Abstract
CD154 (CD40 ligand) expression on CD4 T cells is normally tightly controlled, but abnormal or dysregulated expression of CD154 has been well documented in autoimmune diseases, such as systemic lupus erythematosus. Beyond regulation by NFAT proteins, little is known about the transcriptional activation of the CD154 promoter. We identified a species-conserved purine-rich sequence located adjacent to the CD154 transcriptional promoter proximal NFAT site, which binds early growth response (Egr) transcription factors. Gel shift assays and chromatin immunoprecipitation assays reveal that Egr-1, Egr-3, and NFAT1 present in primary human CD4 T cells are capable of binding this combinatorial site in vitro and in vivo, respectively. Multimerization of this NFAT/Egr sequence in the context of a reporter gene demonstrates this sequence is transcriptionally active upon T cell activation in primary human CD4 T cells. Overexpression of Egr-1, but not Egr-3, is capable of augmenting transcription of this reporter gene as well as that of an intact CD154 promoter. Conversely, overexpression of small interfering RNA specific for Egr-1 in primary human CD4 T cells inhibits CD154 expression. Similarly, upon activation, CD154 message is notably decreased in splenic CD4 T cells from Egr-1-deficient mice compared with wild-type controls. Our data demonstrate that Egr-1 is required for CD154 transcription in primary CD4 T cells. This has implications for selective targeting of Egr family members to control abnormal expression of CD154 in autoimmune diseases such as systemic lupus erythematosus.
Expression of CD154 (CD40 ligand) is restricted to recently activated CD4 T lymphocytes (1). By interacting with its receptor, CD40, which is expressed on B cells and other APCs, CD154 directs a wide array of immunologic events (2). B cells require CD154 stimulation for growth, development, and Ab isotype switching (3). In addition, germinal center formation and CD8 T cell effector function rely on CD154 expression (4). Increased and prolonged expression of CD154 after T cell activation has been documented in systemic lupus erythematosus, the prototype autoimmune disease characterized by chronic Th cell-dependent B cell activation, and contributes to the production of pathogenic class-switched autoantibodies in that disease (5-7). Because of its pleiotropic effects, CD154 expression is normally very tightly regulated in CD4 T cells (8). CD154 expression is controlled at both the levels of transcription and mRNA stability (8, 9). Both the human and murine CD154 promoters have been described, but outside of the regulation by NFAT proteins (10-12), very little is known about the factors that directly regulate CD154 transcription. There has been some suggestion that AP-1 (12), NF-κB (13, 14), and the AT-hook transcription factor AKNA (15) transcriptional activators may also increase CD154 message. Recently, a report proposed that early growth response (Egr)3 transcription factors, but not AP-1 proteins, are involved in the upregulation of CD154 promoter activity in response to T cell co-stimulation through CD28 (16).
CD154 is a member of the TNF superfamily of ligands, including CD178 (Fas ligand) (17). Like CD154, CD178 is also regulated by NFAT binding in the proximal promoter region (18-20). There has also been considerable evidence that CD178 transcription is increased by Egr transcription factors (21). However, the role of the individual Egr family members in CD178 expression remains controversial, with evidence for Egr-1, -2, and -3 each having critical roles in transcription (20, 22-26). In examining the role of Egr factors in CD154 transcription using Jurkat thymoma extracts, Lindgren et al. (16) identified Egr-1, and possibly Egr-3, as proteins bound to a composite NFAT/Egr site in the proximal CD154 promoter. To define the requirements for induction of CD154 expression in primary human peripheral blood T cells, we have investigated the proteins that bind to the proximal 5′ regulatory region of the human CD154 (hCD154) gene. We find that a purine-rich motif near the documented proximal NFAT site associates with Egr-1 and Egr-3, but not Egr-2, in vitro and in vivo in primary CD4 T cells. Moreover, Egr-1 appears to be critical for CD154 transcriptional activity in primary CD4 T cells. Thus, Egr-1 represents a novel and important transcription factor involved in CD154 expression in activated primary CD4 T cells.
Materials and Methods
Construction of plasmids
Firefly luciferase reporter plasmids were prepared by ligating oligonucleotides, each containing two tandem repeats of either the NFAT/Egr (5′ −73, AGCACATTTTCCAGGAAGTGTGGGCTGCAACG-3′) or the AP-1/NFAT (5′ −88, GAGAGAAGACTACGAAGCACATTTTCCAGGAA-3′) sequences from the hCD154 proximal promoter, between the NheI and XhoI multiple cloning sites of the pGL3-promoter luciferase reporter vector (Promega), using T4 DNA ligase. DH5-α-competent clones were isolated, and multiple independent large-scale preparations of the plasmids were performed with Qiagen maxi plasmid isolation kits. Construct identities were confirmed by DNA sequencing of both strands. The pJDM-948 (Egr-1), pJDM-1118 (Egr-2), pJDM-1412 (Egr-3), and pCMVneo (control) expression plasmids were generously provided by Dr. J. Milbrandt (Washington University, St. Louis, MO). The NFAT1 expression plasmid and pREP4 control expression vector were previously described (27).
CD4 T cell isolation and activation
Primary human CD4 T cells were isolated from peripheral blood of healthy donors by negative selection according to the manufacturer's protocol (RosetteSep; StemCell Technologies) as previously described (28). Flow cytometry revealed 90–97% purity of CD4 T cell populations (data not shown). Cells were placed in culture medium containing RPMI 1640 for 2–6 h at 37°C in the presence of phorbol ester (50 ng/ml phorbol dibutyrate (see Fig. 1 only) or 25 ng/ml PMA) and calcium ionophore (1.5 μM ionomycin) (Sigma-Aldrich). Institutional review board approval was obtained for these studies.
FIGURE 1.
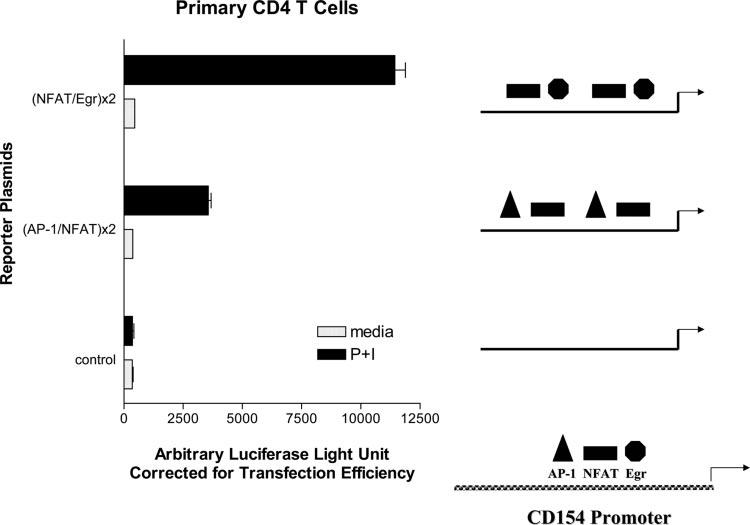
NFAT and Egr cooperate to augment CD154 transcription. Primary human CD4 T cells were transiently transfected (see Materials and Methods) with firefly luciferase reporter plasmids directed by duplications of the hCD154 proximal promoter AP-1/NFAT sites or NFAT/Egr sites (see Materials and Methods for construct design). Cells were then rested (media) or stimulated with phorbol ester and ionomycin (P+I) for 6 h, and cell lysates were analyzed for luciferase activity. Firefly luciferase activity was corrected for transfection efficiency with Renilla luciferase activity from a cotransfected control plasmid. Results are representative of one of three similar experiments. Diagrams depicting the relative positions of the neighboring AP-1, NFAT, and Egr binding sites in the proximal hCD154 promoter, and the combination of transcription factor binding sites in the reporter constructs, are shown at the right.
