Abstract
Structural differences between dynein and kinesin suggest a unique molecular mechanism of dynein motility. Measuring the mechanical properties of a single molecule of dynein is crucial for revealing the mechanisms underlying its movement. We measured the step size and force produced by single molecules of active cytoplasmic dynein by using an optical trap and fluorescence imaging with a high temporal resolution. The velocity of dynein movement, 800 nm/s, is consistent with that reported in cells. The maximum force of 7–8 pN was independent of the ATP concentration and similar to that of kinesin. Dynein exhibited forward and occasional backwards steps of ≈8 nm, independent of load. It is suggested that the large dynein heads take 16-nm steps by using an overlapping hand-over-hand mechanism.
Keywords: microtubules, motor protein, optical tweezers, step size, nanotechnology
Dynein is a molecular motor that moves along microtubules to the direction of the minus end. Cytoplasmic dynein transports cellular organelles toward the minus end of microtubules, whereas most kinesin molecules transport organelles toward the plus end (1–3). Hirakawa et al. (4) have reported that purified axonemal dynein produced maximum forces of 5 pN, and they also showed stepwise displacements of 8 nm. In contrast, another study (5) has demonstrated that single molecules of purified cytoplasmic dynein move with short steps (8 nm) at high loads (>0.8 pN) and long steps (16–32 nm) at low forces (<0.4 pN), and from these findings a molecular gear mechanism was proposed. The values for maximum force and the size of the steps differ between those two reports. Moreover, the very low velocity (<50 nm/s) of movement of the purified cytoplasmic dynein (5) is in direct contrast to the high velocity (≈1 μm/s) of dynein movement in a cell and in an in vitro motility assay (6–8). These discrepancies possibly result from inactive dynein molecules being included with the purified native dynein in the analysis.
In this study a method of coating the beads with dynein was developed to keep dynein active and allow the force and step size produced by single molecules of dynein to be measured. The step size of active cytoplasmic dynein was 8 nm and was independent of both force and ATP concentration. The stall force was 7–8 pN. These values are very similar to those recorded for kinesin (9); however, the stepping manner was different from that of kinesin.
Results
Active Dynein Bound to Protein A-Coated Beads.
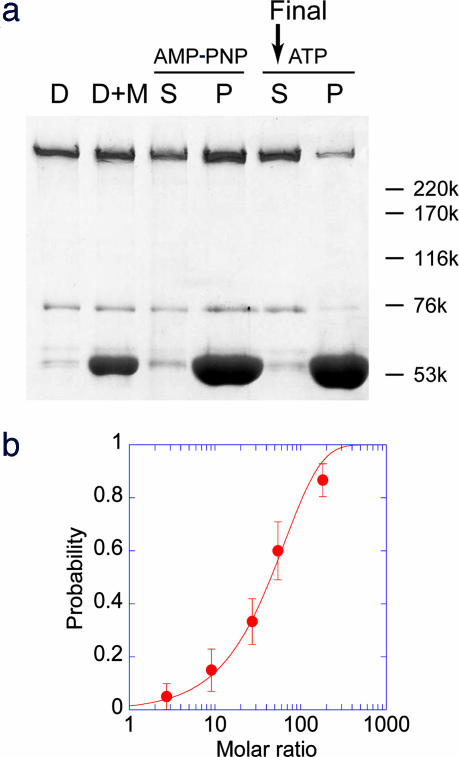
The method used to purify cytoplasmic dynein without its accessory protein, dynactin, has been described (Fig. 1a, lane D) (10, 11). Dynein was further purified by allowing it to bind to the microtubules (Fig. 1a, lane AMP-PNP) (12). Dynein was then released from microtubules in the presence of 0.1–10 mM ATP (Fig. 1a, lane ATP).
Fig. 1.
