sequence comparison of this region shows nearly complete amino acid identity between this mouse fragment and the human gene. Only two amino acid substitutions, Pro1518 → Ser and Asp1527 → Glu, are present. Furthermore, these two amino acid changes have no effect on binding of this region of ABP-280 to the furin cd (Liu, G., and G. Thomas, unpublished results).
Figure 2.
Identification of TP107 as mouse ABP-280. (A) Alignment of TP107 and ABP-280. Shown are the nucleic acid sequence for the TP107 insert (top), the corresponding TP107 open reading frame (middle), and the amino acid sequence of the corresponding region of human ABP-280 (Gorlin et al., 1990). Species-specific amino acid changes are shown in italics. (B) Schematic of ABP-280. ABP-280 is a homodimer composed of two 280-kD subunits, each of which contains an amino-terminal actin binding domain followed by 24 repeats of a structurally conserved 96-amino acid β-sheet structure (rectangles). Hinge regions are located between repeats 15/16 and 23/24. The centrally placed hinge region permits the orthogonal positioning of cortical microfilaments required for the “sol-gel” transition states of cytosol, as well as the formation of lamellapodia necessary for cell crawling. The COOH-terminal 24th repeat contains the ABP-280 dimerization domain. Alignment of TP107 with the reported sequence for human ABP-280 shows that it is contained within the region bridging the 13th and 14th repeats (residues 1490–1607 in human ABP-280).
Interaction of ABP-280 with Furin In Vitro and In Vivo
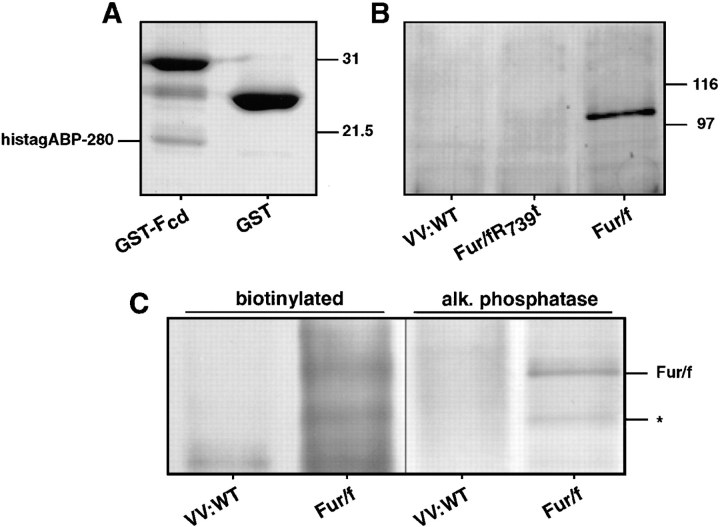
To demonstrate that the region of ABP-280 encoded by TP107 interacts directly with the furin cd, the protein–protein binding was studied in vitro (Fig. 3 A). A His-tagged construct, HistagABP-280, containing the ABP-280 fragment identified by the two-hybrid screen, was incubated with either GST or a GST-furin cd (GST-Fcd) construct. After incubation, the GST constructs were isolated by binding to glutathione-agarose beads and the recovered proteins resolved by SDS-PAGE. Using this method, the interaction between HistagABP-280 and GST-Fcd was readily detected, whereas no interaction was observed with GST alone.
Figure 3.
Interaction of ABP-280 and furin. (A) Binding in vitro of HistagABP-280 to GST-furin cd. 6 μg of a His-tagged construct containing residues 1490–1607 of mouse ABP-280 was combined with 10 μg of GST or a GST fusion protein containing the entire furin cd (GST-Fcd). Glutathione agarose was then added and bound proteins were removed from the washed beads with SDS-sample buffer and separated by SDS-PAGE (10% acrylamide). Proteins were detected by staining the gel with Coomassie Blue R250. The positions of molecular weight standards are shown on the right. (B) Coimmunoprecipitation of ABP-280 and fur/f. Replicate plates of BSC-40 cells were infected with wild-type vaccinia virus or vaccinia recombinants expressing either fur/f or the furin cd truncation mutant fur/fR739t depicted in Fig. 1 (moi = 5). At 16 h after infection, the cells were harvested and the clarified cell extracts were incubated overnight with the anti– filamin(ABP-280) mAb (Gorlin et al., 1990). Samples were then treated with protein G Sepharose and bound proteins dissolved in SDS-sample buffer, separated by SDS-PAGE and transferred to nitrocellulose. Coimmunoprecipitating FLAG epitope-tagged furin constructs were detected by incubating the blot with mAb M1. Parallel plates of total cell extract show that equivalent amounts of mAb M1 immunoreactive fur/f constructs were expressed in each sample (data not shown). The positions of molecular weight standards are shown on the right. (C) Coimmunoprecipitation of ABP-280 with cell surface fur/f. Parallel plates of BSC-40 cells were infected with either wild-type vaccinia virus or the recombinant expressing fur/f. At 6 h after infection, the cells were placed on ice and cell surface molecules were biotinylated as described in Materials and Methods. The cells were then harvested and subjected to immunoprecipitation with the anti–ABP-280 mAb as described in B. The immunoprecipitates were boiled in mRIPA containing 1% SDS, diluted 10-fold in buffer, and furin molecules immunoprecipitated with the anti–furin cd antiserum. The immunoprecipitates were resolved by SDS-PAGE, transferred to nitrocellulose, and biotinylated proteins were detected by chemiluminescence (left). The blot was then reprobed with the anti–furin cd antiserum and developed with alkaline phosphatase (right). 100 kD fur/f is marked. The smaller 82 kD biotinylated furin protein (*) represents a degradation product of the endoprotease, generated under these conditions, that still contains the intact cytosolic domain.
Next, a coimmunoprecipitation experiment was performed to determine whether ABP-280 and furin associate in vivo (Fig. 3 B). Replicate plates of BSC-40 cells, an African Green monkey kidney epithelial line, expressing epitope(FLAG)-tagged constructs encoding either full length furin (fur/f) or the furin cd truncation mutant lacking the cytosolic domain (fur/fR739t, Fig. 1 A), was harvested in a detergent buffer containing DNAase I to depolymerize actin filaments. ABP-280 molecules were immunoprecipitated from this extract with an anti–ABP-280 antibody, and the immunoprecipitates were subjected to SDS-PAGE and immunoblotting. Coimmunoprecipitating FLAG-tagged furin molecules were detected with mAb M1. In agreement with the two-hybrid and bacterial fusion protein analyses, fur/f and ABP-280 can be coimmunoprecipitated in vivo. Furthermore, the coimmunoprecipitation was dependent on furin cd sequences since fur/fR739t was not detected.
ABP-280 is concentrated in the cortical regions of the cytoplasm where, in addition to organizing actin cables, it tethers a number of cell surface receptors to the actin cytoskeleton (Fox, 1985; Ohta et al., 1991; Sharma et al., 1995). By contrast, although furin cycles to the cell surface, its steady state localization is primarily in the TGN. Thus, ABP-280 would be expected to be associated with only a small percentage of the total cellular furin molecules. Consistent with these findings, quantitative immunoprecipitations showed that only 1–3% of the total cellular pool of fur/f was associated with ABP-280 (data not shown). To investigate whether ABP-280 binds furin molecules at the cell surface, cells expressing fur/f were placed on ice and subjected to cell surface biotinylation. The biotinylated cells were washed and ABP-280 molecules were immunoprecipitated. The immunoprecipitates were then denatured in SDS and a second immunoprecipitation was performed with an anti–furin cd antiserum. The immunoprecipitates were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (Fig. 3 C). Biotinylated proteins immunoprecipitated sequentially with antibodies against ABP-280 and furin were detected by chemiluminescence. This analysis revealed a 100-kD biotinylated protein, the size expected for epitope-tagged furin, specifically in extracts from cells expressing fur/f and not cells infected with wild-type vaccinia virus. To confirm that the biotinylated protein is furin, the filter was then incubated with the anti– furin cd antiserum, and total furin molecules were visualized by development of the blot with alkaline phosphatase. As expected, the biotinylated protein comigrates with fur/f. In addition, the biotinylation appeared specific for cell surface molecules since the RER-localized zymogenic form of the endoprotease (Molloy et al., 1994) was not detected by this procedure.