Luciferase reporter assay
Primary human CD4 T cells were cultured with 1 μg/ml PHA and irradiated syngeneic whole mononuclear cells for 19.5 h before transfection as described (29). Five million live cells were transiently transfected with 5 μg of firefly luciferase reporter plasmid along with 1–3 μg of Renilla luciferase transfection efficiency control plasmids, pRL-TK or pRL-null (Promega), by electroporation (Bio-Rad) at 250 V and 960 μF. After 2 h of rest, the cells were plated at 1 × 106 cells/96-well microtiter well with medium alone, or with phorbol ester and ionomycin, and incubated at 37°C for 6 h. Cells were then lysed, and lysates were analyzed in duplicate by luminometry (Lumat 9507; Berthold Technologies) using the Dual-Lucif-erase Reporter Assay System (Promega). For Fig. 4B, 5 million primary human CD4 T cells were transfected by Amaxa electroporation, as previously described (8), with 1–2 μg of control, Egr-1, or NFAT1 expression plasmids, either alone or together. The cells were activated for 6 h with phorbol ester and ionomycin, and cell lysates were analyzed by luminometry as above.
FIGURE 4.
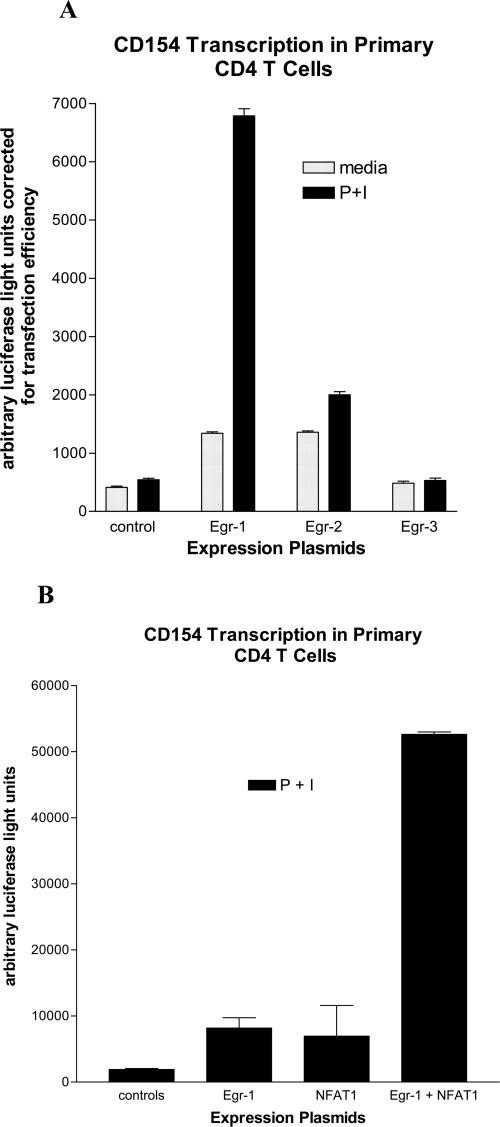
Egr-1 augments CD154 transcription. Primary human CD4 T cells were transiently transfected with a firefly luciferase reporter plasmid driven by the proximal hCD154 promoter (10). A, The cells were cotransfected with expression plasmids for Egr-1, Egr-2, Egr-3, or control (empty vector). After a brief rest, the cells were activated for 6 h with medium alone or phorbol ester and ionomycin (P+I). Cell lysates were analyzed in duplicate for luciferase activity and corrected for transfection efficiency with a cotransfected Renilla luciferase control plasmid. Results are representative of one of five similar experiments. B, Primary CD4 T cells were transfected with expression plasmids for Egr-1, NFAT1, Egr-1+NFAT1, or controls (empty vectors). The cells were activated for 6 h with P+I, and cell lysates were analyzed for luciferase activity. Results are representative of one of four similar experiments.
EMSA
Nuclear extracts were prepared from Jurkat or primary human CD4 T cells, at rest or following stimulation for 2 h with PMA and ionomycin, by the method of Schreiber et al. (30) as previously described (27). Protein concentration was measured by the Bradford assay. Protein (5 μg) was incubated for 20 min at room temperature with 100,000 cpm of 32P-labeled double-stranded oligonucleotide probe. The NFAT/Egr probe, −73 to −41 with respect to the transcription start site, 5′-AGCACATTTTCCAGGAAGTGTGGGCTGCAACGA-3′, corresponds to the hCD154 promoter proximal NFAT site (underlined) and downstream (3′) purine-rich sequence, which is conserved in the murine promoter (12). The samples were resolved on 6% precast polyacrylamide gels according to the manufacturer's (Bio-Rad) instructions as previously described (31). In some experiments, the nuclear extracts were preincubated for 30 min on ice with 1 μl of Ab to STAT1 (control), Egr-1, -2, or -3 (Santa Cruz Biotechnology), or NFAT1 (BD Transduction Laboratories). Specificity of complexes was also determined by preincubating extracts for 30 min at room temperature with 20-fold excess of cold (unlabeled) self-oligonucleotide competitor. After electrophoresis, gels were dried and bands were analyzed by standard autoradiography.
Real-time PCR-based chromatin immunoprecipitation (ChIP) assay
ChIP assays used reagents from Upstate Biotechnology with modifications from the manufacturer's instructions. Briefly, 5 million CD4 T cells per immunoprecipitation sample were isolated from whole blood (rosette gradient separation) and were stimulated for 2 h at 37°C with PMA (25 ng/ml) and ionomycin (1.5 μM). Cells were cross-linked with 1% formaldehyde for 10 min, stopped with 0.125 M glycine, washed several times with PBS containing protease inhibitor mixture (catalogue no. 8340; Sigma-Aldrich), pelleted, and resuspended in lysis buffer for 30 min with 30 s vortexing every 10 min. Each lysate was diluted to 1 ml using Tris/EDTA and sonicated on a Branson 250 sonifier, at a setting of 3, four times for 15 s each with 2 min rest on ice between sonications. Sonicates were precleared with protein A (or protein G for NFAT1 IgG1)-Sepharose beads (prepared with BSA and salmon sperm DNA to inhibit inadvertent binding to beads) and incubated in dilution buffer overnight at 4°C with 10 μl of Egr-1 (C-19), Egr-2 (C-14), Egr-3 (C-24), or rabbit IgG isotype control, polyclonal Abs (Santa Cruz Biotechnology), or NFAT1 mAb (702610; BD Biosciences) and its mouse IgG1 isotype control. Samples were immunoprecipitated with protein A- or protein G-Sepharose for 1 h. The beads were washed and eluted with 1% SDS in 0.1 M sodium bicarbonate, and supernatants were reverse cross-linked by addition of NaCl and heat for 6 h at 65°C. Immunoprecipitated samples were digested with proteinase K for 1 h at 45°C, and DNA was extracted with phenol/chloroform/isoamyl alcohol. The DNA was then treated with RNase for 30 min at 37°C, and equal volumes of immunoprecipitated DNA were analyzed by TaqMan real-time PCR (Applied Biosystems) using primers and probe specific for the proximal hCD154 promoter (forward 5′-TTTGCTGGGAGAGAAGACTACGA-3′, reverse 5′-TGGCCACCTTACTCAGGATTAGTTA-3′, TaqMan probe 5′-fam-TCCAGGAAGTGTGGGCTGCAACG-bhq-3′). Fold differences (between specific Ab and isotype control immunoprecipitations) were calculated using the formula: 2−ΔCt, where ΔCt = Ctsample − Ctcontrol.
