Summary
The Irr protein from the bacterium Bradyrhizobium japonicum is expressed under iron limitation to mediate iron control of haem biosynthesis. The regulatory input to Irr is the status of haem and its precursors iron and protoporphyrin at the site of haem synthesis. Here, we show that Irr controls the expression of iron transport genes and many other iron-regulated genes not directly involved in haem synthesis. Irr is both a positive and negative effector of gene expression, and in at least some cases the control is direct. Loss of normal iron responsiveness of those genes in an irr mutant, as well as a lower total cellular iron content, suggests that Irr is required for the correct perception of the cellular iron status. Degradation of Irr in iron replete cells requires haem. Accordingly, control of Irr-regulated genes by iron was aberrant in a haem-defective strain, and iron replete mutant cells behave as if they are iron-limited. In addition, the haem mutant had an abnormally high cellular iron content. The findings indicate that B. japonicum senses iron via the status of haem biosynthesis in an Irr-dependent manner to regulate iron homeostasis and metabolism.
Introduction
Iron is an essential element for most living organisms. It is required for many cellular processes in catalysis, electron transfer, oxygen metabolism, regulation and signal tduction. Iron availability can be limiting because it is predominantly in the insoluble ferric form in aerobic environments. On the other hand, excessive intracellular iron can be deleterious because it generates reactive oxygen species that damage cellular components (Braun and Killmann, 1999; Touati, 2000). Iron homeostasis is strictly regulated so that iron acquisition, storage and consumption are geared to iron availability, and that intracellular levels of free iron do not reach toxic levels (reviewed in Andrews et al., 2003).
In bacteria, the Fur (ferric-uptake regulator) protein is regarded as a global regulator of iron metabolism and homeostasis. It has been particularly well-studied in the γ-proteobacteria Escherichia coli (Crosa, 1997; Escolar et al., 1999; Hantke, 2001) and Pseudomonas aeruginosa (Ochsner et al., 1995; Ochsner and Vasil, 1996), and the prevailing model of Fur function is based largely on work from those organisms. In the presence of iron, the metal binds Fur directly, which enhances its DNA-binding activity to represses genes under its control.
Rhizobia belong to the α-Proteobacteria, a taxonomic group comprising numerous members that form close or intracellular associations with higher eukaryotes in a symbiotic or pathogenic context. Rhizobia live as free-living soil organisms or in symbiosis with leguminous plants, where they convert atmospheric nitrogen to ammonia to fulfil the nutritional nitrogen requirement of the plant host (Nap and Bisseling, 1990). Recent work with rhizobial species shows that control of iron metabolism differs substantially from the E. coli model. Some rhizobia have iron regulators in addition to Fur, or have structural Fur homologues that appear not to be iron regulators (Hamza et al., 1998; 1999; Todd et al., 2002; Chao et al., 2004; 2005; Diaz-Mireles et al., 2004; Platero et al., 2004; Viguier et al., 2005).
The Irr protein has been characterized in Bradyrhizobium japonicum and Brucella abortus (Hamza et al., 1998; Martinez et al., 2005), and homologues are present in other α-Proteobacterial genomes. Haem is an iron protoporhyrin and is the end product of a multistep biosynthetic pathway. Irr mediates iron control of the haem biosynthetic pathway to prevent the accumulation of toxic porphyrin precursors from exceeding iron availability (Hamza et al., 1998). Irr accumulates under iron limitation, where it negatively regulates the pathway. Irr is a conditionally stable protein that degrades rapidly when cells are exposed to iron, allowing derepression of haem synthesis (Hamza et al., 1998; Qi et al., 1999). This iron-dependent turnover is mediated by haem (Qi et al., 1999), which binds directly to Irr to promote degradation. Accordingly, Irr persists in haem synthesis mutant strains in the presence of iron, and mutations in Irr that affect the haem binding site stabilize the protein in the presence of the metal (Qi et al., 1999; Yang et al., 2005). Thus, haem is an effector molecule in Irr degradation that reflects the availability of iron for haem synthesis. Irr interacts directly with the haem biosynthesis enzyme ferrochelatase (Qi and O’Brian, 2002). In the presence of iron, ferrochelatase inactivates Irr followed by haem-dependent degradation. Under iron limitation, protoporphyrin relieves the inhibition of Irr by ferrochelatase, probably by promoting protein dissociation, allowing genetic repression. Thus, Irr responds to iron via the status of protoporphyrin and haem locally at the site of haem synthesis.
In the present study, we show that Irr controls many iron-regulated genes, and is not restricted to haem biosynthesis. Furthermore, the findings suggest that the status of haem biosynthesis may be a regulatory input signal to control iron metabolism.