ABP-280-dependent Internalization of Furin
On the basis that ABP-280 has been shown to (a) tether several hematopoietic cell surface receptors (Fox, 1985; Ohta et al., 1991; Sharma et al., 1995), and (b) bind cell surface furin (Fig. 3 C), we speculated that ABP-280 might modulate the rate of internalization of furin from the cell surface. To investigate this possibility, we employed two genetically paired cell lines that either lack or express ABP-280. M2 cells are a human melanoma that are ABP-280 deficient. By contrast, A7 cells are a subclone of M2 cells that are rescued for expression of ABP-280 (Cunningham et al., 1992). Furin internalization in M2 and A7 cells was measured quantitatively by determining the amount of endocytosed [125I]-mAb M1 in fur/f-expressing cells (Fig. 4). As a control, we also determined the rate internalization of the transferrin receptor (TfR), a cycling receptor that traffics independently of the microfilament cytoskeleton (Gottlieb et al., 1993; Sandvig and van Deurs, 1990).
Figure 4.
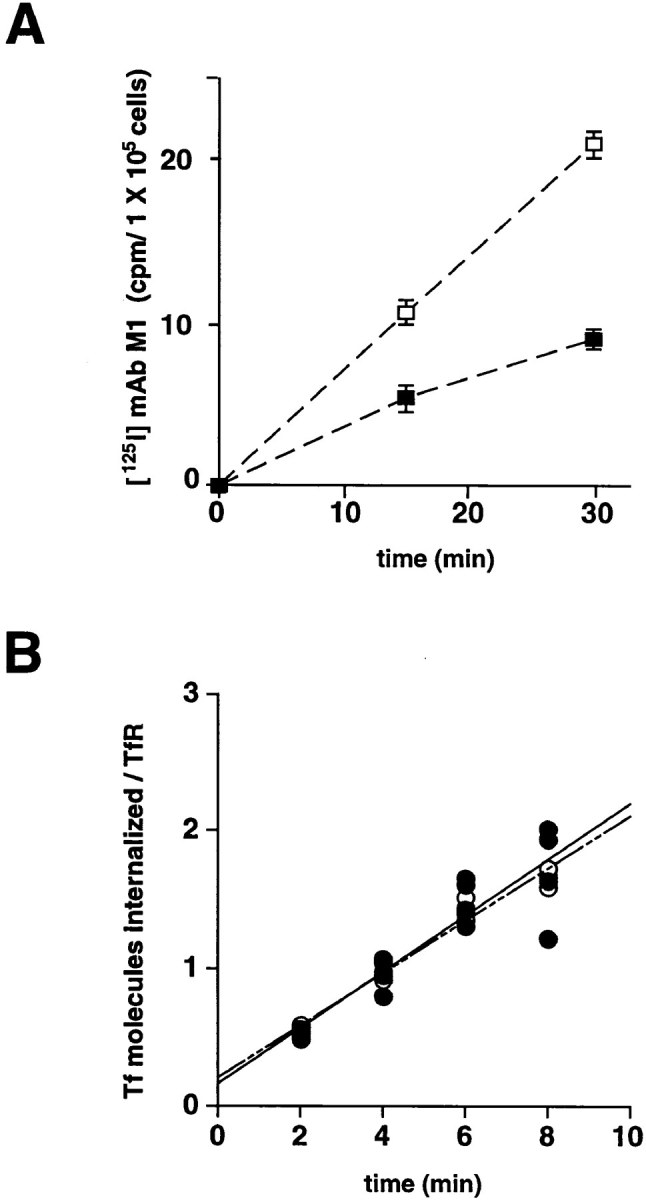
Quantitative internalization of [125I]-mAb M1 and [125I]-transferrin into M2 and A7 cells. (A) Parallel plates of M2 (□) and A7 (▪) cells were infected with VV: hfur/f (moi = 10). 4 h after infection, [125I]-mAb M1 was added to the culture medium. Control plates were placed on ice immediately and the remaining samples were incubated at 37°C for the indicated times, and then transferred to ice. At the indicated times, the supernatants were removed, the cells washed twice, and then incubated with Proteinase K. The detached cells were suspended in fresh medium, pelleted through a serum pad, and the amount of internalized [125I] was determined. For each time point, the counts from the control sample were subtracted from the value of the assay sample. Experiments using non– epitope-tagged furin showed no detectable increase in signal over background. All time points were measured in triplicate. Bars indicate standard deviations. (B) The measurement of [125I]-transferrin uptake into M2 (○) and A7 (•) cells was performed as described in Materials and Methods. Cells were incubated with 35 nM [125I]-transferrin for the indicated times at 37°C. At the end of each time point, cells were transferred to 4°C and washed with mild acid to remove cell surface transferrin. The cells were solubilized with NaOH and counted in a gamma counter. Binding assays at 4°C were performed simultaneously with each assay to determine the number of transferrin receptors on the cell surface.
The rate of furin internalization was cell line dependent. The ABP-280-deficient M2 cells internalized 2.3-fold more [125I]-mAb M1 than did A7 cells (Fig. 4 B), despite the equivalent levels of fur/f expression in the two cell lines (data not shown). By contrast, the rate of internalization of the [125I]-transferrin (Tf) was similar in both cell lines (0.17 and 0.19 Tf molecules per TfR molecule per min in A7 and M2 cells, respectively, Fig. 4, A). These results, coupled with those of Fig. 3, argue that ABP-280, by tethering the endoprotease to the cell surface, modulates the rate of furin internalization. The lack of effect of ABP-280 on the rate of TfR internalization is consistent with reports demonstrating TfR internalization to be unaffected by drug-induced alterations in cytoskeletal function (i.e., the lack of effect of cytochalasin D on the internalization of TfR, Gottlieb et al., 1993; Sandvig and van Deurs, 1990). Furthermore, these results demonstrate that the role of this actin-binding protein on furin trafficking is selective for only a subset of membrane proteins.
ABP-280-dependent Routing of Furin
An immunofluorescence assay was performed to determine whether ABP-280 is necessary for steps in furin routing in addition to regulating internalization of the endoprotease from the cell surface (Fig. 5). Parallel plates of M2 and A7 cells grown on coverslips were infected with the vaccinia recombinant expressing fur/f. Both the TGN concentration of fur/f as well as its cycling between this compartment and the cell surface in each of the cell lines was determined using the anti–FLAG mAbs M1 and M2 (see Fig. 1 A). As a control, we also performed this study in BSC-40 cells, known to contain ABP-280 (Fig. 3 B) and used previously to study the TGN localization of furin as well as its cycling between this compartment and the cell surface (Jones et al., 1995; Molloy et al., 1994).
Figure 5.
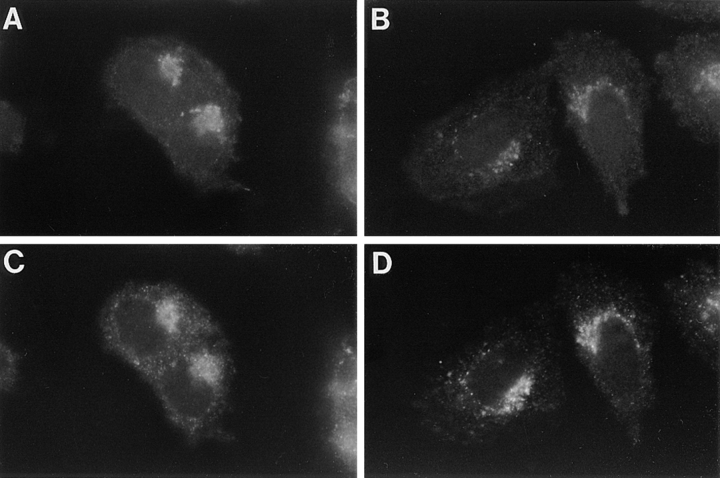
Localization and recycling of epitope-tagged furin. Parallel plates of BSC-40 (A and B), M2 (C and D), and A7 cells (E and F) grown on glass coverslips were infected with VV:hfur/f (moi = 10). At 4 h after infection, mAb M1 (6 μg/ml final concentration) was added to the culture media for an additional hour. The cells were then fixed and permeabilized, and the samples were incubated with mAb M2 to detect the steady state distribution of furin staining. The samples were then processed for immunofluorescence microscopy. The mAb M1 was detected using a goat anti–mouse IgG2b-FITC secondary antibody (A, C, and E) and mAb M2 was visualized using a goat anti–mouse IgG1-TXR antibody (B, D, and F). The exposure time in C was adjusted to equal the exposure time in E to compare directly the intensity of fluorescence staining between samples.