Small interfering RNA (siRNA)
DNA vectors to be used for transient transfection of siRNA sequences were designed essentially as described (32). In short, human genomic DNA was isolated from CEM-SS cells (33) using a DNeasy tissue kit (Qiagen). The human U6 promoter sequence was amplified using genomic DNA as a template. The primers used in the PCR flanked the U6 sequence (5′ U6 universal primer, 5′-CGGAATTCCCCCAGTGGAAAGACGCGCAG-3′; 3′ primer, 5′-CGGTTTTCGTCCTTTCCACAAG-3′). PCR primers were designed based on human small nuclear RNA gene sequence (GenBank accession no. M14486). PCRs were conducted as follows: 30 s at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, then extended at 72°C for 5 min. PCR products were cloned into a TA cloning vector (Invitrogen Life Technologies), and the U6 sequence was confirmed by sequencing in the nucleic acid core facility at the Children's Hospital of Philadelphia.
The selection of siRNA sequences was based on web-based programs (〈www.dharmacon.com〉 and 〈www.ambion.com/techlib/misc/siRNA_finder.html〉) and our own institution's software. Every sequence was analyzed by basic local alignment search tool searching (〈www.ncbi.nlm.nih.gov/BLAST〉) to ensure that there were no significant sequence homologies with other genes. Four unique siRNA sequences specific for Egr-1 (accession number NM_001964) were selected for testing. Scrambled siRNA sequences for each corresponding siRNA sequence were designed using our institutional software, and these sequences were also screened by basic local alignment search tool analysis. These siRNA sequences and their counterparts are listed below. Oligonucleotide primers were then designed for each individual siRNA sequence into one large primer possessing a HindIII digestion site, a termination site for RNA polymerase III, the specific sense siRNA sequence, a 9-bp spacer, the anti-sense siRNA sequence, and the 3′ U6 primer sequence. PCRs were the same as in the human U6 promoter cloning above. PCR products were cloned into pCR2.1-TOPO cloning vectors (TOPO TA Cloning Kit; Invitrogen Life Technologies) and confirmed using enzyme digestion and gel electrophoresis.
The numbers correspond to the sequence positions based on the accession number above: siRNA 590 for Egr-1, 5′-ACATCTCTCTGAACAACGA-3′; random 590 for Egr-1, 5′-CTCTAGCGAAACCAATCAT 3′; siRNA 746 for Egr-1, 5′-GTGGCCTAGTGAGCATGAC-3′; random control 746 for Egr-1, 5′-GATCAGGCGACGGCTTGAT-3′; siRNA 900 for Egr-1, 5′-CACGCCGAACACTGACATT-3′; random 900 for Egr-1, 5′-CCATTACAACCGTCGAACG-3′; siRNA 1517 for Egr-1, 5′-AGATCCACTTGCGGCAGAA-3′; and random control 1517 for Egr-1, 5′-AGTAAACATGCGCGCAGTC-3′.
Primary human CD4 T cells were transfected with increasing concentrations (2.5, 5, and 7.5 μg) of the Egr-1-specific siRNA expression plasmids or the associated scrambled random control vectors by Amaxa electroporation as described (8). The cells were rested overnight at 37°C and stimulated for 2 h with PMA and ionomycin as described above. Expression of CD154 on the cell surface was detected by flow cytometry as previously detailed (34). Egr-1 mRNA was evaluated along with GAPDH as a control gene by real-time RT-PCR as previously described (14). The Egr-1 primers used were 5′-GGCGAGCAGCCCTACGAG-3′ on the coding strand and 5′-GGTCTCCACCAGCACCTTCTC-3′ on the noncoding strand. The annealing temperature used during PCR was 60°C, and products were detected by SYBR green incorporation.
Northern blot analysis
Spleen cells were isolated from Egr-1+/+ and Egr-1−/− mice (35) that were both transgenic for the 3A9 TCR (36). 3A9 is an MHC class II-restricted TCR that recognizes a hen egg lysozyme (HEL) peptide presented by I-Ak. For freshly isolated T cells, CD4 T cells were purified from single cell suspensions of RBC-lysed splenocytes using magnetic beads (Dynal Biotech) as previously described (37). For T cell stimulation, whole splenocytes were placed in culture with the HEL peptide at a 13 mM concentration. After incubation for 5 h at 37°C, CD4 cells were isolated using magnetic beads and RNA prepared using TRIzol (Invitrogen Life Technologies) (27). Samples were run on a gel, and the Northern blot was probed with a 596-bp, 32P-labeled murine CD154 cDNA and analyzed by autoradiography as described (27). The blot was then stripped and probed for the housekeeping gene GAPDH. Institutional approval for the animal studies was obtained.
Results
Cooperative transcriptional activation of an NFAT/Egr site in the proximal CD154 promoter
Comparing the nucleotide sequences of the human and murine CD154 proximal promoters (38, 39), a 37-bp region of identity was noted, including and flanking the most proximal NFAT binding site (10, 12). Because many NFAT sites cooperate with neighboring transcription factors, this was originally proposed to be an NFAT-AP-1 site using murine EL-4 thymoma extracts (12). More recently, this region was studied using human Jurkat thymoma cells, and NFAT was found to interact with Egr proteins and not AP-1 in response to CD28 costimulation (16). To explore this in primary human CD4 T cells, luciferase reporter genes were generated, which were driven by duplications of the proposed AP-1/NFAT site (12) or the NFAT/Egr site (16) (Fig. 1). Primary human CD4 T cells were transiently transfected with the AP-1/NFAT, NFAT/Egr, or empty control reporter plasmids and were subsequently polyclonally activated in vitro for 6 h with phorbol ester and calcium ionophore. Both the AP-1/NFAT and the NFAT/Egr reporter plasmids responded to polyclonal activation, but the NFAT/Egr construct consistently responded 2- to 4-fold better than the NFAT/AP-1 plasmid (Fig. 1). Thus, an NFAT/Egr interaction was explored.