Results
The Irr protein controls ferric iron transport genes
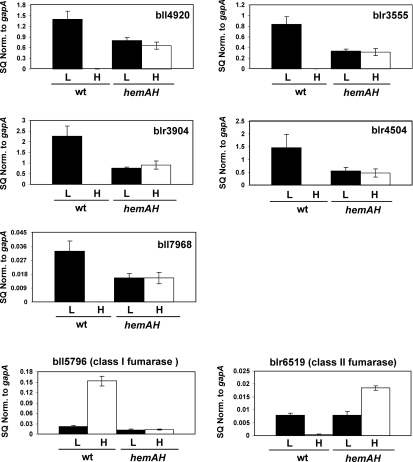
The Irr protein negatively controls haem biosynthesis when iron is limiting. Irr accumulates in cells grown under iron deprivation where it is needed, but degrades when cells are exposed to iron. We found previously that an irr mutant is deficient in an inducible ferric iron transport activity (Hamza et al., 1998), suggesting that Irr may have a broader role in regulating iron-dependent processes. Iron deficiency induces high-affinity transport systems in bacteria to scavenge available iron (Andrews et al., 2003). The B. japonicum genome encodes five ferric iron chelate receptors. We used quantitative real time polymerase chain reaction (qRT-PCR) to measure mRNA levels of the five ferric iron chelate receptor genes in the parent and irr strains grown in high or low-iron media (Fig. 1). These data were normalized to the gapA gene, which was not iron-regulated or affected in the irr strain (This was corroborated in the microarray experiments described below). The data showed strong induction of these genes in response to iron limitation in the parent strain. However, induction of the iron transport genes was abolished or diminished in the irr strain (Fig. 1), showing that Irr normally has a positive affect on their expression. These findings agree with the previous observation that an irr strain is defective in iron transport (Hamza et al., 1998). Induction of blr3555 and bll4920 appeared to be strictly dependent on Irr because there was little transcript in the mutant in low-iron cells. Levels of blr3904 and blr4504 mRNA were diminished but measurable in the irr strain (Fig. 1). This remaining activity was iron-responsive in the mutant as judged by the approximately threefold decrease in mRNA levels in the presence of iron. This suggests some Irr-independent iron regulation of those genes. A similar result was observed previously using reporter fusions of the hmuR haem receptor gene (Nienaber et al., 2001). In that study, promoter activity was diminished in an irr strain, but the remaining activity was iron responsive.
Fig. 1.
Iron-dependent expression of the ferric receptor genes and fumarase genes in the wild-type strain LO (Wt) or the irr mutant LODTM5 (irr). mRNAs were analysed by quantitative real-time PCR from cells grown in media supplemented with no iron (L) or with 12 µM FeCl3 (H). The data are expressed as the relative starting quantity (SQ) of the respective mRNAs normalized to the housekeeping gene gapA. The data are expressed as the average of three replicates ± the standard deviation.
The Irr protein affects many genes within the iron regulon
Iron transport is central to iron homeostasis. The observation that Irr controls the expression of iron transport genes led us to ask whether Irr affects iron metabolism more generally. To do this, we first characterized the iron regulon in parent strain LO by using whole genome microarray analysis to identify changes in gene expression in cells in response to iron limitation. (Table S1) Then, we compared strain LO to the irr mutant LODTM5 grown in iron-deficient media using microarrays (Table S2), and focused on those genes that were also iron-regulated in the wild type (Table 1).
Table 1.
Genes within the iron regulon that are aberrantly regulated in irr strain LODTM5 gn in iron-limited media compared with parent strain LO gn in iron-limited media.a
| Genes that are downregulated in the irr mutant strain compared with the parent strain | |||
|---|---|---|---|
| Geneb | irr/irr+ | SD | Gene product |
| blr0056 | −2.41 | 0.10 | Unknown protein |
| bll0233 | −2.11 | 0.32 | Hypothetical protein |
| bll0620 | −2.02 | 0.39 | Two-component response regulator |
| blr0697 | −5.68 | 1.63 | Hypothetical protein |
| blr0698 | −8.86 | 3.44 | Putative hydroxymethylglutaryl-CoA lyase |
| bsr0858 | −3.10 | 1.64 | Hypothetical protein |
| bsr0859 | −3.20 | 1.82 | Hypothetical protein |
| bll1076 | −3.50 | 0.43 | Hypothetical protein |
| blr1180 | −2.66 | 0.37 | Two-component response regulator |
| bll1555 | −2.99 | 0.51 | Unknown protein |
| bll2216 | −2.77 | 0.11 | Transcriptional regulatory protein TetR family |
| blr3445 | −3.78 | 0.72 | Putative enoyl-CoA hydratase |
| blr3553 | −11.51 | 3.77 | Unknown protein |