The staining patterns of the fur/f-dependent internalized mAb M1 were dramatically different and depended on the expression of ABP-280. In BSC-40 cells, the internalized mAb M1 accumulated largely in the TGN, where its staining pattern overlapped with that of the mAb M2 postfixation pattern. Internalized mAb M1 was also detected in a population of endosomes/lysosomes (Fig. 5 A). Surprisingly, ABP-280-deficient M2 melanoma cells displayed strikingly less staining of internalized mAb M1 (Fig. 5 C). Much reduced levels of TGN staining were observed. In addition, the accumulation of internalized mAb M1 in the peripheral punctate endosome/lysosome structures was also significantly reduced. By contrast, when the mAb M1 internalization was examined under identical conditions in the ABP-280-rescued melanoma cell line, A7, the robust paranuclear staining that overlapped with the TGN-localized mAb M2 was restored. In addition, the presence of internalized antibody in peripheral endosomes/lysosomes was also readily apparent (Fig. 5 E).
Unlike the striking differences in staining patterns of internalized furin, the steady state staining patterns of the endoprotease were remarkably consistent and were not affected by the presence or absence of ABP-280. Characteristic of its known localization to the TGN in many cell types (Molloy et al., 1994; Schafer et al., 1995; Voorhees et al., 1995), furin has a predominant paranuclear steady state staining pattern (mAb M2 post-fix staining) when expressed in each of the cell lines (Fig. 5, B, D, and F). To confirm that fur/f is localized to the TGN in the M2 and A7 cells, its immunofluorescence staining pattern was compared with that of γ-adaptin, an integral component of the AP-1 adaptin complex associated with the TGN (Fig. 6). In agreement with previous studies, the fur/f staining pattern was virtually indistinguishable from that of γ-adaptin in both cell lines. Furthermore, the similar mAb M2 post-fix staining patterns independent of the expression of ABP-280 suggest that ABP-280 is not essential for the concentration of furin in the TGN. These results are consistent with those in Fig. 1 B showing that the furin cd acidic cluster, the region on the furin cd necessary for TGN localization, is not important for binding to ABP-280.
Figure 6.
Colocalization of fur/f and γ-adaptin. Replicate plates of M2 (A and C) and A7 (B and D) cells were infected with the vaccinia recombinant expressing fur/f (moi = 10). At 4 h after infection, the cells were fixed with paraformaldehyde, permeabilized, and incubated with mAb M2 to detect fur/f (A and B) and 100/3 to detect γ-adaptin (C and D). The mAb M2 was visualized with anti–mouse IgG1-TXR and mAb 100/3 was visualized with IgG2b-FITC.
The strikingly weaker staining of internalized mAb M1 in the M2 cells compared with A7 cells, despite their greater internalization of [125I]-mAb M1 (Fig. 4 B), suggested that either the internalized mAb M1 is rapidly degraded in the ABP-280-deficient melanoma cells or internalized furin is missorted and does not accumulate in discrete compartments amenable to analysis by immunofluorescence microscopy. To examine the possibility that [125I]-mAb M1 is degraded more rapidly in the M2 cells, the stability of internalized [125I]-mAb M1 was determined in both cell lines after a 1-h uptake. Extracts from both cell lines contained similar amounts of protein A-bound [125I]-mAb M1, demonstrating that internalized mAb M1 was not aberrantly degraded in M2 cells (data not shown).
To examine the possibility that internalized furin is missorted in the ABP-280-deficient M2 cells, a time course study was performed. When either M2 or A7 cells treated with mAb M1 for 5 min were analyzed by immunofluorescence microscopy, a largely peripheral punctate pattern of similar staining intensity was observed in both cell types (Fig. 7, A and B). These results raised the possibility that, although the kinetics of furin internalization are different in the two cell types (Fig. 4 B), the efficiency by which the endoprotease enters early endosomes may be very similar and thus independent of ABP-280. To examine this possibility, the staining pattern of internalized mAb M1 was compared to that of rhodamine-transferrin (r-Tf). Previously, we showed that accumulation of internalized furin into early endosomes can be achieved by treatment of cells with tautomycin, an inhibitor of the furin phosphatase that modulates transit of furin from the early endosomes to the TGN (Jones et al., 1995). Using these conditions to impede exit of furin from early endosomes, the staining pattern of internalized mAb M1 remained similar in the M2 and A7 cells and colocalized largely with that of internalized r-Tf (Fig. 7, C–F). Despite the similar staining patterns of internalized mAb M1 at these early time points, by 60 min, the patterns were again strikingly different; little staining of internalized mAb M1 could be observed in the M2 cells, whereas in A7 cells a robust staining of the paranuclear region and peripheral punctate compartments was observed (Fig. 7, G and H, respectively).
Figure 7.
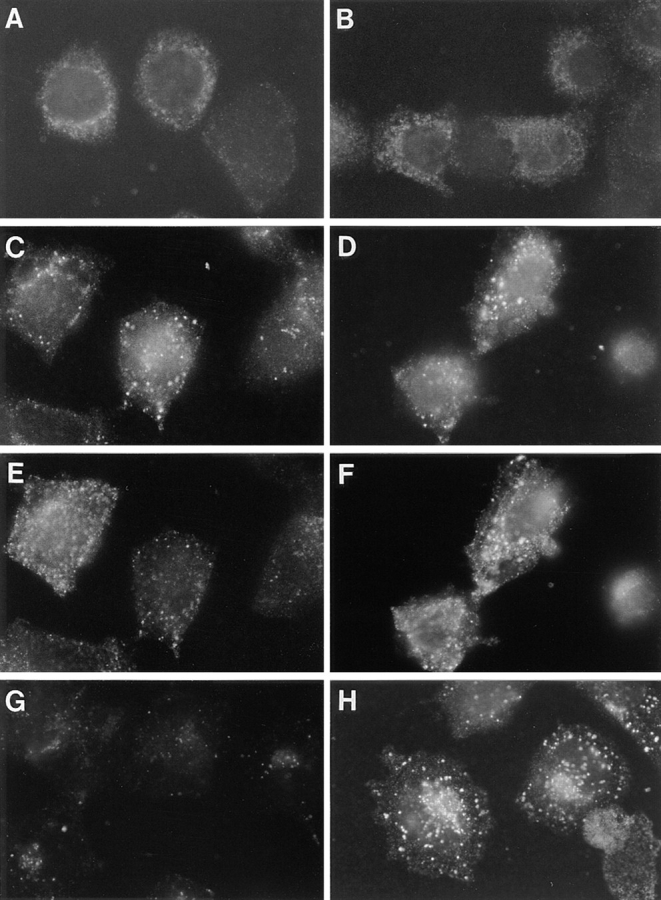
Time course of fur/f internalization. Parallel plates of M2 (A, C, E, and G) and A7 (B, D, F, and H) cells grown on glass coverslips were infected with VV:hfur/f (moi = 10). At 4 h after infection, mAb M1 (6 μg/ml final concentration) was added to the culture media for either 5 min (A and B), 15 min (C–F) or 60 min (G and H). Cells in C–F were also incubated with 40 ng/ml r-Tf and 100 nM tautomycin (to accumulate internalized furin in the early endosomes). C and D show mAb M1 and E and F show r-Tf in double-labeled cells. Samples were processed for immunofluorescence microscopy as described in Fig. 5. Internalized mAb M1 was visualized with anti–mouse IgG2b-TXR antibody.
Although ABP-280 binds cell surface furin and modulates quantitatively the rate of internalization of the endoprotease from the cell surface (Figs. 3 and 4), the immunocytochemical time course study (Fig. 7) suggested that efficiency of furin sorting in the early endocytic pathway, including (a) the trafficking of furin from the cell surface to the early endosomes, and (b) the tautomycin-dependent accumulation of furin in early endosomes, is at least qualitatively independent of ABP-280. Rather, expression of ABP-280 (a) affects the availability of furin for internalization (Fig. 4), and (b) is required for the efficient sorting of furin from early endosomes to later compartments within the endocytic pathway, including the TGN (Figs. 5 and 7). To test this hypothesis, a temperature-shift experiment was conducted (Fig. 8). Previous studies showed that sorting from early endosomes to compartments later in the endocytic pathway is temperature dependent (Gruenberg and Howell, 1989). In this experiment, replicate plates of M2 cells were incubated with mAb M1 for 1 h at 23°C. Under these conditions, the internalized mAb M1 was readily detectable and accumulated in a perinuclear staining pattern (Fig. 8 A). When the applied mAb M1 was removed and the washed cells were shifted to 37°C for an additional hour, the internalized mAb M1 remained readily detectable and mobilized to a more paranuclear location characteristic of the TGN (Fig. 8 B). To confirm that the temperature shift rescued the efficient retrieval of furin in the ABP-280-deficient M2 melanoma cells to the TGN, the paranuclear staining pattern of internalized furin was compared to γ-adaptin. However, because mAb M1 and the γ-adaptin mAb (100/3) are the same subtype, the experiment was repeated with mAb M2 in place of mAb M1. After the temperature shift, the internalized mAb M2 showed a TGN-like paranuclear staining pattern that overlapped with that of γ-adaptin (Fig. 8, C and D). Furthermore, and characteristic of a TGN-localized protein, the internalized mAb M2 staining pattern was resistant to brefeldin A, whereas the γ-adaptin staining pattern dispersed after treatment with the drug (Fig. 8, E and F). These results, together with those in Fig. 7, show that the missorting of furin from early endosomes to the TGN in the ABP-280-deficient cells can be overcome, at least to some extent, by temperature shift. Although the biochemical basis for the ability of the temperature shift to rescue the sorting of internalized furin in the absence of ABP-280 is not known, possible mechanisms are presented in Discussion.