Purification and force generation of single cytoplasmic dynein molecules. (a) SDS/PAGE of dynein purification. D lane, dynein purified by the anion-exchange column. D+M lane, dynein mixed with the microtubules. S and P indicate supernatant and pellet, respectively, after dynein mixed with the microtubules was centrifuged in the presence of 0.1 mM adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP). After the pellet was suspended, it was centrifuged in the presence of 10 mM ATP. The supernatant was then used for the experiment. Numbers on the right indicate molecular mass in kDa. (b) The probability of the moving beads relative to the total beads was calculated. The molar ratio is the ratio of the bead concentration to the dynein concentration (10–20 nM) at the time of mixing. The probability was fitted to the curve, 1 − exp(−x/65), indicating that single molecules produced the force.
The method used to bind dynein to the coverslips for in vitro motility assay and beads for optical trap assay was modified from previously reported methods (5, 8). Beads coated with protein A were suitable for motility and force generation. With dynein attached to glass coverslips and bound to microtubules marked at the minus end, the microtubules moved with the marked end trailing. This result indicated that dynein moved toward the minus end of the microtubule. Polystyrene beads, 0.2 μm in diameter, were coated with protein A. Freshly purified dynein was bound to the beads coated with protein A. Beads were trapped and brought into contact with the microtubule. Displacement and force were measured with very high temporal resolution by using small beads (9). Experiments were performed within 2 h after protein purification to ensure that active dynein was used in the measurements.
The fraction of beads that moved processively increased exponentially with the increase in the ratio of dynein to the beads, indicating that single molecules of dynein move processively (Fig. 1b) (4, 13, 14). The fraction of beads that bound to microtubules in the presence of 1 mM adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) was almost the same as the fraction that moved in the presence of 1 mM ATP. Dynein binds tightly to microtubule in the presence of AMP-PNP (12). This finding also provides support for the conclusion that single molecules move processively (4). The ratio of beads-to-dynein molecules required to move 50% of the beads was ≈40:1, which was considerably lower than the value obtained by using dynein on a noncoated bead (5) (Fig. 5, which is published as supporting information on the PNAS web site). The ratio was also lower than the value (>120:1) when the beads were coated with BSA or casein. This result indicates that beads coated with protein A provide a suitable medium for dynein to bind. Experiments measuring step size and force were performed at a ratio of 0.1:0.3 of the mobile-to-immobile beads, where >95% of the beads that moved and produced force were caused by single molecules of dynein (9).
Force–Velocity Relation of Single Dynein Molecules.
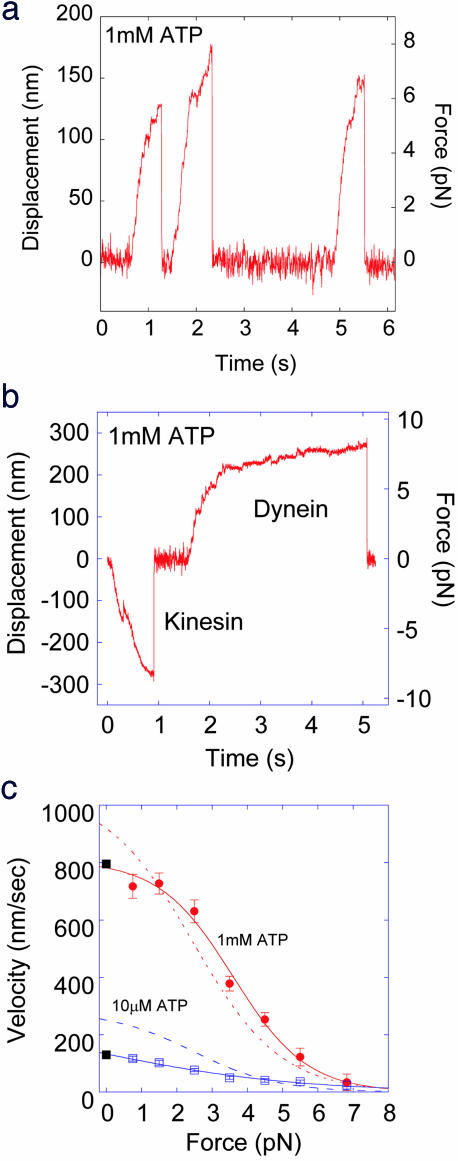
Single molecules of dynein moved processively and produced forces up to 7–8 pN in the presence of 1 mM ATP (Fig. 2a). The dynein bound to beads coated with BSA or casein also produced forces of ≈7 pN, indicating that protein A coating did not modify the motility of dynein. However, when the beads were not coated with the proteins low forces and velocities were often observed similar to those reported in a previous study (5) (Fig. 5). This result suggests that the presence of protein A is important for keeping dynein active.