| blr3554 | −6.36 | 1.71 | Unknown protein |
| blr3555 | −42.60 | 12.93 | Ferric-siderophore receptor precursor |
| bsr3556 | −101.67 | 39.67 | Hypothetical protein |
| bll3557 | −9.66 | 1.00 | Putative cytochrome b561 |
| bll3558 | −3.90 | 0.96 | Two-component hybrid sensor and regulator |
| bll3559 | −2.71 | 1.12 | Two-component response regulator |
| blr3561 | −18.14 | 14.14 | Hypothetical protein |
| blr3562 | −22.00 | 17.63 | Hypothetical protein |
| blr3904 | −2.32 | 0.44 | Ferric-siderophore receptor precursor |
| blr3905 | −2.17 | 0.41 | Hypothetical protein |
| blr3906 | −2.01 | 0.28 | Biopolymer transport protein (ExbB) |
| blr4146 | −5.94 | 1.59 | Hypothetical protein |
| bll4177 | −6.76 | 7.15 | Hypothetical protein |
| blr4504 | −4.14 | 0.73 | Ferric-siderophore receptor precursor |
| blr4505 | −4.31 | 0.53 | Hypothetical protein |
| bll4708 | −5.61 | 1.22 | Probable ATP-binding protein |
| bll4920 | −31.91 | 11.85 | Ferric-siderophore receptor precursor (FegA) |
| blr5540 | −8.19 | 0.33 | Hypothetical protein |
| blr5541 | −2.65 | 0.31 | Hypothetical protein |
| bll5595 | −2.78 | 1.34 | Hypothetical protein |
| blr6519 | −3.27 | 0.45 | Fumarase, class II (FumC) |
| blr6801 | −5.53 | 2.16 | Unknown protein |
| blr6802 | −2.92 | 0.49 | Hypothetical protein |
| bll6994 | −3.33 | 0.13 | Putative phosphatidylethanolamine N-methyltransferase |
| bll6995 | −2.60 | 0.40 | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase |
| bll7074 | −2.30 | 0.16 | Hypothetical protein |
| bll7076 | −2.06 | 0.34 | Haemin receptor precursor (HmuR) |
| blr7077 | −2.73 | 0.48 | Haemin ABC transporter haemin-binding protein (HmuT) |
| blr7078 | −2.11 | 0.31 | Haemin ABC transporter permease protein (HmuU) |
| blr7079 | −2.19 | 0.09 | Haemin ABC transporter ATP-binding protein (HmuV) |
| blr7296 | −20.34 | 6.07 | Hypothetical protein |
| blr7297 | −26.70 | 2.87 | Unknown protein |
| bll7310 | −4.01 | 3.46 | Probable ArcA2 arginine deiminase |
| bll7311 | −3.33 | 2.75 | Probable ArcD2 arginine-ornithine antiporter |
| blr7314 | −4.81 | 4.40 | Unknown protein |
| blr7315 | −3.78 | 2.79 | Unknown protein |
| blr7321 | −2.33 | 0.81 | Hypothetical protein |
| bll7322 | −2.67 | 1.69 | Hypothetical protein |
| bsr7328 | −2.80 | 1.60 | Unknown protein |
| blr7329 | −2.45 | 1.12 | Putative multidrag resistance protein |
| bll7414 | −2.66 | 0.37 | Probable Elongation factor G |
| blr7418 | −4.99 | 1.52 | Hypothetical glutathione S-transferase like protein |
| blr7471 | −2.52 | 0.10 | Phosphatidylglycerophosphate synthase |
| bsr7472 | −2.16 | 0.30 | Molybdopterin converting factor small subunit |
| blr7730 | −5.20 | 0.79 | Hypothetical protein |
| blr7746 | −2.77 | 0.11 | Capsule expression protein |
| bll7967 | −4.65 | 2.02 | Uncharacterized iron-regulated protein |
| bll7968 | −12.75 | 1.33 | Ferric-siderophore receptor precursor |
| Genes that are upregulated in the irr mutant strain compared with the parent strain | |||
| Geneb | irr/;irr+ | SD | Gene product |
| blr0095 | 4.75 | 0.84 | Hypothetical protein |
| blr0488 | 8.90 | 1.81 | 3-isopropylmalate dehydratase large subunit |
| blr1080 | 2.79 | 0.48 | Hypothetical protein |
| bll1200 | 2.84 | 0.35 | 5-aminolevulinic acid synthase |
| blr2581 | 3.12 | 2.24 | Putative D-fructose-1,6-bisphosphatase protein |
| bll2737 | 2.30 | 0.16 | Oxidoreductase with iron-sulphur subunit |
| bll3190 | 2.53 | 0.27 | ABC transporter ATP-binding protein |
| bll3192 | 2.54 | 0.35 | ABC transporter substrate-binding protein |
| bll3193 | 2.41 | 0.25 | Unknown protein |
| blr4582 | 14.38 | 7.22 | Acetylornithine aminotransferase |
| blr5037 | 5.07 | 0.71 | Delta-aminolevulinic acid dehydratase |
| bll5796 | 3.35 | 0.47 | Fumarase, class I |
| bll5913 | 4.35 | 0.90 | Cytochrome c |
| blr5962 | 2.17 | 0.41 | ABC transporter ATP-binding protein |
| blr6742 | 4.02 | 0.50 | Putative glutamate synthase small subunit |
| blr6743 | 3.52 | 0.67 | Putative ferredoxin oxidoreductase alpha subunit |
| blr6744 | 3.43 | 0.51 | Putative ferredoxin oxidoreductase beta subunit |
The list is a subset of genes regulated in the irr strain compared with the parent strain under iron limitation. Only genes that were also upregulated in the parent strain in iron limited cells compared with the parent strain gn in iron replete media were included.