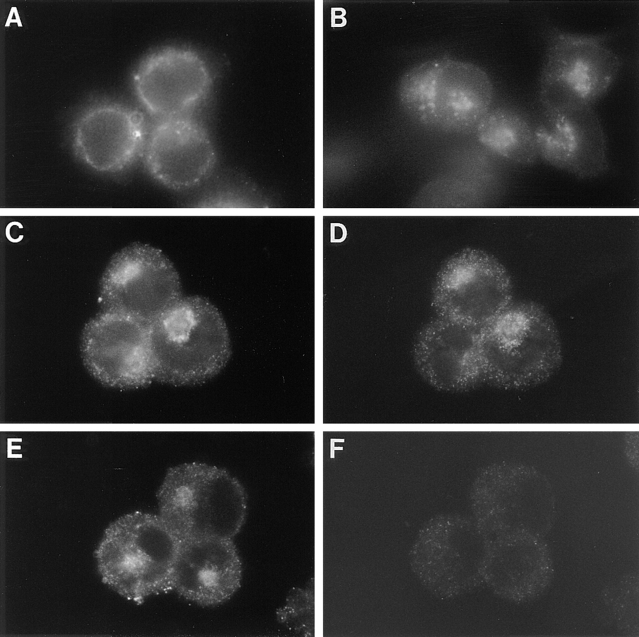
Figure 8.
Temperature dependence of fur/f retrieval to the TGN. Replicate plates of M2 cells were infected with VV:hFur/f as above. At 4 h after infection, the cells were placed at 23°C, and then treated with mAb M1 (A and B) or mAb M2 (C–F) for 1 h and either fixed immediately (A) or washed to remove the mAb (B–F) and shifted to 37°C for an additional hour before fixation. Before fixation, the samples in E and F were treated with 5 μg BFA/ml for 20 min. After fixation, the samples in C–F were incubated with mAb 100/3 to visualize both γ-adaptin (D and F) and internalized fur/f (mAb M2, C and E). Internalized mAbs M1 and 100/3 were visualized with anti–mouse IgG2b-TXR antibody. Internalized mAb M2 was visualized with anti–mouse IgG1-FITC.
Localization of Late but Not Early Endocytic Compartments is Dependent on ABP-280
To determine whether the aberrant routing of furin in the ABP-280-deficient M2 cells is specific for this endoprotease or rather results from a generalized defect in the sorting of all itinerant membrane proteins in the TGN/endosomal system, the distribution of molecules internalized into early and late endocytic compartments was compared in A7 and M2 cells (Fig. 9). Addition of r-Tf to parallel cultures of M2 and A7 cells resulted in incorporation of the marker into identical early/recycling endosome populations (Fig. 9, A and B, respectively). These results are in agreement with the tautomycin studies performed in Fig. 7 demonstrating that sorting from the cell surface into early endosomes is independent of ABP-280.
Figure 9.
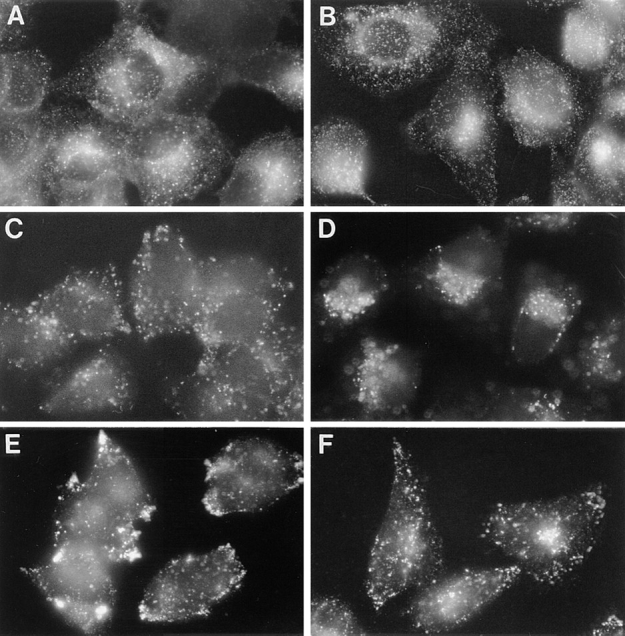
Importance of ABP-280 to the localization of endocytic compartments. Parallel plates of M2 (A, C, and E) and A7 (B, D, and F) cells were grown on coverslips. Early endocytic compartments were visualized by treating the cultures with rhodamine-transferrin (r-Tf, 40 ng/ml) for 30 min at 37°C before fixation (A and B). Late endosomes were visualized by addition of Texas Red-dextran beads (10,000 mol wt, 10 μg/ml) for 30 min at 37°C before fixation (C and D). To visualize lysosomes, cells were fixed, permeabilized with detergent, and incubated with a LAMP-1 antibody followed by incubation with anti–mouse IgG1-FITC (E and F).
Internalization of molecules into late endocytic compartments, however, was strikingly different between the two cell lines. When M2 cells were incubated with FITC-conjugated dextran beads, a fluid phase marker, the beads accumulated into endosomal compartments dispersed throughout the cell (Fig. 9 C). By contrast, in A7 cells, internalized dextran beads accumulated into paranuclear compartments characteristic of late/recycling endosomes (Fig. 9 D). Furthermore, staining of M2 and A7 cells for LAMP-1, a lysosomal marker, also revealed a differental localization of this antigen. In M2 cells, the LAMP-1 staining pattern was prominent in the cell periphery, where it was concentrated largely in cell processes (Fig. 9 E). By contrast, the LAMP-1 staining pattern in A7 cells was largely paranuclear, with populations of lysosomes distributed throughout the cytoplasm (Fig. 9 F). Thus, in addition to the requirement of ABP-280 for the efficient sorting of furin from early endosomes to the TGN, expression of this actin binding protein is also necessary for the proper localization of lysosomes and late endosomes.
ABP-280 Modulates Furin Processing Compartments
The requirement of ABP-280 for the correct routing of furin in the TGN/endosomal system suggested that this actin binding protein may modulate the processing of furin substrates in vivo. Because furin processes a large number of proprotein molecules in multiple compartments within the TGN/endosomal system, the role of ABP-280 in the proteolytic maturation of furin substrates in either the TGN/ biosynthetic pathway or the endocytic pathway was investigated.
The importance of ABP-280 to the maturation of furin substrates in the biosynthetic pathway was examined by determining the processing of pro-β-nerve growth factor (Fig. 10 A). Pro-βNGF is synthesized initially as a 35–40-kD proprotein and, during transit through the biosynthetic pathway, it is cleaved at a consensus furin site (-Arg-Ser-Lys-Arg↓ -) to generate the 13 kD bioactive βNGF (Bresnahan et al., 1990). Parallel plates of M2 and A7 cells were infected with a vaccinia recombinant expressing pro-βNGF. The processing of pro-βNGF by both cell lines was complete, resulting in the secretion of the processed 13-kD βNGF peptide. The lack of effect of ABP-280 on the processing of pro-βNGF is consistent with the above immunofluorescence studies showing that ABP-280 is unnecessary for localization of furin to the TGN.
Figure 10.