Fig. 2.
Force generation by single cytoplasmic dynein molecules. (a) Bead movement in the presence of 1 mM ATP at a trap stiffness of 45 fN/nm. Force reached 6–8 pN before the dynein molecule dissociated from the microtubule. (b) Dynein beads were mixed with kinesin at a ratio of 10:1 in the presence of 1 mM ATP at a trap stiffness of 30 fN/nm. (c) Force–velocity dependence at 1 mM (red circles) and 10 μM ATP (violet squares). Velocity was calculated from the displacement (20–30 nm) divided by the time taken to cover this distance. Each symbol is the average value of the velocities from 20–30 individual force traces. Velocities at zero force were measured by using an in vitro motility assay (closed squares). Dotted line indicates the force–velocity relation of bovine kinesin-1 from a previous study (9).
To confirm the direction of movement, beads with dynein attached were mixed with kinesin-1 molecules. Directional movement in both the forward and backward directions was observed, confirming that the direction of movement by single molecules of dynein is opposite to that of kinesin (Fig. 2b).
The force trace of dynein movement is very similar to that of kinesin other than the direction (Fig. 2b). To confirm the similarity, the relationship of force to velocity was obtained by analysis of 20–30 traces (Fig. 2c). The velocity of dynein decreased with increased force. The velocity at low forces and high ATP concentrations of ≈800 nm/s is consistent with that observed in cells and in vitro assays (0.5–1.5 μm/s) (6–8). At low ATP concentrations (10 μM), the velocity decreased and the stall force of ≈7 pN was independent of the concentration of ATP (Fig. 2c). The velocity at low forces is about half that obtained for kinesin-1 [Fig. 2c, dashed blue line (9)]. This distinction explains the difference in Km values, i.e., ATP concentration at the half-maximal activated ATPase activity between kinesin-1 and dynein. The Km value of cytoplasmic dynein was measured to be 89 μM, which is 2.5-fold larger than that of kinesin-1 [36 μM (9)].
Step Size of Single Dynein Molecules.
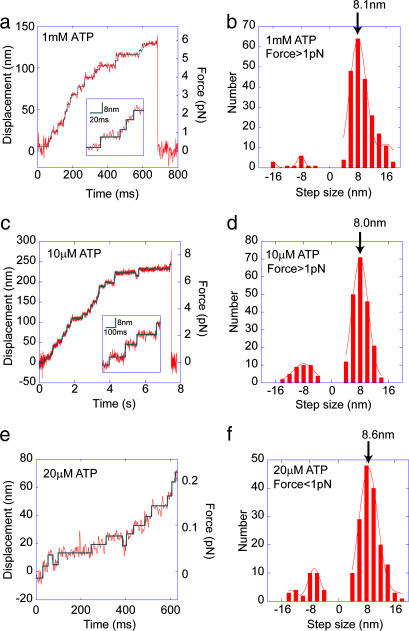
Single molecules of cytoplasmic dynein moved stepwise at ATP concentrations of 1 mM and 10 μM (Fig. 3a and c). Steps of 8 nm were clearly detected in the expanded traces (Fig. 3 a and c Insets). Steps of 8 nm were also observed at very low forces of <0.3 pN as measured with a trap laser at low power (Fig. 3e). Backward 8-nm steps were also occasionally detected (Fig. 3 c and e).