Genes in bold were analysed further in the article.
Comparison of the parent strain grown under high- or low-iron conditions detected 343 genes upregulated or downregulated in response to iron limitation (Table S1). Thus, B. japonicum has a large iron regulon, which is similar to what has been observed in other bacteria (Baichoo and Helmann, 2002; Ochsner et al., 2002; Grifantini et al., 2003; McHugh et al., 2003). Many of the regulated genes are organized in clusters in the genome (indicated by shading in Table S1), which may give some insight into the roles of hypothetical or unknown genes that are proximal to genes of known or putative function. Here, we refer to a cluster as simply a group of adjacent genes regardless of orientation. Moreover, 64 of the 176 genes induced by iron limitation in the wild type were found in clusters that include transporter genes.
About one-third (61 of 176) of the genes that increased in response to iron limitation in the wild type were downregulated in the irr mutant (Table 1), suggesting that Irr is normally involved in the positive regulation of those genes. Furthermore, 30 of the 45 genes that show at least a 10-fold increase in expression in the wild type in response to iron limitation are regulated by Irr. These findings further indicate that regulation by Irr is not restricted to genes involved in haem biosynthesis. In agreement with the quantitative RT-PCR analysis (Fig. 1), the microarray study detected the five ferric iron chelate receptor genes. We measured 59Fe transport in cells of the parent and mutant strains prepared in the same manner as were cells for the microarray analysis (Fig. 2A). High-affinity iron transport activity was induced in cells of the parent strain grown in low-iron media. However, this activity was greatly diminished in the irr strain. Thus, the transport activity is in good agreement with the quantitative RT-PCR and microarray studies.
Fig. 2.
Phenotypes of the irr mutant.
A. Ferric iron transport. Parent strain LO (circles) or irr mutant LODTM5 (squares) were grown in media containing no added iron (filled symbols) or 12 µM iron (open symbols). At time zero, 0.05 µM 59Fe was added to the assay medium, and aliquots were subsequently taken at various time points and counted. Each point is the average of triplicate samples, and the standard deviations were less than 10%.
B. Immunoblot analysis of the hemB product ALA dehydratase (ALAD) and Irr in cells of parent strain LO and irr mutant LODTM5 grown in low- or high-iron media as described above. Fifty micrograms of protein were loaded per lane.
C. Measurement of the iron content of the parent strain LO and irr strain LODTM5 by atomic absorption spectrometry. Cells grown in media supplemented with no iron (L) or with 12 µM FeCl3 (H). The data were based on five to six replicates with error bars within 95% confidence window. The data are shown in two panels to expand the y-axis for the low-iron cells.
Among the 167 genes that were downregulated in response to iron limitation in the wild type (Table S1), 17 of them were identified that involve Irr (Table 1). In these cases, mRNA levels were higher in the irr mutant than in the wild type, suggesting that Irr normally represses those genes. The class I fumarase, 3-isopropylmalate dehydratase and glutamate synthase are iron sulphur proteins (Flint et al., 1992; Prodromou et al., 1992; Vanoni and Curti, 1999), and genes encoding three additional iron-sulphur proteins were identified in this analysis as well (bll2737, blr6743, blr6744). Haem is an iron-containing cofactor; genes encoding the haem biosysnthetic enzymes δ-aminolevulinic acid (ALA) synthase and ALA dehydratase were affected in the irr strain, as was a cytochrome c. We measured ALA dehydratase protein by immunoblotting in the parent strain and the irr mutant grown under the same conditions employed for the microarray analysis (Fig. 2B). ALA dehydratase accumulated to high levels in iron replete cells of the wild type, but not under iron limitation. However, in the irr strain, ALA dehydratase was expressed highly independently of the iron status. These observations agree with the microarray analysis (Table 1), and are similar to previous findings (Hamza et al., 1998). It makes physiological sense for iron-containing proteins to be downregulated when iron is scarce, and Irr mediates this control for the genes described above.