NFAT1, Egr-1, and Egr-3 bind to the proximal CD154 promoter
Nuclear extracts were generated from resting or polyclonally activated primary human CD4 T cells or Jurkat T cells and were reacted with a radiolabeled oligonucleotide probe corresponding to the CD154 promoter proximal NFAT site and 3′ flanking sequence. Using Jurkat T cell extracts, which are known to express Egr proteins following activation (40), two difficult-to-discern gel-retarded complexes were identified by the mobility shift assay from activated but not resting T cell extracts (Fig. 2A, lanes 1 and 2). Abs to NFAT1 (lane 3) and Egr-1 (lane 4), and to a variable degree Egr-3 (lane 6), inhibited formation of these complexes, whereas Abs to Egr-2 (lane 5) and STAT1 (lane 7) as negative controls had no effect. Similar results were obtained using activated primary human CD4 T cell nuclear extracts (Fig. 2, B and C). The two complexes were more easily discernible, particularly when electrophoresed for a longer period of time (Fig. 2C), and they were specific in that they were partially or completely competed away by 20-fold (Fig. 2C, lane 2) and 200-fold (Fig. 2B, lane 3) excess of unlabeled (cold) self-probe, respectively. Competition with cold consensus Egr or NFAT oligonucleotides partially inhibited both complexes, and anti-NFAT1 Ab occasionally led to a faint supershift (data not shown), suggesting that NFAT and Egr proteins bound to the proximal CD154 promoter. To characterize the Egr proteins bound to this probe, nuclear extracts were first incubated with specific anti-Egr Abs before incubation with the oligonucleotide probe. Ab directed to Egr-1 consistently blocked formation of the upper band (Fig. 2C, lane 3), whereas Ab to Egr-2 had no effect (Fig. 2C, lane 4). In some experiments, Ab to Egr-3 partially blocked formation of the upper complex (data not shown), but, in most gel shifts, there was no effect of anti-Egr-3 Ab (Fig. 2C, lane 5). In nuclear extracts from activated primary CD4 T cells, but not Jurkat T cells, the lower complex was preferentially inhibited by anti-NFAT1 Ab (Fig. 2B, lane 4), but using a probe including sequence 5′ (upstream) of the NFAT site, Abs to AP-1 components, c-fos and c-Jun, yielded no consistent evidence for AP-1 binding in conjunction with NFAT to the proximal CD154 promoter (data not shown). All of these in vitro binding results are entirely consistent with the findings of Lindgren et al. (16) and suggest that Egr-1, and Egr-3 variably, complexes with NFAT on the proximal CD154 promoter, but AP-1 from human T cell extracts does not. Moreover, NFAT1 and Egr-1 may form a complex depicted by the upper band on the EMSA.
FIGURE 2.
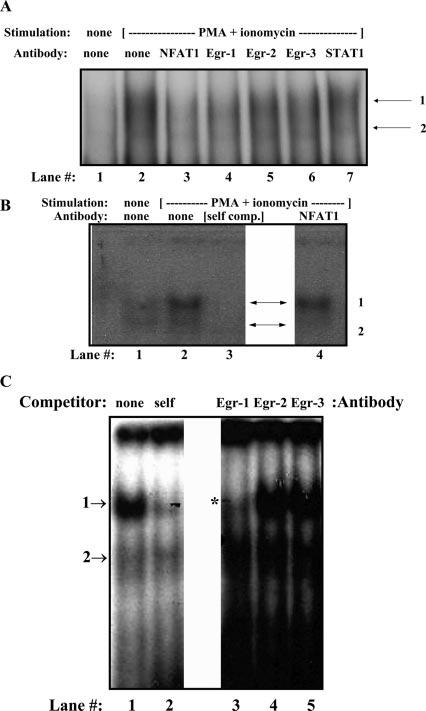
Egr-1 binds the proximal hCD154 promoter in vitro. Nuclear extracts from resting (A, lane 1) or polyclonally activated Jurkat (A) and primary human CD4 T cells (B and C) were incubated with an oligo-nucleotide probe encompassing the hCD154 proximal promoter NFAT and Egr sites. This resulted in the formation of two complexes (arrows) in activated extracts as seen in A (lane 2), B (lane 2), and C (lane 1). Both complexes were partially inhibited by excess self, cold probe (B, lane 3; C, lane 2). The upper complex (C, *) was specifically inhibited by an anti-Egr-1 Ab (A, lane 4; C, lane 3) but not by Abs to Egr-2 (A, lane 5; C, lane 4), Egr-3 (A, lane 6; C, lane 5), or STAT1 (A, lane 7). Anti-NFAT1 also inhibited upper complex formation in Jurkat extracts (A, lane 2) but primarily blocked lower complex formation in primary CD4 T cell extracts (B, lane 4).
To explore NFAT and Egr binding to the proximal hCD154 promoter in vivo, ChIP assays were conducted. Primary human CD4 T cells were polyclonally activated for 2 h, DNA-bound protein was cross-linked, and the cells were then lysed. Chromatin was extracted and sonicated to a range of 200-1000 bp in length. Transcription factors bound to DNA were immunoprecipitated with specific Abs, or isotype controls, directed against NFAT1, Egr-1, Egr-2, and Egr-3. Immunoprecipitated chromatin was amplified and quantified by real-time PCR using primers and a TaqMan probe specific to the proximal hCD154 promoter. Abs to NFAT1, Egr-1, and Egr-3, but not to Egr-2, repeatedly demonstrated significantly fewer cycles to reach threshold levels (CT) compared with isotype controls (Fig. 3A). Averaging results from five experiments (Fig. 3B) clearly established NFAT1, Egr-1, and Egr-3 binding to the proximal hCD154 in vivo in primary human CD4 T cells following stimulation capable of inducing CD154 transcription (10). Therefore, similar to the in vitro binding studies (Fig. 2 and Ref. 16), NFAT1, Egr-1, and Egr-3 are capable of binding the proximal hCD154 promoter in vivo in activated primary human CD4 T cells.
FIGURE 3.
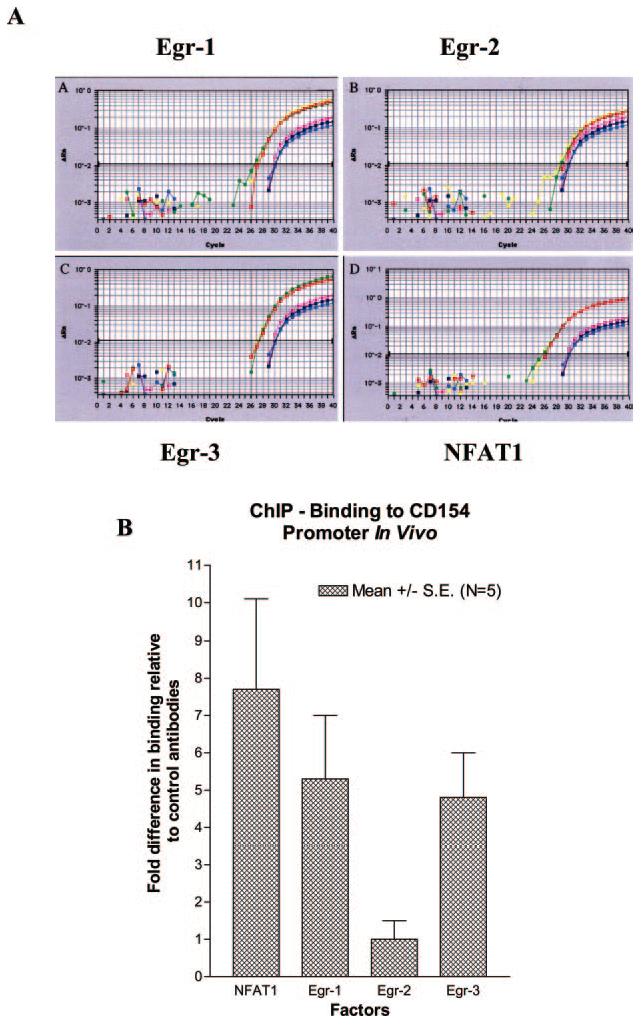
Egr-1 and NFAT1 bind the proximal hCD154 promoter in vivo. Primary human CD4 T cells were activated for 2 h with phorbol ester and ionomycin and underwent chromatin immunoprecipitation with iso-type control and anti-Egr-1, -Egr-2, -Egr-3, and -NFAT1 Abs as described in Materials and Methods. A, Immunoprecipitated DNA was amplified and quantified by real-time PCR using primers and a probe specific to the proximal hCD154 promoter (see Materials and Methods). Samples were analyzed in triplicate for the test (red, green, and yellow curves) and control (pink, blue, and purple curves) Abs. B, Fold differences in binding relative to control Abs were calculated (see Materials and Methods), and averages and SEs of the means from five independent experiments are depicted for each test transcription factor analyzed.