Fig. 3.
Step size of single dynein molecules. (a and c) Stepwise movement at high concentration (1 mM, a), and low concentration (10 μM, c) of ATP. (a and c Insets) Expanded traces of the stepwise movement. (e) Bead movement in 20 μM ATP at a trap stiffness of 3.0 fN/nm. (a, c, and e) Step sizes of >4 nm and dwell times of >5 ms (a) and >20 ms (c and e) were fitted to a rectangular curve (30) (blue line). (b, d, and f) Histograms of step sizes. (b) ATP concentration of 1 mM and forces >1 pN. (d) ATP concentration of 10 μM and forces >1 pN. (f) ATP concentration of 20 μM and forces <1 pN. The histograms a–c were fitted to a Gaussian curve with the center at a multiple of the unit step size of 8.1, 8.0, and 8.6 nm, respectively.
Step sizes of >4 nm were analyzed over the range of 1 to 7 pN. A major peak was observed at ≈8 nm and minor peaks were seen at 16 and −8 nm (Fig. 3 b and d). The 16-nm steps were assumed to be rapid double steps of 8 nm generated with a temporal resolution of 4 ms or less. The observed step size was ≈8 nm and was independent of force in the range of 1–7 pN and the ATP concentration.
A previous study by Mallik et al. (5) found that the step size of single dynein molecules increased to 16–24 nm at low (<0.8 pN) or zero force. Here, the step size at low forces of <1 pN was measured at a low laser trap stiffness (Fig. 3e). A histogram of step size at forces in the range 0.0 to 0.9 pN indicated a major peak at 8 nm (Fig. 3f). The step size at low forces was 8–9 nm and was independent of force.
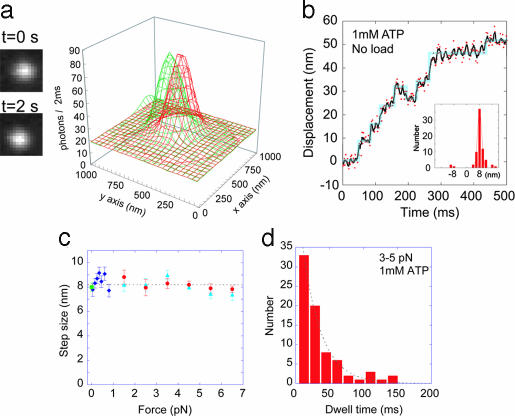
The movement of single dynein molecules was measured at zero load without a laser trap by using fluorescence imaging with 1-nm accuracy (15). The intensity of organic fluorescence dyes such as Cy3 is too weak to be used for measurements at high temporal resolutions <100 ms (15). The step dwell time of cytoplasmic dynein was ≈10 ms. To improve the temporal resolution and lifetime of the fluorescence, dynein was bound to CdSe quantum dots, which are very stable and have an intense fluorescence (16). To slow the movement of dynein, the temperature was decreased to 16°C. At an exposure time of 2 ms, clear images were obtained (Fig. 4a and Fig. 6, which is published as supporting information on the PNAS web site). The position of dynein was determined by fitting a profile of the fluorescence intensity to a 2D Gaussian curve (Fig. 4a). These methods showed that the step size of dynein is 8 nm (Fig. 4b). The histogram of step sizes showed a major peak of 8.0 nm and a minor peak at 16 nm (Fig. 4b Inset).
Fig. 4.