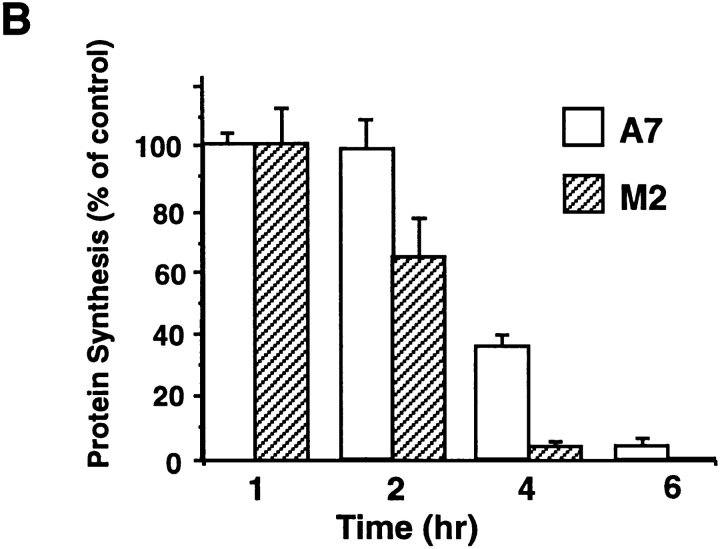
Dependence of ABP-280 on processing of furin substrates. (A) Processing of pro-βNGF. Parallel plates of A7 and M2 cells were infected with a vaccinia recombinant expressing mouse pro-βNGF (moi = 10). At 3 h after infection, the cells were metabolically labeled with 35S-[Met/Cys]. Secreted βNGF proteins were immunoprecipitated from the culture media with pooled rat anti–βNGF antibodies. The washed immunoprecipitates were then resolved by SDS-PAGE and pro-βNGF-derived proteins visualized by phosphoimage analysis. (B) Processing of Pseudomonas exotoxin. Parallel cultures of M2 and A7 cells were incubated in culture medium containing 100 nM PE for the indicated times. The rinsed cells were then metabolically labeled with 35S-[Met/Cys] for 30 min, harvested in mRIPA, and the incorporation of [35S]-amino acids into TCA-precipitable material was quantified by scintillation counting. A7 cells, white bars; M2 cells, hatched bars.
To examine the processing of furin substrates in the endocytic pathway, the furin-dependent activation of Pseudomonas exotoxin (PE) was studied. Inactive protoxin bound to cell surface α2-macroglobulin receptors/low density lipoprotein receptor-related protein (α2MR/LRP) is internalized into endocytic compartments (Kounnas et al., 1992). Subsequently, PE is cleaved by furin at a consensus furin site (-Arg-Gln-Pro-Arg↓ -) to release an active 37-kD fragment that translocates into the cytosol and catalyzes the ADP-ribosylation of elongation factor-2 to inhibit translation. Indeed, cleavage by furin is the rate limiting step in PE toxicity (Chiron et al., 1994).
Parallel plates of M2 and A7 cells were treated with 10 nM PE for increasing periods of time. Generation of the active PE fragment by the furin-dependent cleavage of the protoxin was quantified by measurement of the processing-dependent inhibition of protein synthesis in cells metabolically labeled with [35S]-Met/Cys (Fig. 10 B). Although the toxin inhibited protein synthesis in both the M2 and A7 cells, the kinetics of inhibition were markedly different between these cell lines. Treatment of the M2 cells with PE for 2 h resulted in a nearly 50% inhibition of protein synthesis, whereas the same treatment in a parallel culture of A7 cells showed a negligible effect. This differential sensitivity to PE continued to be evident at 4 h after toxin addition. These results show that ABP-280 modulates the formation of furin-containing processing compartments in the endocytic pathway. Furthermore, the increased sensitivity of M2 cells to PE is consistent with an increased rate of internalization of furin in these ABP-280-deficient cells (Fig. 4).
Discussion
The multiple roles of actin filaments in cells include membrane organization, maintenance of cell polarity, and cell locomotion (Bretscher, 1993; Luna and Hitt, 1992; Mays et al., 1994). Diverse actin-associated functions are achieved by a retinue of actin binding proteins (Hitt and Luna, 1994). Here, we report one actin binding protein, ABP-280, is the first identified furin sorting protein. ABP-280 tethers cell surface–localized furin molecules by binding to the membrane proximal region of furin's cytosolic domain. ABP-280 modulates the rate of internalization of furin from cell surface and is necessary for the efficient sorting of furin from early endosomes to the TGN. We also show that ABP-280 is necessary for localization of lysosomes and late but not early endosomes. Finally, proprotein processing analyses show that ABP-280 modulates the formation of furin processing compartments in the endocytic pathway.
Discrimination between Resident and Transiently Presented Membrane Proteins at the Cell Surface
ABP-280 is a multi-functional protein. Through its NH2-terminal actin binding domains and COOH-terminal dimerization domain (see Fig. 2 B), it directs the position of cortical actin, forming orthogonal filament networks involved in the sol-gel state transitions of the cytosol necessary for lamellapod extension and cell crawling (Cunningham et al., 1992). In addition, ABP-280 serves to link resident cell surface proteins to underlying actin filaments. In platelets, constitutive linkage of the von Willebrand's receptor is achieved by attachment of the GP1bα cd to the second rod domain of ABP-280 between repeats 17–19 (Meyer et al., 1997). Constitutive linkage of the neutrophil β2-integrin chain and the high affinity immunoglobulin receptor, FcγRI, are also mediated by ABP-280, although the specific ABP-280 domain(s) involved have not been defined (Ohta et al., 1991; Sharma et al., 1995). Our results showing that ABP-280 directs the intracellular trafficking of furin, an itinerant TGN/endosomal membrane protein, represents a new role for this actin-binding protein. The binding of the furin cd to the 13th and 14th repeats of ABP-280 (Figs. 1 and 2) is the first demonstration of a protein tethering site in rod domain 1 of ABP-280 and suggests that this rod domain is capable of binding additional membrane proteins.
Tethering of resident cell surface molecules (e.g., GP1bα, CD18, and FcγRI) by ABP-280 is apparently required for their ligand-dependent rearrangement of the cytoskeleton associated with leukocyte activation (Cunningham et al., 1996). For example, binding IgG Fc domains to the FcγRI receptor induces cell surface changes required for phagocytosis. By contrast, the tethering of cell surface furin by ABP-280 (Fig. 3) does not appear to be involved in gross cytoskeletal rearrangements. Rather, ABP-280 modulates the rate of internalization of furin from the cell surface (Fig. 4 B), perhaps by transiently tethering the endoprotease. How binding of ABP-280 to furin is regulated remains to be determined. In platelets, ligand binding apparently causes a dissociation of FcγRI from ABP-280, thus facilitating internalization (Harrison et al., 1994; Ohta et al., 1991). It is possible that substrate binding induces internalization of furin. Alternatively, dissociation may be regulated by signaling cascades. Both furin and ABP-280 are phosphorylated in vivo (Jones et al., 1995; Ohta et al., 1995; Ohta and Hartwig, 1996). However, phosphorylation of the furin cd acidic cluster does not affect binding of ABP-280 (Fig. 1, and data not shown). Conversely, phosphorylation of ABP-280 does affect interaction with actin cables (Ohta et al., 1995), raising the possibility that protein kinase cascades may modulate ABP-280 binding to furin.
Importance of ABP-280 and the Cytoskeleton on Protein Sorting and Organelle Localization
The tethering of furin to the cell surface by ABP-280 (Fig. 3) and the importance of this actin binding protein to the rate of furin, but not TfR, internalization (Fig. 4) are consistent with its established properties. Although ABP-280 modulates the rate of furin internalization, the efficiency of furin sorting to the early endosome, as well as the tautomycin-dependent accumulation of the endoprotease in this compartment, is independent of this actin binding protein (Fig. 7). Surprising, therefore, was the finding that ABP-280 is also important for (a) the efficient retrieval of internalized furin from early endosomes to the TGN (Figs. 4–8), and (b) the localization of late endosomal (dextran bead uptake) and lysosomal (LAMP-1 staining) compartments (Fig. 9).
The stability of the internalized mAb M1 and the lack of a readily discernible furin(mAb M1)-containing compartments 60 min after uptake in M2 (ABP-280-deficient) cells suggests that the endoprotease simply fails to concentrate in a compartment readily amenable to detection by microscopy. Since furin at steady state concentrates principally in the TGN (Fig. 6, and Molloy et al., 1994; Schafer et al., 1995; Voorhees et al., 1995), we would not expect to see the internalized protein accumulate in other compartments— except under controlled conditions such as temperature shift (Fig. 8) or treatment with tautomycin (to accumulate furin in early endosomes, see Fig. 7). Note that the population of endosome/lysosome staining seen after mAb M1 uptake in Fig. 5, A and E, and Fig. 7 H represents primarily post-TGN compartments, as previously demonstrated (Jones et al., 1995). Importantly, internalized furin eventually returns to the TGN since the steady state level of endogenous furin in both M2 and A7 cells is localized to the TGN (data not shown).
Together, our results argue that routing of membrane proteins through early endocytic compartments is independent of the actin filament system. However, the efficient retrieval of furin from early endosomes to the TGN requires ABP-280. Our findings are in agreement with previous studies that used cytochalasin D to show that delivery of molecules to mature endosomes and lysosomes, but not the internalization of TfR to early endosomes, is dependent on the microfilament network (Sandvig and van Deurs, 1990). Unlike studies with pharmacologic agents, however, the effects described here are apparently specific for ABP-280 since reintroduction of this actin binding protein by stable expression of the cDNA (A7 cells) fully corrects these varied trafficking defects.