Egr-1 is capable of augmenting CD154 transcriptional reporter gene activity
We previously demonstrated the requirement of NFAT binding to the proximal NFAT binding site in the hCD154 promoter for CD154 transcription (10). To evaluate the ability of the various Egr proteins to transactivate the CD154 promoter, expression vectors coding for the individual Egr family members were cotransfected along with a hCD154 promoter-driven reporter plasmid (10) into primary human CD4 T cells. Cells were subsequently rested or polyclonally activated for 6 h in vitro. Compared with the empty vector control, the Egr-1 and Egr-2, but not Egr-3, expression plasmids modestly directed transcription of the hCD154 proximal promoter (Fig. 4A). Upon T cell stimulation, Egr-1 markedly increased CD154 transcriptional reporter gene activity, and this was dose-dependent (data not shown). Egr-2 only modestly increased CD154 transcriptional reporter gene activity, whereas Egr-3 had no effect (Fig. 4A). Similarly, overexpression of Egr-1 notably augmented transcription of the NFAT/Egr plasmid studied in Fig. 1 (data not shown). Although both Egr-1 and Egr-3 are capable of binding the proximal hCD154 promoter, only Egr-1 is able to transactivate CD154 transcription in primary human CD4 T cells. In addition, coexpression of Egr-1 and NFAT1 augmented CD154 transcription in a synergistic fashion (Fig. 4B).
Egr-1 is required for CD154 expression
Overexpression of Egr-1 clearly augmented CD154 transcription (Fig. 4), but this approach does not address the role of endogenous Egr-1 in CD154 expression. To study the role of endogenous Egr-1 in CD154 expression, siRNAs specific to Egr-1 were created. Using software design, four pairs of siRNA specific to Egr-1 and corresponding scrambled sequence control expression cassettes were generated (see Materials and Methods). Primary human CD4 T cells were transfected with increasing concentrations of the siRNA or matched scrambled control expression cassettes and rested overnight. The next morning, the cells were polyclonally stimulated for 2 h and analyzed for cell surface expression of CD154 by flow cytometry. Two (nos. 590 and 1517) of the four siRNA sequences were capable of inhibiting CD154 expression relative to their matched scrambled sequence controls.
One of six representative experiments performed using siRNA 590 and its matched scrambled control is depicted in Fig. 5A. At the lowest concentrations of siRNA expression cassettes, there was minimal (∼4%) inhibition of CD154 expression (data not shown) relative to the scrambled control. However, at the highest concentration tested, CD154 expression, as measured by mean fluorescence intensity, in the presence of the Egr-1-specific siRNA was about half (54%; for all six experiments, 66.5% ± 10.1 SD) that of the control expression plasmid (Fig. 5A). Similarly, Egr-1 mRNA levels, as detected by real-time RT-PCR, were reduced by ∼50% (Fig. 5B). Although these results were not dramatic, they were reproducible. However, the approach was limited by nonspecific inhibition of CD154 expression by the process of transfection, as well as by cell toxicity at doses of expression cassettes > 7.5 μg/transfection (data not shown). Therefore, an alternative genetic approach to studying CD154 expression in primary CD4 T cells in the absence of Egr-1 was required.
FIGURE 5.
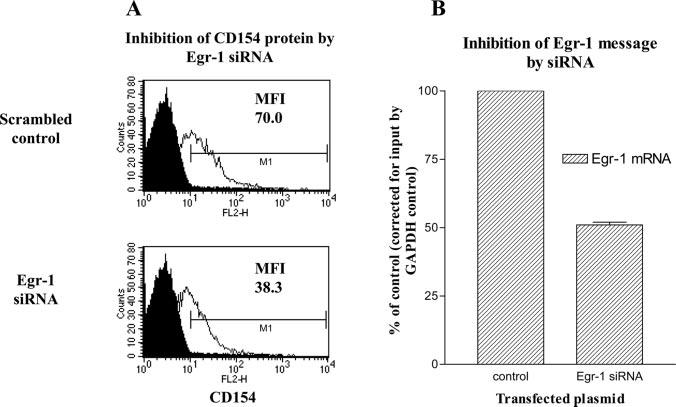
Inhibition of endogenous Egr-1 lowers CD154 surface expression. Primary human CD4 T cells were transiently transfected with increasing amounts of expression plasmids for Egr-1 siRNA (no. 590; see Materials and Methods) or scrambled sequence control. Cells were then activated for 2 h with phorbol ester and ionomycin and analyzed for CD154 surface expression (A) by flow cytometry or Egr-1 mRNA (B) by real-time RT-PCR. Results are representative of one of six similar experiments. The black filled area under the curve represents isotype control Ab staining. The unfilled curves depict CD154 Ab staining in the presence of 7.5 μg of Egr-1 siRNA (bottom) or scrambled control (top) expression plasmids. The mean fluorescence intensities (MFI) of CD154 expression are noted.
Because the proximal NFAT/Egr binding sequences are identical between mouse and human, the role of Egr-1 in CD154 expression was analyzed using Egr-1-deficient mice. The 3A9 TCR transgene, which recognizes a HEL peptide in the context of MHC class II, I-Ak (41), was bred onto the Egr-1 knockout (−/−) (35) and wild-type (WT), otherwise genetically identical, backgrounds. CD4+ splenocytes were purified and used to isolate RNA directly, or whole splenocytes were stimulated in culture with HEL peptide for 5 h, followed by purification of CD4 T cells and RNA isolation. CD154 expression was analyzed at the mRNA level because re-agents for evaluating cell surface expression of murine CD154 are suboptimal. The transcripts were analyzed at 5 h following stimulation because CD154 message in CD4 T cells is optimal at this time point following TCR stimulation (compared with 2 h for phorbol ester and ionomycin activation) (34). In the absence of the appropriate HEL peptide stimulation, there was a minimal level of background CD154 expression in both WT and Egr-1−/− CD4 T cells (Fig. 6, left lanes). However, upon stimulation with HEL peptide there was strong induction of CD154 message relative to the GAPDH control in WT Egr-1, but not in Egr-1−/−, CD4 T cells (Fig. 6, right lanes). These results nicely confirm the requirement of Egr-1 for CD154 induction in primary CD4 T cells. This is not likely to be the result of a general defect in Egr-1−/− T cells, because these cells proliferate equally well, compared with WT cells following T cell stimulation (42), and CD69 induction is also intact (data not shown).