The B. japonicum genome encodes both a class I and class II fumarase, a tricarboxylic acid cycle enzyme (Acuna et al., 1991), and both genes are iron-regulated in an Irr-dependent manner (Table 1, Fig. 1). Class I fumarases (bll5796) are iron-sulphur proteins (Flint et al., 1992) whereas class II fumarases (blr6519) do not contain iron (Ueda et al., 1991). Quantitative RT-PCR showed that both of these genes are regulated by iron, but in an opposite manner (Fig. 1). The class II fumarase mRNA was highly induced under iron limitation in the wild type, but not in the irr mutant, indicating that Irr is necessary for activation in the parent strain. The iron-containing class I fumarase gene was also controlled by iron, but was downregulated in iron-limited cells, showing 15-fold less mRNA compared with iron replete cells. This level was elevated 10-fold in the irr strain in low iron compared with the parent strain, indicating that Irr normally downregulates this gene. Some control by iron was observed in the irr mutant, suggesting some Irr-independent regulation. It is plausible that the iron-independent fumarase substitutes for the iron-sulphur protein when iron is limiting. We suggest that Irr is involved in control of energy metabolism at the level of the tricarboxylic acid cycle.
Evidence that the irr mutant cannot correctly perceive the cellular iron status
The data presented suggest that an irr mutant does not correctly perceive the iron status based on the loss of normal responses to iron limitation with respect to many genes (Table 1, Fig. 1). Specifically, the mutant behaves as if there is sufficient available iron when grown in iron-deficient media. To address this directly, we measured the iron content of parent strain LO and irr strain LODTM5 in cells grown under iron limitation by atomic absorption spectroscopy (Fig. 2C). The irr strain is even more iron deficient than the parent strain when grown in low-iron media, containing about 40% less iron. This decrease is likely to be significant because the parent strain is already iron-limited. The lower iron levels in the irr mutant are probably due to the lack of induction of iron transport (Table 1, Figs 1 and 2A). Furthermore, the microarray data suggest that downregulation of chemotaxis genes is indicative of iron deficiency (Table S1). We found that many of these chemotaxis genes were downregulated in the irr strain compared with the wild type when the cells were grown under iron limitation (Table S2), supporting the conclusion that the mutant is more iron deficient than the parent strain. We suggest that Irr is necessary for perception of the iron status to regulate iron metabolism and homeostasis.
Evidence for both positive and negative regulatory activities for Irr
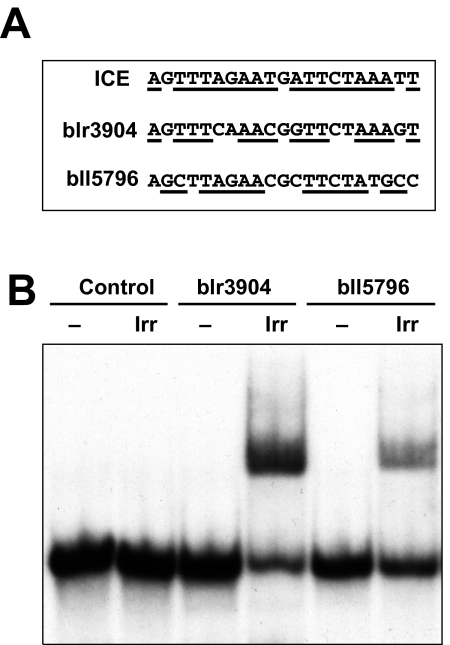
Inactivation of the irr gene results in both increases and decreases in genes compared with wild-type cells, and thus Irr can have both a positive and negative role in gene expression. Therefore, it is plausible that Irr has two regulatory activities. Previously, an iron control element (ICE) was identified upstream of the divergent haem transport genes hmuR and hmuT (Nienaber et al., 2001), and it is a target for Irr (Rudolph et al., 2006). An ICE motif is an AT-rich imperfect palindrome (Fig. 3A). Here, we identified putative ICE motifs upstream of several Irr-regulated genes. We chose to assess the putative elements upstream of bll5796 (class I fumarase), which is downregulated by iron limitation in the wild type, and blr3904 (iron transport protein), which is normally upregulated in those cells. Double stranded DNA probes containing the respective elements were radiolabelled and analysed by electrophoretic mobility shift assays (EMSA) using purified recombinant Irr (Fig. 3B). Irr bound both elements, but not a non-specific DNA control, which agrees with mRNA analyses showing that those genes are controlled by Irr. Collectively, these findings suggest that Irr can be a direct positive and negative regulator of gene expression. Numerous genes affected in the irr strain do not contain an obvious ICE motif, suggesting either that Irr can recognize disparate sequences, or that the control by Irr is indirect.