Step sizes and dwell times. (a) Fluorescence images and profiles of the images of a quantum dot dynein interacting with a microtubule. The green-colored profile was taken at time 0 and the red profile was taken at t = 2 s. (b) Displacements of single dynein molecules at 2-ms intervals were determined by the intensity profiles fitted to 2D Gaussian curves (red dots). Three points were averaged to reduce noise (black line). Step sizes were automatically detected by using computer programming (blue line). (Inset) A histogram of the step size with a peak at 8.0 nm. (c) Step sizes were measured at high trap stiffness and ATP concentrations of 1 mM (circles) and 10 μM (triangles). Step sizes at low trap stiffness and 20 μM ATP (diamonds) and zero load (squares) were obtained. The average step size was 8.1 nm. (d) Dwell time between adjacent steps. The time was measured at 3- to 5-pN force in the presence of 1 mM ATP. The dwell time was fitted to a single exponential curve to obtain a time constant of 27 ms.
The step sizes at various forces and ATP concentrations are summarized in Fig. 4c. The step size of 8 nm was independent of force over the range 0 to 7 pN and ATP concentration over the range 10 μM to 1 mM. The average of all of the data was 8.1 nm.
The time taken from the beginning of one step to the beginning of the next step was analyzed as the dwell time and was used to determine the step reaction rate and the number of rate-limiting steps. The histogram of the dwell time at forces of 3–5 pN showed an exponential decay with a time constant of 27 ms (Fig. 4d), indicating that a rate-limiting state is one with a rate of 37 per s.
Discussion
The stall force of single molecules of cytoplasmic dynein in the laser trap assays was 7–8 pN and was independent of the ATP concentration and load. The stall force and step size of cytoplasmic dynein were consistent with the results for axonemal dynein [≈6 pN (17); ≈5 pN and 8 nm (4)] and cytoplasmic dynein in a cell (8 nm) (18). However, these results are inconsistent with previous findings of low force, low velocity, and a long step size for cytoplasmic dynein (5). These discrepancies are most likely caused by the active state of dynein. Improvements of the bead-coating method and temporal resolution of the optical trap made it possible to measure the fast movement of the active dynein molecules. This idea is supported by the fact that the low forces and velocity are similar to those in a previous study (5) when the bead was not coated (Fig. 5).
The single exponential decay of the dwell time distribution indicates only one rate-limiting state in an 8-nm step (14). Maximum ATPase rate and the sliding velocity of recombinant single-headed dynein was 160 per s and 1,200 nm per s, respectively (12). From these values, the sliding distance of single-headed dynein was calculated to be 7.5 nm per ATP molecule hydrolyzed. These results suggest that 8-nm steps of single molecules of dynein are coupled with the hydrolysis of ATP molecules under our experimental conditions. Dynein has four potential ATP binding and hydrolysis sites, and one site, P1, is the primary site for ATPase activity (19). Thus, 8-nm steps would be coupled with ATP hydrolysis mainly in the P1 site.
The maximum velocity, stall force, and step size of the cytoplasmic dynein are strikingly similar to those reported for kinesin-1 (9, 13, 14, 20). Hand-over-hand and inchworm models for kinesin have been proposed previously (14, 21–24). The hand-over-hand mechanism was supported by the results that kinesin-1 has an asymmetrical dwell time and the head takes 16-nm steps (21–23). The result that dynein has one rate-limiting state at the 8-nm step supports the hand-over-hand model because the simple inchworm model requires two step reactions to produce one 8-nm step (21, 22). Therefore, each dynein head steps by 16 nm, whereas dynein molecule steps by 8 nm.
The dynein head differs greatly from kinesin-1 in structure, size, and amino acid sequence. The diameter of the ring-shaped dynein head is 15 nm, which is almost 2-fold greater than the step size. How then does dynein step by 8 nm with a hand-over-hand mechanism? Electron microscopy of dynein shows a Φ-shaped structure, indicating that the tails of the two heads are close to each other and the ring domains partially overlap (10, 25). When the heads of dynein bind to the microtubule, they also overlap (25–27). A possible model to explain the stepping movement is that the dynein heads are positioned axially along the microtubule similar to kinesin. Thus, the ring regions representing the dynein heads should overlap.