FIGURE 6.
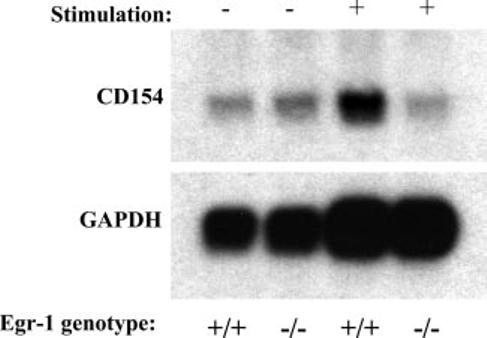
Egr-1 is required for CD154 message induction. T cell transgenic primary murine CD4 T cells from Egr-1 WT (+/+) or knockout (−/−) mice were rested (−) or stimulated (+) in vitro for 5 h with the appropriate peptide/MHC combination (see Materials and Methods). mRNA was isolated and analyzed by Northern blot with a probe specific to murine CD154 cDNA. The blot was then stripped and probed for the GAPDH housekeeping gene as a loading control. Results are representative of one of two similar experiments.
Discussion
CD154 expression by CD4 T cells is critical for a wide variety of immunologic responses and, thus, its expression is tightly regulated (8). Even subtle degrees of CD154 dysregulation can contribute to disease pathology in autoimmune disorders, such as systemic lupus erythematosus (6). To understand CD154 dysregulation, it is important to first be familiar with its regulation under normal circumstances. Surprisingly, for such a critically important and closely regulated molecule, there has been a relative paucity of studies analyzing CD154 regulation (8). In CD4 T cells, CD154 expression is activation dependent and largely regulated at the level of transcription (10, 12, 14, 16, 34, 43), although control of mRNA stability is clearly important for the duration of CD154 expression (9, 44).
In studying CD154 transcription in CD4 T cells, it has been clearly shown that NFAT proteins are critical for activation-dependent expression and responsible for the cyclosporin-sensitive nature of induction (10, 12). This is not surprising because at least three functional NFAT binding sites have been identified in the hCD154 promoter (10, 11). Multiple NFAT family members are present in primary human CD4 T cells. However, the bulk of NFAT proteins are made up of NFAT1 and NFAT2, which is inducible (45, 46), with a small amount of NFAT4 present (27, 47, 48). NFAT1 has obviously been shown to bind to the hCD154 promoter (10, 11) but, interestingly, NFAT4 appears to inhibit CD154 expression (49). This is similar to the inability of NFAT4 to augment IL-4 transcription in primary CD4 T cells (27). Thus, NFAT1, and to some degree NFAT2, appear to be the critical NFAT family members required for CD154 expression in CD4 T cells. Similarly, NFAT1 seems to be required for optimal CD154 expression in NFAT1-deficient mice (50). In this study, we have shown for the first time the binding of NFAT1 to the CD154 promoter in vivo following activation of primary CD4 T cells (Fig. 3).
Similar to the NFAT family of transcription factors, multiple Egr proteins are used in T cells. In particular, there are a variety of target genes regulated by Egr-1 in T cells. These include, but are not restricted to, IL-2 (51, 52), IL-2Rβ-chain (53), and TNF-α (54, 55). In the current study, we show that Egr-1 and Egr-3, but not Egr-2, are capable of binding the hCD154 promoter in vivo upon activation of primary human CD4 T cells (Fig. 3). This is similar to what we (Fig. 2) and others have seen in vitro (16). This is also reminiscent of what others have noted for another TNF superfamily protein, Fas ligand (CD178), promoter. Li-Weber et al. (24) noted that Egr-1 and Egr-3 present in human Jurkat thymoma extracts bound with NFAT to a similar combinatorial site in the hCD178 promoter. However, there has been some controversy as to the precise Egr family members involved in the regulation of the murine CD178 promoter. In one study, Egr-1, Egr-2, and Egr-3 all bound the murine CD178 promoter in Th2 cells, but only Egr-1 and Egr-2 from Th1 extracts bound the promoter because Egr-3 was lacking in these cells (20). By contrast, Egr-2 and Egr-3 were shown to be potent activators of CD178 expression, and Egr-3 expression was enriched in Th1 cells (22). Because Egr-3 expression may vary depending on the presence or absence of Th2 cells, this may explain the variable results seen in binding of Egr-3 to the hCD154 promoter in vitro using extracts derived from different donors of human peripheral blood CD4 T cells (Fig. 2). Nevertheless, only Egr-1, and not Egr-3, seems capable of augmenting CD154 transcription (Fig. 4A). Moreover, Egr-1 cooperates with NFAT1 to augment CD154 promoter activity (Fig. 4B) and may even form a complex on the proximal promoter (Fig. 2).
Not only is Egr-1 capable of binding the CD154 promoter, in vitro and in vivo, and augmenting transcription, its expression is required for optimal activation-dependent expression of CD154 in human (Fig. 5) and murine (Fig. 6) CD4 T cells. The importance of the Egr site in the proximal hCD154 promoter was suggested by earlier mutational studies (16), but why exogenous Egr-3 does not seem to augment CD154 transcription remains unclear. It is not surprising, however, that Egr-1 plays such a critical role because Egr-1 has been known to be an important factor in the regulation of lymphocyte-mediated immune responses for quite some time (56).
Like NFAT2, Egr-1 can be up-regulated in an autocrine fashion (57). However, Egr-1, unlike NFAT2, Egr-2, and Egr-3, does not appear cyclosporin-sensitive nor does it appear to be regulated by NFAT (22). In terms of CD154 regulation, Egr-1 may be acting as the major signaling mediator of CD28 costimulation (16). This is consistent with recent gene array findings showing that Egr-1 is one of the most highly up-regulated genes in response to CD28 activation (58). It will be of interest to explore Egr-1 levels in patients with autoimmune disorders associated with increased CD154 expression. This may allow for selective targeting of expression in these disorders.
Acknowledgments
We thank Dr. Terri Laufer (University of Pennsylvania, Philadelphia, PA) for critical review of the manuscript. We also thank Dr. Jeffrey Milbrandt (Washington University, St. Louis, MO) for the generous gift of the Egr expression plasmids.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This work was supported, in part, by grants from the Arthritis Foundation (to R.Q.C.), the Arthritis National Research Foundation (to R.Q.C.), the Dorough Lupus Foundation (to R.Q.C.), the Kahn Foundation for Lupus Research (to R.Q.C.), the American College of Rheumatology Research and Education Foundation (to R.Q.C.), the Joseph L. Hollander Chair of Pediatric Rheumatology (to T.H.F.), and the National Institutes of Health (Grants R01-AR48257, R21-AR49335, P30-HH2815, M01-RR240 (to R.Q.C.); T32-AI007621 (to R.B.); R01-AI35513, R21-AI054233 (to T.H.F.); and R01-AI42185 (to M.K.C.)).