Fig. 3.
Irr binds to ICE-like motifs upstream of Irr-regulated genes.
A. ICE-like motif upstream of blr3904 and bll5796.
B. Electrophoretic mobility shift assays (EMSA) of Irr with ICE-like motifs. The model ICE motif is the nearly perfect inverted repeat found upstream of blr7895. EMSA analysis was carried out using 500 nM purified recombinant Irr and 100 pM 32P-labelled probes. Complexes were resolved on 5% non-denaturing gels and visualized by autoradiography. A non-specific DNA sequence from the multiple cloning site of plasmid pSK was used as a negative control. ‘–’ denotes free probe.
Evidence that Irr-regulated genes depend on the status of haem
Our findings indicate that Irr is involved in the control of many iron-regulated genes, and not only those directly involved in haem biosynthesis. We showed previously that Irr responds to iron via the status of haem and protoporphyrin by interacting with the enzyme ferrochelatase at the site of haem synthesis (Qi and O’Brian, 2002). Therefore, the current work suggests that the control of iron-regulated genes not directly related to haem synthesis may also be dependent on the status of haem.
To test this idea, we measured iron-dependent expression of several Irr-regulated genes in the haem-defective mutant ΔhemAH, and in its parent strain I110 by quantitative RT-PCR (Fig. 4). Irr accumulates to high levels independently of iron in haem synthesis mutants because haem is required for its degradation, and the protein is active under both iron conditions (Qi et al., 1999; Qi and O’Brian, 2002). Here, we found that the five ferric iron transporter genes strongly upregulated by Irr in the wild type were aberrantly expressed in the haem mutant (Fig. 4). In all cases, the respective mRNAs were partially activated in the haem mutant regardless of the iron status. Similarly, the class II fumarase gene (blr6519) that was induced under iron limitation in the parent strain was highly expressed in the haem mutant in the presence or absence of iron (Fig. 4). We do not know why blr6519 mRNA is greater in high-iron cells than low-iron cells in ΔhemAH.
Fig. 4.
Iron-dependent expression of the five ferric receptor genes and fumarase genes in the wild-type strain I110 (wt) or the hemA hemH mutant strain ΔhemAH. mRNAs were analysed by quantitative real-time PCR from cells grown in media supplemented with no iron (L) or with 12 µM FeCl3 (H). The data are expressed as the relative starting quantity (SQ) of the respective mRNAs normalized to the housekeeping gene gapA. The data are expressed as the average of three replicates ± the standard deviation.
Whereas the iron transport genes and the class II fumarase are normally activated under iron limitation, expression of the class I fumarase gene (bll5796) is normally downregulated in iron deficient cells. In the haem mutant, bll5796 mRNA was low irrespective of the iron status (Fig. 4). Thus, mutant strain ΔhemAH has a low-iron phenotype with respect to genes that are normally upregulated or downregulated in the wild type, which is consistent with a constitutively active Irr. The findings suggest that these iron-regulated genes are affected by the status of haem even though they are not directly involved in haem biosynthesis.
A haem biosynthesis mutant is defective in maintaining iron homeostasis
The low-iron phenotype of mutant strain ΔhemAH led us to ask whether those cells are in fact iron-deficient compared with the wild type. Therefore, we measured cellular iron levels in those cells (Fig. 5). We found that the haem mutant was not iron-deficient, but rather contained almost four times more iron than the parent strain when grown in low-iron media. The reason for the higher iron was not determined, but the constitutively expressed iron transporters may result in greater iron accumulation in the mutant compared with the wild type. Regardless of the reason, the findings show that the haem-defective strain cannot maintain normal iron homeostasis or respond correctly to the cellular iron status. Collectively, the data suggest that B. japonicum senses iron through the status of haem biosynthesis to regulate iron metabolism.
Fig. 5.
Measurement of the iron content of the parent strain I110 and the haem synthesis mutant strain ΔhemAH by atomic absorption spectrometry. Cells grown in media supplemented with no iron (L) or with 12 µM FeCl3 (H). The data were based on five replicates with error bars within 95% confidence window.