Force generation can be explained by structural changes between dynein head and tail that are predicted to occur at the different chemical states; one state is at the power stroke and the other at the recovery stroke (28, 29). Other than specific structural changes, the similarity of the mechanical properties of dynein, e.g., backward steps, step size, and stall force, to those of kinesin-1 predicts that Brownian motion and possibly biased binding of the heads are essential components in the molecular mechanism underlying the dynein steps (9, 20, 30).
Materials and Methods
Preparation of Dynein and Kinesin.
Cytoplasmic dynein and kinesin-1 were purified from porcine brain as described (9–11). Purified dynein in 25% sucrose was rapidly frozen and stored in liquid nitrogen. Just before an experiment, dynein was further affinity-purified by binding to the microtubules (12). Microtubules used in motility assays, and laser trap measurements were prepared from tubulin purified from porcine brains and labeled with tetramethylrhodamine.
Laser Trap Measurements.
Polystyrene beads of 0.2 μm in diameter (9) were coated with 1.4 mg/ml protein A (Sigma; P-6031) for >30 min in solution A containing 10 mM Pipes, 25 mM K-acetate, 4 mM MgSO4, and 1 mM EGTA (pH 7.0). The bead was then mixed with dynein in solution A containing 3 mg/ml BSA and 0.2 M KCl for 10 min. Dynein-coated beads were trapped, and their position was measured by using an optical trapping system (9). The assay solution was solution A, 10 μM–1 mM ATP, 5 mg/ml BSA, and an oxygen scavenger system at 25 ± 1°C. The trap stiffness was 3–45 fN/nm, determined by the amplitude of the Brownian motion of the beads (9). At a trap stiffness of 40–45 fN/nm (Figs. 2 and 3a), displacements to determine the step size and velocity were attenuated by a factor of 1.4 at forces of 1–2 pN to 1.1 at 6–7 pN, because of the stiffness of motor-bead linkage (13, 14). The attenuated displacements were consistent with data where no attenuation was performed at low trap stiffness or zero load (Figs. 3 e and f and 4b).
Individual steps in the movements and the step sizes were automatically detected by using computer programming. Each step was recognized when the step size was >4 nm, and the standard error of noise for 2–16 ms was <1.4 nm. The step size was calculated from the difference between the displacements before and after the step.
An in vitro motility assay was performed with some modifications (10). Coverslips were coated with the solution containing protein A as described for coating the beads. Dynein (1–30 nM) was introduced onto coverslips.
Quantum Dot Fluorescence Imaging with 1-nm Accuracy.
When imaging quantum dots, the CdSe particles (Evi-dots SG-ETC-H20-600) were cross-linked by using 10 mg/ml 1-ethyl-3-(3-dimethylaminopropyle)carbodiimide (EDC, Pierce) and 2 mg/ml protein A. CdSe–protein A particles (180 nM) were mixed with 30 nM dynein for >15 min. Dynein–CdSe complexes were observed under a fluorescence microscope (IX-71; Olympus, Shinjuku-ku, Tokyo) by evanescent illumination (green laser 0.1 mW/μm2 at a specimen; Big Sky Laser Technologies, Bozeman, MT) through an objective (Olympus ×60 Planapo, 1.45 numerical aperture, oil) (31). Fluorescence images were captured by using a cooled electron multiplier–charge-coupled device (Ixon DV860; Andor Technology, Tokyo) at 2-ms intervals. The centroid of the bead in the images was determined by fitting the images to 2D Gaussian curves (15). The CdSe particles have significant advantages over other fluorophores in that they have a much higher fluorescence intensity and a longer lifetime. To minimize the blinking of the CdSe particles, three to five CdSe particles with an attached carboxyl group were cross-linked with the amino group of protein A by EDC. Less than 10% of these complexes interacted with the microtubules, indicating that each complex binds only single dynein molecules. The experiments were performed at 16°C.