Abbreviations used in this paper: Egr, early growth response; hCD154, human CD154; ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; HEL, hen egg lysozyme.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, van Kooten C, Liu YJ, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu. Rev. Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 2.van Kooten C, Banchereau J. CD40-CD40 ligand. J. Leukocyte Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 3.Grewal IS, Flavell RA. The CD40 ligand: at the center of the immune universe? Immunol. Res. 1997;16:59–70. doi: 10.1007/BF02786323. [DOI] [PubMed] [Google Scholar]
- 4.Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu. Rev. Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 5.Cron RQ. CD154 and lupus. Pediatr. Rheumatol. Online J. 2003;1:172–181. [Google Scholar]
- 6.Crow MK, Kirou KA. Regulation of CD40 ligand expression in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2001;13:361–369. doi: 10.1097/00002281-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Datta SK, Kalled SL. CD40-CD40 ligand interaction in autoimmune disease. Arthritis Rheum. 1997;40:1735–1745. doi: 10.1002/art.1780401002. [DOI] [PubMed] [Google Scholar]
- 8.Cron RQ. CD154 transcriptional regulation in primary human CD4 T cells. Immunol. Res. 2003;27:185–202. doi: 10.1385/IR:27:2-3:185. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol. Cell. Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert LA, King G, Cron RQ, Lewis DB, Aruffo A, Hollenbaugh D. The human gp39 promoter: two distinct nuclear factors of activated T cell protein-binding elements contribute independently to transcriptional activation. J. Biol. Chem. 1995;270:29624–29627. doi: 10.1074/jbc.270.50.29624. [DOI] [PubMed] [Google Scholar]
- 11.Lobo FM, Xu S, Lee C, Fuleihan RL. Transcriptional activity of the distal CD40 ligand promoter. Biochem. Biophys. Res. Commun. 2000;279:245–250. doi: 10.1006/bbrc.2000.3914. [DOI] [PubMed] [Google Scholar]
- 12.Tsytsykova AV, Tsitsikov EN, Geha RS. The CD40L promoter contains nuclear factor of activated T cells-binding motifs which require AP-1 binding for activation of transcription. J. Biol. Chem. 1996;271:3763–3770. doi: 10.1074/jbc.271.7.3763. [DOI] [PubMed] [Google Scholar]
- 13.Srahna M, Remacle JE, Annamalai K, Pype S, Huylebroeck D, Boogaerts MA, Vandenberghe P. NF-κB is involved in the regulation of CD154 (CD40 ligand) expression in primary human T cells. Clin. Exp. Immunol. 2001;125:229–236. doi: 10.1046/j.1365-2249.2001.01601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert LA, Cron RQ, Cleary AM, Brunner M, Song A, Lu L-S, Jullien P, Krensky AM, Lewis DB. A T cell-specific enhancer of the human CD40 ligand gene. J. Biol. Chem. 2002;277:7386–7395. doi: 10.1074/jbc.M110350200. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqa A, Sims-Mourtada JC, Guzman-Rojas L, Rangel R, Guret C, Madrid-Marina V, Sun Y, Martinez-Valdez H. Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature. 2001;410:383–387. doi: 10.1038/35066602. [DOI] [PubMed] [Google Scholar]
- 16.Lindgren H, Axcrona K, Leanderson T. Regulation of transcriptional activity of the murine CD40 ligand promoter in response to signals through TCR and the costimulatory molecules CD28 and CD2. J. Immunol. 2001;166:4578–4585. doi: 10.4049/jimmunol.166.7.4578. [DOI] [PubMed] [Google Scholar]
- 17.Van Kooten C, Banchereau J. CD40-CD40 ligand: a multifunctional receptor-ligand pair. Adv. Immunol. 1996;61:1–77. doi: 10.1016/s0065-2776(08)60865-2. [DOI] [PubMed] [Google Scholar]
- 18.Li-Weber M, Laur O, Hekele A, Coy J, Walczak H, Krammer PH. A regulatory element in the CD95 (APO-1/Fas) ligand promoter is essential for responsiveness to TCR-mediated activation. Eur. J. Immunol. 1998;28:2373–2383. doi: 10.1002/(SICI)1521-4141(199808)28:08<2373::AID-IMMU2373>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Latinis KM, Norian LA, Eliason SL, Koretzky GA. Two NFAT transcription factor binding sites participate in the regulation of CD95 (Fas) ligand expression in activated human T cells. J. Biol. Chem. 1997;272:31427–31434. doi: 10.1074/jbc.272.50.31427. [DOI] [PubMed] [Google Scholar]
- 20.Dzialo-Hatton R, Milbrandt J, Hockett RD, Jr., Weaver CT. Differential expression of Fas ligand in Th1 and Th2 cells is regulated by early growth response gene and NF-AT family members. J. Immunol. 2001;166:4534–4542. doi: 10.4049/jimmunol.166.7.4534. [DOI] [PubMed] [Google Scholar]
- 21.Li-Weber M, Krammer PH. Function and regulation of the CD95 (APO-1/Fas) ligand in the immune system. Semin. Immunol. 2003;15:145–157. doi: 10.1016/s1044-5323(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 22.Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, Glimcher LH. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 2000;12:293–300. doi: 10.1016/s1074-7613(00)80182-x. [DOI] [PubMed] [Google Scholar]
- 23.Yoo Y-G, Lee M-O. Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J. Biol. Chem. 2004;279:36242–36249. doi: 10.1074/jbc.M401290200. [DOI] [PubMed] [Google Scholar]
- 24.Li-Weber M, Laur O, Krammer PH. Novel Egr/NF-AT composite sites mediate activation of the CD95 (APO-1/Fas) ligand promoter in response to T cell stimulation. Eur. J. Immunol. 1999;29:3017–3027. doi: 10.1002/(SICI)1521-4141(199909)29:09<3017::AID-IMMU3017>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 25.Droin NM, Pinkoski MJ, Dejardin E, Green DR. Egr family members regulate nonlymphoid expression of Fas ligand, TRAIL, and tumor necrosis factor during immune responses. Mol. Cell. Biol. 2003;23:7638–7647. doi: 10.1128/MCB.23.21.7638-7647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgado M, Ganea D. Vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide inhibit expression of Fas ligand in activated T lymphocytes by regulating c-Myc, NF-κB, NF-AT, and early growth factors 2/3. J. Immunol. 2001;166:1028–1040. doi: 10.4049/jimmunol.166.2.1028. [DOI] [PubMed] [Google Scholar]
- 27.Cron RQ, Bort SJ, Wang Y, Brunvand MW, Lewis DB. T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells. J. Immunol. 1999;162:860–870. [PubMed] [Google Scholar]
- 28.Cron RQ. HIV-1, NFAT, and cyclosporin: immunosuppression for the immunosuppressed? DNA Cell Biol. 2001;20:761–767. doi: 10.1089/104454901753438570. [DOI] [PubMed] [Google Scholar]