Discussion
Previous work demonstrated that Irr interacts directly with the haem biosynthetic enzyme ferrochelatase, and responds to iron via the status of haem and its precursors at the site of synthesis (Qi et al., 1999; Qi and O’Brian, 2002). Thus, the finding that Irr affects many genes within the iron regulon strongly suggests that B. japonicum senses iron via the status of haem biosynthesis to control iron transport, the tricarboxylic acid cycle, expression of iron-containing proteins, and other processes not directly related to haem metabolism. Mutants defective either in Irr or in haem synthesis resulted in loss of normal iron-dependent control, and in aberrant total cellular iron content. The irr mutant has a high-iron phenotype with respect to genes normally regulated by iron, but those cells are iron-deficient. On the other hand, a haem synthesis mutant, which expresses Irr constitutively, has a low-iron phenotype with respect to Irr-regulated genes, and contains more cellular iron than the wild type. These findings indicate that iron metabolism and homeostasis are linked to the status of haem in an Irr-dependent manner. This mechanism differs substantially from iron-dependent control of gene expression described in other well-studied bacteria. In those cases, iron homeostasis is mediated to a large extent by Fur (Baichoo et al., 2002; Ochsner et al., 2002; Grifantini et al., 2003; McHugh et al., 2003), which binds iron directly to regulate its activity. We propose that B. japonicum senses iron by the status an iron-dependent process. We are not aware of a similar control mechanism in other bacteria, but it is interesting to note that a haem biosynthesis mutant of the yeast Saccharomyces cerevisiae cannot induce transcription of iron transport genes (Crisp et al., 2003). Although this phenotype is essentially the opposite of what we observed for B. japonicum (Fig. 4), it nevertheless suggests a link between iron transport and haem. It would be interesting to learn the basis of that control in yeast, and whether other iron-regulated processes are affected in the haem synthesis mutant.
The irr strain was shown to contain less iron than the parent strain when grown under iron limitation (Fig. 2C), which is likely due to the low expression levels of iron transport in the mutant (Figs 1 and 2A). Ironically, a fur mutant of E. coli expresses iron transporter genes constitutively, but is also iron deficient (Abdul-Tehrani et al., 1999). This was shown to be the result of the repression of many genes encoding iron-containing proteins (McHugh et al., 2003). By contrast, numerous genes involved in the expression of iron-containing proteins are upregulated in the irr mutant (Table 1). Thus, in B. japonicum the cellular iron deficiency of the irr strain appears to be caused by defective iron acquisition, whereas in the E. coli fur mutant, it is the result of low incorporation of the metal into proteins. The findings suggest diverse strategies for maintaining iron homeostasis in bacteria.
The iron regulon of B. japonicum is large, and Irr appears to control only a subset of those genes. B. japonicum contains a functional fur gene as well, but preliminary data suggest that Fur cannot account for all of the Irr-independent regulation (J. Yang and M.R. O’Brian, unpubl. obs.). Furthermore, the genome contains a gene encoding a protein that is homologous in its C-terminus to Irr (blr1216). However, we have been unable to detect blr1216 mRNA in wild-type cells grown under the conditions used in the present study (data not shown). Thus, we speculate the presence of a yet unidentified iron regulator in B. japonicum. Interestingly, blr1216 was upregulated in the irr strain in microarray analysis (Table S2). It is possible that conditions created by the irr deletion, such as a low-iron content, can induce that gene.
Experimental procedures
Strains and media
Bradyrhizobium japonicum I110 and LO were parent strains used in the present work. Strain LODTM5 is a mutant derivative of LO that contains a transposon Tn5 within the irr gene (Hamza et al., 1998). Strain ΔhemAH is a double mutant of I110 that is defective in hemA and hemH genes, encoding δ-aminolevulinic acid synthase and ferrochelatase respectively (Qi et al., 1999). B. japonicum strains were routinely grown at 29°C, 150 r.p.m. in GSY medium as described previously (Frustaci et al., 1991). Strain LODTM5 was grown with 50 µg ml−1 kanamycin and 50 µg ml−1 streptomycin. Strain ΔhemAH was grown with 50 µg ml−1 kanamycin, 50 µg ml−1 streptomycin, 100 µg ml−1 spectinomycin and 15 µM haemin to fulfil its haem auxotrophy. For low-iron conditions, modified GSY medium was used, which contains 0.5 g l−1 yeast extract instead of 1 g l−1 and no exogenous iron source. The actual iron concentration of the medium was 0.3 µM, as determined with a Perkin-Elmer model 1100B atomic absorption spectrometer The parent strain LO and irr strain LODTM5 have doubling times of about 8 and 9.5 h under low-iron conditions. Strain ΔhemAH was grown with 0.05 µM haemin for low-iron conditions The mutant grows similarly to the wild type. The medium was supplemented with 12 µM FeCl3.6H2O for high-iron conditions.