The ability of the temperature shift to rescue the trafficking of furin (Fig. 8) provided further evidence that the loss of mAb M1 staining after antibody uptake in M2 cells (Fig. 5 C and Fig. 7 G) is due to a defect in vesicle sorting caused by the absence of ABP-280. However, the mechanism(s) underlying the rescue of furin trafficking by the temperature shift are not known. Perhaps the reduced temperature induces a local redistribution of the actin cables that compensate for the lack of ABP-280, or perhaps may compensate for an ABP-280-dependent sorting step. One possibility is that the temperature shift facilitates the transfer of transport vesicles from the actin-based cytoskeleton to microtubules. Indeed, such a duality in cytoskeletal components to direct vesicle traffic has been reported in yeast (Lille and Brown, 1992) and neuronal cells (Kuznetsov et al., 1992). Finally, the role of ABP-280 in endosome to TGN sorting appears to be distinct from its role in localization of late endocytic compartment since (a), unlike internalized mAb M1 (Figs. 5 and 7), the intensity of the fluorescent signal from internalized dextran was independent of expression of ABP-280 (Fig. 9), and (b) the mislocalization of dextran beads in M2 cells failed to be rescued by the temperature shift (data not shown).
ABP-280-dependent Localization of Cellular Compartments
Whereas most studies have demonstrated the importance of microtubules to the positioning of cellular organelles (for review see Cole and Lippincott-Schwartz, 1995), our findings demonstrate directly an important role for an identified component of the microfilament-based cytoskeleton in the localization of late endosomes and lysosomes. To date, ABP-280 has not been reported to interact directly with cellular organelles. However, its association with mRNA molecules (Bassell et al., 1994) suggests ABP-280 may indeed have multiple roles in organization of cellular components. Thus, the ABP-280-dependent localization of late endosomes and lysosomes (Fig. 9) may be achieved by a direct interaction between ABP-280 and these compartments. More likely, however, the importance of ABP-280 to the localization of late endocytic compartments and lysosomes may be an indirect effect due to the importance of this cytoskeletal protein to the organization of actin cables and their likely role in organizing cellular compartments via binding to other organelle-associated actin binding proteins (Beck et al., 1994; Devarajan et al., 1996; Jung et al., 1996; Ceccaldi et al., 1995).
Our findings are in agreement with earlier studies (van Deurs et al., 1995) which demonstrated that, after treatment with cytochalasin D, lysosomes are located primarily in the periphery and often in cell processes. The authors speculated that microfilaments are required for the normal movement and distribution of endocytic compartments— actin filaments are involved in bringing mature endosomes and lysosomes together in the paranuclear cytoplasm and facilitating fusions, whereas microtubules pull them apart and direct them towards the cell periphery. Interestingly, the homogeneous ABP-280-dependent high-angle branching of short actin filaments results in the formation of a microfilament web (∼100-nm spacing) that excludes larger cellular organelles but permits movement of vesicle traffic (Hartwig and Shevlin, 1986). Thus, consistent with this possibility, the absence of ABP-280 (or cells treated with cytochalasin D) may cause a breakdown of the spatial constraint on the diffusion of large vesicles imposed by the ABP-280-induced orthogonal microfilament networks.
Role of ABP-280 in the Formation of Furin Processing Compartments
Our results point to an important role for ABP-280 in regulating the formation of furin-processing compartments. The more efficient activation of Pseudomonas exotoxin, as determined by inhibition of protein translation, in M2 cells compared with A7 cells (Fig. 10) demonstrates that ABP-280 modulates the formation of furin processing compartments in the endocytic pathway. Furthermore, this assay of the activity of endogenous furin is consistent with our finding that furin is internalized from the cell surface at a greater rate in M2 cells compared with A7 cells (Fig. 4). It remains a formal possibility that the differential effect observed on PE toxicity between M2 and A7 cells may result from altered processing of the α2MR/LRP, also a furin substrate (Gu et al., 1996), in these two cell lines. However, the equally efficient processing of pro-βNGF in both M2 and A7 cells argues that ABP-280 does not affect the formation of processing compartments in the TGN/biosynthetic pathway. Furthermore, this assay of the activity of endogenous furin is consistent with our findings that (a) furin is concentrated in the TGN (by virtue of its paranuclear staining pattern) in both M2 and A7 cells (Fig. 4), and (b) ABP-280 does not interact with the region of the furin cd that contains the TGN localization signal.
To date, most identified furin substrates are cleaved in the TGN/biosynthetic pathway. Furin processing at the cell surface and in endosomes has only been shown for the activation of bacterial pathogens (see Introduction). Nonetheless, these findings, together with our demonstration of the tethering of furin molecules to the cell surface by ABP-280 (Fig. 3) and the importance of this cytoskeletal-associated protein in modulating the rate of internalization of furin (Fig. 4), suggest important roles for this endoprotease in the activation of endogenous proprotein substrates both at the cell surface and in endocytic compartments. The tethering of cell surface furin by ABP-280 provides a mechanism to regulate the concentration of this low abundance endoprotease to regions of the plasma membrane where efficient processing can occur. Although no endogenous substrates for furin have been unequivocally shown to be cleaved at the cell surface, several candidates exist. For example, the membrane type 1 matrix metalloprotease, MT-MMP1, is a furin substrate (Pei and Weiss, 1996; Sato et al., 1996) that, by virtue of its cytosolic domain, localizes to regions of the cell surface by an as yet unidentified docking system (Nakahara et al., 1997). The zymogenic form of MT-MMP1 appears at the cell surface (Cao et al., 1994; Sato et al., 1994), suggesting that furin may cleave this substrate, and possibly others, subsequent to their delivery to this compartment.
A Model for the Role of ABP-280 in the Routing of Furin
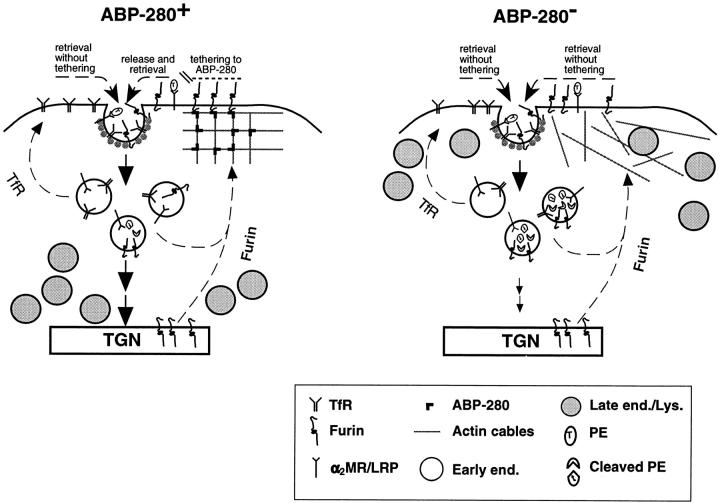
Based on the binding of ABP-280 to the furin cd, we propose a simple model to describe the trafficking and compartmentalization of itinerant membrane proteins in the TGN/endosomal system (Fig. 11). This model predicts the interplay of two classes of cytosolic binding proteins involved in either (a) protein kinesis (e.g., clathrin-associated adapters), or (b) stabilization/tethering of membrane proteins to specific compartments (e.g., ABP-280). In the case of furin, binding of ABP-280 to the region of the furin cd near the tyrosine-based and di-leucine-like internalization signals would stabilize furin at the cell surface and simultaneously mask the binding of cellular machinery involved in the internalization of the endoprotease. In addition, binding of furin to ABP-280 provides a mechanism to concentrate this endoprotease to regions of the cell surface involved in proprotein processing (e.g., the furin-catalyzed cleavage of anthrax protective antigen [Klimpel et al., 1992] and potentially endogenous substrates such as MT-MMP1). Dissociation of furin from ABP-280 by an as yet undetermined mechanism would permit the binding of the exposed internalization motifs of the endoprotease to the cellular internalization machinery (e.g., μ subunits of the AP-2 clathrin-associated adapters) (Ohno et al., 1995) and hence retrieval of the enzyme to intracellular compartments (endosomes and TGN). Whereas the efficiency of sorting of furin from the cell surface to the early endsomes is independent of ABP-280, efficient sorting of the endoprotease from early endosomes to the TGN is dependent on expression of this actin binding protein. By contrast to furin, the routing of TfR and, presumably, other cycling receptor molecules would not be modulated by binding to ABP-280 or other stabilization-class binding proteins. For such constitutively cycling membrane proteins, binding to the cellular internalization machinery would proceed directly. Thus, our data showing that furin is internalized at a greater rate in the M2 cells is consistent with the continued accessibility of the furin cd internalization signals in the absence of ABP-280. The sorting of furin between other cellular compartments is thus predicted to result similarly from the dynamic interplay between additional sets of cytosolic binding proteins. Our demonstration of ABP-280 as the first identified furin cd binding protein will now enable the testing of this model not only for this endoprotease but also for the trafficking of other itinerant membrane proteins in the TGN/endosomal system.