Supplementary Material
Acknowledgments
We thank Jan M. West and Sharyn A. Endow for critically reading this manuscript and Ken’ya Furuta for valuable discussions. This work was supported by Grants-in-Aid for Scientific Research in Priority Areas from the Japan Ministry of Education, Culture, Sports, Science, and Technology (to H.H.); Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation; and Special Coordination Funds for Promoting Science and Technology of Japan Ministry of Education, Culture, Sports, Science, and Technology (to Y.Y.T. and H.H.).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vale R. D. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa N., Takemura R. Curr. Opin. Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Koonce M. P., Samso M. Trends Cell Biol. 2004;14:612–619. doi: 10.1016/j.tcb.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Hirakawa E., Higuchi H., Toyoshima Y. Y. Proc. Natl. Acad. Sci. USA. 2000;97:2533–2537. doi: 10.1073/pnas.050585297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallik R., Carter B. C., Lex S. A., King S. J., Gross S. P. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 6.Paschal B. M., Shpetner H. S., Vallee R. B. J. Cell Biol. 1987;105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 8.King S. J., Schroer T. A. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 9.Nishiyama M., Higuchi H., Yanagida T. Nat. Cell Biol. 2002;4:790–797. doi: 10.1038/ncb857. [DOI] [PubMed] [Google Scholar]
- 10.Toba S., Toyoshima Y. Y. Cell Motil. Cytoskeleton. 2004;58:281–289. doi: 10.1002/cm.20018. [DOI] [PubMed] [Google Scholar]
- 11.Bingham J. B., King S. J., Schroer T. A. Methods Enzymol. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- 12.Nishiura M., Kon T., Shiroguchi K., Ohkura R., Shima T., Toyoshima Y. Y., Sutoh K. J. Biol. Chem. 2004;279:22799–22802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- 13.Svoboda K., Block S. M. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 14.Kojima H., Muto E., Higuchi H., Yanagida T. Biophys. J. 1997;73:2012–2022. doi: 10.1016/S0006-3495(97)78231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 16.Warshaw D. M., Kennedy G. G., Work S. S., Krementsova E. B., Beck S., Trybus K. M. Biophys. J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shingyoji C., Higuchi H., Yoshimura M., Katayama E., Yanagida T. Nature. 1998;393:711–714. doi: 10.1038/31520. [DOI] [PubMed] [Google Scholar]
- 18.Kural C., Kim H., Syed S., Goshima G., Gelfand V. I., Selvin P. R. Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 19.Kon T., Nishiura M., Ohkura R., Toyoshima Y. Y., Sutoh K. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 20.Carter N. J., Cross R. A. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 21.Kaseda K., Higuchi H., Hirose K. Nat. Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 22.Asbury C. L., Fehr A. N., Block S. M. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildiz A., Tomishige M., Vale R. D., Selvin P. R. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 24.Hua W., Chung. J., Gelles J. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 25.Amos L. A. J. Cell Sci. 1989;93:19–28. doi: 10.1242/jcs.93.1.19. [DOI] [PubMed] [Google Scholar]
- 26.Goodenough U. W., Heuser J. E. J. Cell Biol. 1982;95:798–815. doi: 10.1083/jcb.95.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupetti P., Lanzavecchia S., Mercati D., Cantele F., Dallai R., Mencarelli C. Cell Motil. Cytoskeleton. 2005;62:69–83. doi: 10.1002/cm.20084. [DOI] [PubMed] [Google Scholar]
- 28.Burgess S. A., Walker M. L., Sakakibara H., Knight P. J., Oiwa K. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 29.Kon T., Mogami T., Ohkura R., Nishiura M., Sutoh K. Nat. Struct. Mol. Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 30.Okada Y., Higuchi H., Hirokawa N. Nature. 2003;424:574–577. doi: 10.1038/nature01804. [DOI] [PubMed] [Google Scholar]
- 31.Tokunaga M., Kitamura K., Saito K., Iwane A. H., Yanagida T. Biochem. Biophys. Res. Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.