- 29.Cron RQ, Schubert LA, Lewis DB, Hughes CC. Consistent transient transfection of DNA into non-transformed human and murine T-lymphocytes. J. Immunol. Methods. 1997;205:145–150. doi: 10.1016/s0022-1759(97)00065-3. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M, Genin A, Cron RQ. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology. 2004;321:323–331. doi: 10.1016/j.virol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Wu MT, Wu RH, Hung CF, Cheng TL, Tsai WH, Chang WT. Simple and efficient DNA vector-based RNAi systems in mammalian cells. Biochem. Biophys. Res. Commun. 2005;330:53–59. doi: 10.1016/j.bbrc.2005.02.129. [DOI] [PubMed] [Google Scholar]
- 33.Nara PL, Hatch WC, Dunlop NM, Robey WG, Arthur LO, Gonda MA, Fischinger PJ. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res. Hum. Retroviruses. 1987;3:283–302. doi: 10.1089/aid.1987.3.283. [DOI] [PubMed] [Google Scholar]
- 34.Jullien P, Cron RQ, Dabbagh K, Cleary A, Chen L, Tran P, Stepick-Biek P, Lewis DB. Decreased CD154 expression by neonatal CD4+ T cells is due to limitations in both proximal and distal events of T cell activation. Int. Immunol. 2003;15:1461–1472. doi: 10.1093/intimm/dxg145. [DOI] [PubMed] [Google Scholar]
- 35.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J. Biol. Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 36.Ho WY, Cooke MP, Goodnow CC, Davis MM. Resting and anergic B cells are defective in CD28-dependent costimulation of naive CD4+ T cells. J. Exp. Med. 1994;179:1539–1549. doi: 10.1084/jem.179.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi H, Kersh GJ. Induction of the early growth response gene 1 promoter by TCR agonists and partial agonists: ligand potency is related to sustained phosphorylation of extracellular signal-related kinase substrates. J. Immunol. 2003;170:315–324. doi: 10.4049/jimmunol.170.1.315. [DOI] [PubMed] [Google Scholar]
- 38.Tsitsikov EN, Ramesh N, Geha RS. Structure of the murine CD40 ligand gene. Mol. Immunol. 1994;31:895–900. doi: 10.1016/0161-5890(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 39.Hollenbaugh D, Grosmaire LS, Kullas CD, Chalupny NJ, Braesch-Andersen S, Noelle RJ, Stamenkovic I, Ledbetter JA, Aruffo A. The human T cell antigen gp39, a member of the TNF gene family, is a ligand for the CD40 receptor: expression of a soluble form of gp39 with B cell costimulatory activity. EMBO J. 1992;11:4313–4321. doi: 10.1002/j.1460-2075.1992.tb05530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skerka C, Decker EL, Zipfel PF. Coordinate expression and distinct DNA-binding characteristics of the four EGR-zinc finger proteins in Jurkat T lymphocytes. Immunobiology. 1997;198:179–191. doi: 10.1016/S0171-2985(97)80039-3. [DOI] [PubMed] [Google Scholar]
- 41.Ho AM, Jain J, Rao A, Hogan PG. Expression of the transcription factor NFATp in a neuronal cell line and in the murine nervous system. J. Biol. Chem. 1994;269:28181–28186. [PubMed] [Google Scholar]
- 42.Schnell FJ, Kersh GJ. Control of recent thymic emigrant survival by positive selection signals and early growth response gene 1. J. Immunol. 2005;175:2270–2277. doi: 10.4049/jimmunol.175.4.2270. [DOI] [PubMed] [Google Scholar]
- 43.Mishra N, Brown DR, Olorenshaw IM, Kammer GM. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-γ gene and protein expression in lupus T cells. Proc. Natl. Acad. Sci. USA. 2001;98:2628–2633. doi: 10.1073/pnas.051507098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 45.Chuvpilo S, Jankevics E, Tyrsin D, Akimzhanov A, Moroz D, Jha MK, Schulze-Luehrmann J, Santner-Nanan B, Feoktistova E, Konig T, et al. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity. 2002;16:881–895. doi: 10.1016/s1074-7613(02)00329-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhou B, Cron RQ, Wu B, Genin A, Wang Z, Liu S, Robson P, Baldwin HS. Regulation of the murine Nfatc1 gene by NFATc2. J. Biol. Chem. 2002;277:10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- 47.Timmerman LA, Healy JI, Ho SN, Chen L, Goodnow CC, Crabtree GR. Redundant expression but selective utilization of nuclear factor of activated T cells family members. J. Immunol. 1997;159:2735–2740. [PubMed] [Google Scholar]
- 48.Lyakh L, Ghosh P, Rice NR. Expression of NFAT-family proteins in normal human T cells. Mol. Cell. Biol. 1997;17:2475–2484. doi: 10.1128/mcb.17.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Amasaki Y, Kamogawa Y, Nagoya M, Arai N, Arai K, Miyatake S. Role of NFATx (NFAT4/NFATc3) in expression of immunoregulatory genes in murine peripheral CD4+ T cells. J. Immunol. 2003;170:3109–3117. doi: 10.4049/jimmunol.170.6.3109. [DOI] [PubMed] [Google Scholar]
- 50.Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- 51.Skerka C, Decker EL, Zipfel PF. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J. Biol. Chem. 1995;270:22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- 52.Decker EL, Skerka C, Zipfel PF. The early growth response protein (EGR-1) regulates interleukin-2 transcription by synergistic interaction with the nuclear factor of activated T cells. J. Biol. Chem. 1998;273:26923–26930. doi: 10.1074/jbc.273.41.26923. [DOI] [PubMed] [Google Scholar]
- 53.Lin JX, Leonard WJ. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor β-chain promoter through noncanonical Egr and Sp1 binding sites. Mol. Cell. Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decker EL, Nehmann N, Kampen E, Eibel H, Zipfel PF, Skerka C. Early growth response proteins (EGR) and nuclear factors of activated T cells (NFAT) form heterodimers and regulate proinflammatory cytokine gene expression. Nucleic Acids Res. 2003;31:911–921. doi: 10.1093/nar/gkg186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer B, Meichle A, Hensel G, Charnay P, Kronke M. Characterization of an Krox-24/Egr-1-responsive element in the human tumor necrosis factor promoter. Biochim. Biophys. Acta. 1994;1219:413–421. doi: 10.1016/0167-4781(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 56.McMahon SB, Monroe JG. The role of early growth response gene 1 (Egr-1) in regulation of the immune response. J. Leukocyte Biol. 1996;60:159–166. doi: 10.1002/jlb.60.2.159. [DOI] [PubMed] [Google Scholar]
- 57.Schwachtgen JL, Campbell CJ, Braddock M. Full promoter sequence of human early growth response factor-1 (Egr-1): demonstration of a fifth functional serum response element. DNA Seq. 2000;10:429–432. doi: 10.3109/10425170009015615. [DOI] [PubMed] [Google Scholar]
- 58.Riley JL, Mao M, Kobayashi S, Biery M, Burchard J, Cavet G, Gregson BP, June CH, Linsley PS. Modulation of TCR-induced transcriptional profiles by ligation of CD28, ICOS, and CTLA-4 receptors. Proc. Natl. Acad. Sci. USA. 2002;99:11790–11795. doi: 10.1073/pnas.162359999. [DOI] [PMC free article] [PubMed] [Google Scholar]