Figure 11.
A model for ABP-280 on trafficking within the TGN/endosomal system. Shown are the routing of TfR, furin, the α2MR/ LRP-dependent internalization of PE, the proteolytic activation of the toxin in early endosomes, and the localization of late endosomes and lysosomes in the TGN/endosomal system. In ABP-280+ cells (e.g., A7), this actin binding protein directs formation of the cortical microfilaments into an orthogonal lattice, whereas, in ABP-280− cells (e.g., M2), the actin cables form disorganized arrays. In ABP-280+ cells, lysosomes and late endocytic compartments are concentrated primarily in the paranuclear region, whereas, in ABP-280− cells, these compartments are mislocalized (Fig. 9). Whether the localization of these compartments is dependent on their direct interaction with ABP-280 or microfilaments is not known. Conversely, the localization of early endosomes and the TGN is independent of ABP-280 (Figs. 5–9). TfR cycles constitutively between the early endosomes and the plasma membrane, and neither the cell surface receptor number, the cycling time of the receptor, nor the subcellular distribution of early endosomes is directly affected by the presence or absence of ABP-280 (Figs. 4 and 9). By contrast, the trafficking of furin is directed by ABP-280. In A7 cells, furin present at the cell surface is tethered by ABP-280 (Fig. 3). It is conceivable that this tethering gathers the proprotein convertase into patches to form furin processing compartments at the cell surface (e.g., activation of the anthrax toxoid [Klimpel et al., 1992] and potential MT-MMP1 [see Discussion]). Whether the α2MR/LRP is similarly tethered is not known. Release of furin from ABP-280 by an as yet unidentified mechanism allows the endoprotease to be retrieved into clathrin-coated pits where the tyrosine-based and/or di-leucine-like motifs within the furin cd are available to interact with the cellular internalization machinery (e.g., AP-2 complexes). In the ABP-280-deficient M2 cells, furin delivered to the cell surface is not tethered to the cortical microfilaments and instead is rapidly internalized into clathrin-coated pits. This unrestricted internalization of furin enhances the probability that it encounters substrates (e.g., PE) in endocytic compartments. Although ABP-280 modulates the rate of furin internalization (Fig. 4), it does not effect the efficiency of sorting of the endoprotease from the cell surface to early endosomes (Fig. 7). By contrast, the efficient retrieval of the internalized furin from the early endosomes to the TGN requires ABP-280 (Figs. 5 and 7). Large arrows represent efficient sorting, whereas small arrows denote inefficient sorting.
Acknowledgments
We thank members of the Thomas lab for many helpful discussions during the course of this work. We thank M. Sanders for assistance with the yeast two-hybrid analyses. We thank S. Hollenberg for the two-hybrid reagents, E. Ungewickell for the 100/3 mAb, and R. Draper for the PE and helpful discussions. We are grateful to S. Rybak, S. Molloy, S. Arch, and R. Kelly for critically reading the manuscript and for helpful advice.
Abbreviations used in this paper
- α2MR/LRP
macroglobulin receptors/ low density lipoprotein receptor-related protein
- cd
cytosolic domain
- PE
Pseudomonas exotoxin
- Tf
transferrin
- TGN
trans-Golgi network
- TXR
Texas red
Note Added in Proof
The DNA sequence reported in this paper is available from GenBank/DDBJ/EMBL under accession number AF034129.
Footnotes
This was supported by National Institutes of Health (NIH) grants DK-37274 and DK-44629 (G. Thomas) and DK-40608 (C.A. Enns). G. Liu was supported by NIH Training Grants DK-07680 and DK-07674, and also National Research Service Award. DK-09394.
Address all correspondence to Gary Thomas, Vollum Institute and Department of Cell and Developmental Biology, Oregon Health Sciences University, Portland, OR 97201. Tel.: 503-494-6955. Fax: 503-494-4534. E-mail: thomasg@ohsu.edu
References
- Bartel PL, Fields S. Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Powers CM, Taneja KL, Singer RH. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J Cell Biol. 1994;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo DA, Gleason JB, Sanchez RI, Roth RA, Fuller RS. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J Biol Chem. 1994;269:25830–25837. [PubMed] [Google Scholar]
- Beck KA, Buchanan JA, Malhotra V, Nelson WJ. Golgi spectrin: identification of an erythroid beta-spectrin homolog associated with the Golgi complex. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan PA, Leduc R, Thomas L, Thorner J, Gibson HL, Brake AJ, Barr PJ, Thomas G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-β-NGF In Vivo. . J Cell Biol. 1990;111:2851–2859. doi: 10.1083/jcb.111.6.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Microfilaments and membranes. Curr Opin Cell Biol. 1993;5:653–660. doi: 10.1016/0955-0674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- Cao J, Sato H, Takino T, Seiki M. The C-terminal region of membrane type matrix metalloproteinase is a functional transmembrane domain required for pro-gelatinase A activation. J Biol Chem. 1994;270:801–805. doi: 10.1074/jbc.270.2.801. [DOI] [PubMed] [Google Scholar]
- Carpen O, Pallai P, Staunton DE, Springer TA. Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and alpha-actinin. J Cell Biol. 1992;118:1223–1234. doi: 10.1083/jcb.118.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi PE, Grohovaz F, Benfenati F, Chieregatti E, Greengard P, Valtorta F. Dephosphorylated synapsin I anchors synaptic vesicles to actin cytoskeleton: an analysis by videomicroscopy. J Cell Biol. 1995;128:905–912. doi: 10.1083/jcb.128.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO (Eur Mol Biol Organ) J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron, M.F., C.M. Fryling, and D.J. Fitzgerald. 1994. Cleavage of pseudomonas exotoxin and diphtheria toxin by a furin-like enzyme prepared from beef liver. J. Biol. Chem. 269, 18167–18176. [PubMed]
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Costa de Beauregard MA, Pringault E, Robine S, Louvard D. Suppression of villin expression by antisense RNA impairs brush border assembly in polarized epithelial intestinal cells. EMBO (Eur Mol Biol Organ) J. 1995;14:409–421. doi: 10.1002/j.1460-2075.1995.tb07017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- Cunningham JG, Meyer SC, Fox JE. The cytoplasmic domain of the alpha-subunit of glycoprotein (GP) Ib mediates attachment of the entire GP Ib-IX complex to the cytoskeleton and regulates von Willebrand factor-induced changes in cell morphology. J Biol Chem. 1996;271:11581–11587. doi: 10.1074/jbc.271.19.11581. [DOI] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds beta I sigma spectrin and associates with the Golgi apparatus. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Dubois CM, Laprise MH, Blanchette F, Gentry LE, Leduc R. Processing of transforming growth factor beta 1 precursor by human furin convertase. J Biol Chem. 1995;270:10618–10624. doi: 10.1074/jbc.270.18.10618. [DOI] [PubMed] [Google Scholar]
- Fox JE. Identification of actin-binding protein as the protein linking the membrane skeleton to glycoproteins on platelet plasma membranes. J Biol Chem. 1985;260:11970–11977. [PubMed] [Google Scholar]
- Garred O, van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J Biol Chem. 1995;270:10817–10821. doi: 10.1074/jbc.270.18.10817. [DOI] [PubMed] [Google Scholar]
- Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect Immun. 1995;63:82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh B, Ohnishi Y, Inocencio NM, Esaki E, Nakayama K, Barr PJ, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66:6391–6397. doi: 10.1128/jvi.66.11.6391-6397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TA, Ivanov IE, Adesnik M, Sabatini DD. Actin microfilaments play a critical role in endocytosis at the apical but not the basolateral surface of polarized epithelial cells. J Cell Biol. 1993;120:695–710. doi: 10.1083/jcb.120.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Simons K. The transgolgi network: sorting at the exit site of the golgi complex. Science. 1986;234:438–443. doi: 10.1126/science.2945253. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Howell KE. Membrane traffic in endocytosis: insights from cell-free assays. Annu Rev Cell Biol. 1989;5:453–481. doi: 10.1146/annurev.cb.05.110189.002321. [DOI] [PubMed] [Google Scholar]
- Gu M, Gordon VM, Fitzgerald DJ, Leppla SH. Furin regulates both the activation of Pseudomonas exotoxin A and the quantity of the toxin receptor expressed on target cells. Infec Immun. 1996;64:524–527. doi: 10.1128/iai.64.2.524-527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- Harrison PT, Davis W, Norman JC, Hockaday AR, Allen JM. Binding of monomeric immunoglobulin G triggers Fc gamma RI-mediated endocytosis. J Biol Chem. 1994;269:24396–24402. [PubMed] [Google Scholar]
- Hartwig JH, Shevlin P. The architecture of actin filaments and the ultrastructural location of actin-binding protein in the periphery of lung macrophages. J Cell Biol. 1986;103:1007–1020. doi: 10.1083/jcb.103.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt AL, Luna EJ. Membrane interactions with the actin cytoskeleton. Curr Opin Cell Biol. 1994;6:120–130. doi: 10.1016/0955-0674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- Hu RJ, Moorthy S, Bennett V. Expression of functional domains of beta G-spectrin disrupts epithelial morphology in cultured cells. J Cell Biol. 1995;128:1069–1080. doi: 10.1083/jcb.128.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inocencio NM, Moehring JM, Moehring TJ. Furin activates Pseudomonas exotoxin A by specific cleavage in vivo and in vitro. J Biol Chem. 1994;269:31831–31835. [PubMed] [Google Scholar]
- Jones BG, Thomas L, Molloy SS, Thulin CD, Fry MD, Walsh KA, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO (Eur Mol Biol Organ) J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E, Fucini P, Stewart M, Noegel AA, Schleicher M. Linking microfilaments to intracellular membranes: the actin-binding and vesicle- associated protein comitin exhibits a mannose-specific lectin activity. EMBO (Eur Mol Biol Organ) J. 1996;15:1238–1246. [PMC free article] [PubMed] [Google Scholar]
- Klimpel KR, Molloy SS, Thomas G, Leppla SH. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Hatsuzawa K, Shibamoto S, Ito F, Nakayama F, Kitamura N. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett. 1993;328:25–29. doi: 10.1016/0014-5793(93)80958-w. [DOI] [PubMed] [Google Scholar]
- Kounnas MZ, Morris RE, Thompson MR, FitzGerald DJ, Strickland DK, Saelinger CB. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J Biol Chem. 1992;267:12420–12423. [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Lille SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;365:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokeshwar VB, Fregien N, Bourguignon LY. Ankyrin-binding domain of CD44(GP85) is required for the expression of hyaluronic acid-mediated adhesion function. J Cell Biol. 1994;126:1099–1122. doi: 10.1083/jcb.126.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton—plasma membrane interactions. Science. 1992;258:955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Mays RW, Beck KA, Nelson WJ. Organization and function of the cytoskeleton in polarized epithelial cells: a component of the protein sorting machinery. Curr Opin Cell Biol. 1994;6:16–24. doi: 10.1016/0955-0674(94)90111-2. [DOI] [PubMed] [Google Scholar]
- Meyer SC, Zuerbig S, Cunningham CC, Hartwig JH, Fox JEB. Identification of the region in actin-binding protein that binds to the cytoplasmic domain of glycoprotein Iba. J Biol Chem. 1997;272:2914–2919. doi: 10.1074/jbc.272.5.2914. [DOI] [PubMed] [Google Scholar]
- Misumi Y, Oda K, Fujiwara T, Takami N, Tashiro K, Ikehara Y. Functional expression of furin demonstrating its intracellular localization and endoprotease activity for processing of proalbumin and complement pro-C3. J Biol Chem. 1991;266:16954–16959. [PubMed] [Google Scholar]
- Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO (Eur Mol Biol Organ) J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H, Howard L, Thompson EW, Sato H, Seiki M, Yeh Y, Chen WT. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc Natl Acad Sci USA. 1997;94:7959–7964. doi: 10.1073/pnas.94.15.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Stossel TP, Hartwig JH. Ligand-sensitive binding of actin-binding protein to immunoglobulin G Fc receptor I (Fc gamma RI) Cell. 1991;67:275–282. doi: 10.1016/0092-8674(91)90179-3. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH. Actin filament cross-linking by chicken gizzard filamin is regulated by phosphorylation in vitro. Biochemistry. 1995;34:6745–6754. doi: 10.1021/bi00020a020. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH. Phosphorylation of actin-binding protein 280 by growth factors is mediated by p90 ribosomal protein S6 kinase. J Biol Chem. 1996;271:11858–11864. doi: 10.1074/jbc.271.20.11858. [DOI] [PubMed] [Google Scholar]
- Pei D, Weiss SJ. Transmembrane-deletion mutants of the membrane-type matrix metalloproteinase-1 process progelatinase A and express intrinsic matrix-degrading activity. J Biol Chem. 1996;271:9135–9140. doi: 10.1074/jbc.271.15.9135. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR, Rothman JE. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Powell SK. Polarity of epithelial and neuronal cells. Annu Rev Cell Biol. 1992;8:395–427. doi: 10.1146/annurev.cb.08.110192.002143. [DOI] [PubMed] [Google Scholar]
- Roth J, Taatjes DJ, Lucocq JM, Weinstein J, Paulson JC. Demonstration of an extensive trans-tubular network continuous with the Golgi apparatus stack that may function in glycosylation. Cell. 1985;43:287–295. doi: 10.1016/0092-8674(85)90034-0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. 2nd Edition, Volume 2. C. Nolan, editor. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 14.1–15.113.
- Sandvig K, van Deurs B. Selective modulation of the endocytic uptake of ricin and fluid phase markers without alteration in transferrin endocytosis. J Biol Chem. 1990;265:6382–6388. [PubMed] [Google Scholar]
- Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Sato H, Kinoshita T, Takino T, Nakayama K, Seiki M. Activation of a recombinant membrane type 1-matrix metalloproteinase (MT1-MMP) by furin and its interaction with tissue inhibitor of metalloproteinases (TIMP)-2. FEBS Lett. 1996;393:101–104. doi: 10.1016/0014-5793(96)00861-7. [DOI] [PubMed] [Google Scholar]
- Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO (Eur Mol Biol Organ) J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CP, Ezzell RM, Arnaout MA. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Smeekens SP, Ohagi S, Chan SJ. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk HD, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO (Eur Mol Biol Organ) J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- Thorne BA, Caton LW, Thomas G. Expression of mouse proopiomelanocortin in an insulinoma cell line. J Biol Chem. 1989;264:3545–3552. [PubMed] [Google Scholar]
- Tooze SA, Weiss U, Huttner WB. Requirement for GTP hydrolysis in the formation of secretory vesicles. Nature. 1990;347:207–208. doi: 10.1038/347207a0. [DOI] [PubMed] [Google Scholar]
- Trowbridge IS, Collawn JF. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- van Deurs B, Holm PK, Kayser L, Sandvig K. Delivery to lysosomes in the human carcinoma cell line HEp-2 involves an actin filament- facilitated fusion between mature endosomes and preexisting lysosomes. Eur J Cell Biol. 1995;66:309–323. [PubMed] [Google Scholar]
- Vey M, Schafer W, Reis B, Ohuchi R, Britt W, Garten W, Klenk HD. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology. 1995;206:746–749. doi: 10.1016/s0042-6822(95)80002-6. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO (Eur Mol Biol Organ) J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RA, Enns CA. Overexpression of the human transferrin receptor slows its endocytosis but not the endocytosis of the EGF receptor. J Biol Chem. 1997;272:2116–2121. doi: 10.1074/jbc.272.4.2116. [DOI] [PubMed] [Google Scholar]
- Walker JA, Molloy SS, Thomas G, Sakaguchi T, Yoshida T, Chambers TM, Kawaoka Y. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J Virol. 1994;68:1213–1218. doi: 10.1128/jvi.68.2.1213-1218.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasley LC, Rehemtulla A, Bristol JA, Kaufman RJ. Pace/furin can process the vitamin K-dependent pro-factor IX precursor within the secretory pathway. J Biol Chem. 1993;268:8458–8465. [PubMed] [Google Scholar]
- Watanabe M, Hirano A, Stenglein S, Nelson J, Thomas G, Wong TC. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J Virol. 1995;69:3206–3210. doi: 10.1128/jvi.69.5.3206-3210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RJ, Barr PJ, Wong PA, Kiefer MC, Brake AJ, Kaufman RJ. Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc Natl Acad Sci USA. 1990;87:9378–9382. doi: 10.1073/pnas.87.23.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]