Microarray analysis. Sample preparation, RNA isolation, cDNA synthesis, fragmentation, terminal labelling and hybridization
Three biological replicates of LO and LODTM5 grown under either high- (12 µM FeCl3.6H2O) or low-iron conditions were used for microarray procedures. Forty millilitres of cultures grown to mid-exponential phase (OD540 0.4–0.6) were quickly transferred to chilled centrifuge tubes containing 8 ml of RNA protect (Qiagen), mixed by inversion and centrifuged at 7000 r.p.m., 4°C for 10 min to pellet cells. Supernatants were discarded and pellets were quickly frozen in liquid nitrogen and stored at −80°C. To isolate RNA, frozen cell pellets were resuspended in 1.5 ml of cold buffer A [0.02 M NaOAc (pH 5.3), 1 mM EDTA (pH 8.0)] and additional 200 µl of RNA protect (50 µl per 10 ml of culture) was added. The cell suspension was transferred to the acid phenol solution (160 µl of 10% SDS, 2 ml of buffer A and 3.5 ml of acid phenol) that was preheated at 65°C for 5 min, vortexed for 30 s, incubated at 65°C for 2 min, vortexed for 1 min, and incubated at 65°C for additional 5 min. The mixture was centrifuged at 7000 r.p.m., 4°C for 5 min and the upper aqueous phase was extracted with 3 ml of phenol: chloroform: isoamyl alcohol (25:24:1) and 3 ml of chloroform successively. RNA was precipitated from the aqueous phase by addition of 1/10 vol. of 3M sodium acetate and 2 vols of 100% ethanol at −80°C overnight; followed by centrifugation at 13 000 r.p.m., 4°C for 30 min. The pellet was washed once with 70% ethanol, and then it was dissolved in 100 µl of RNase-free H2O. RNA samples were treated with RQ1 Rnase-free DNase I (Promega) and purified using RNeasy bacterial RNA purification kits (Qiagen). The genomic DNA contamination was examined by PCR using 500 ng of total RNA as templates.
cDNA synthesis, fragmentation, terminal labelling and hybridization were carried out using GeneChip P. aeruginosa Genome Array Expression Analysis protocol (Affymetrix) with slight modifications indicated below. One thousand five hundred units of M-MLV reverse transcriptase (Promega) were used for 10 µg of total RNA in a 60 µl cDNA synthesis reaction. A mixture of in vitro transcripts was spiked into RNA samples as internal controls. Subsequently, 3 µg of synthesized cDNA was fragmented by DNase I (0.2 U µg−1 cDNA) (Amersham Biosciences) at 37°C for 2.5–3 min and the reaction was immediately inactivated at 98°C for 10 min 2.8 µg of cDNA fragments were then labelled with biotinylated GeneChip® DNA labelling reagent (Affymetrix) using the terminal deoxynucleotidyl transferase (Promega) at 37°C for 75 min and the reaction was stopped by the addition of 2 µl of 0.5 M EDTA. The target is ready to be hybridized onto probe arrays.
Labelled cDNA fragments (2.5 µg) and an internal standard (spike-in) hybridization control were used as targets in the microarray analysis. B. japonicum probe arrays (BJAPETHa520090, Affymetrix) were incubated in the 140 µl of hybridization cocktail in a hybridization oven at 60 r.p.m., 48°C for 16 h. Arrays were washed and stained with streptavidin, biotinylated antistreptavidin antibody and streptavidin phycoerythrin successively in a fluidics station 450 (Affymetrix) and scanned in a GeneChip® scanner 3000 (Affymetrix) according to Affymetrix protocols.
GeneChip data analysis
Signals from scanned images were processed using GeneChip Operating Software (GCOS) version 1.1 (Affymetrix). The Affymetrix GeneChip used represents the 8317 annotated open reading frames as well as 51 RNA genes and intergenic regions. For single array analysis, global scaling was applied by scaling signals to a target signal intensity of 500 using all probe sets. To compare expression under different conditions, we used comparison array analysis where an experimental array was compared with a baseline array. The comparison array data were exported from GCOS and analysed in Microsoft Excel. A decrease was deemed only when signal log ratio was ≤ −1 and the comparison was statistically evaluated as significantly changed (P ≥ 0.98) for all three replicates, or an increase was deemed only when signal log ratio was ≥ 1 and the comparison was statistically evaluated as significantly changed (P ≤ 0.02) for all three replicates. The fold change was calculated from the signal log ration (FC = 2SLR) and the average fold change of three biological replicates were shown in the results table. The intergenic regions were filtered thus not included in the results. Microsoft Access was further used to compare data from different comparison array analysis. Gene names and annotations are from the published genome database (Kaneko et al., 2002, http://www.kazusa.or.jp/rhizobase/index.html).
Quantitative real-time PCR
We determined expressions of selected genes by real-time qPCR with iQ™ SYBR Green supermix (Bio-Rad) using iCycler thermal cycler (Bio-Rad). RNA was isolated and treated with DNase I as above for microarray analysis. Total RNA of 2.5–5 µg was used for cDNA synthesis using Superscript first-strand synthesis system (Invitrogen). cDNA was purified using MinElute PCR purification kit (Qiagen) as above for microarray analysis and various dilutions of cDNA were used as PCR templates. Each PCR reaction contained 10 µl of 2 × SYBR Green supermix, 0.2 µM primers (IDT DNA Technology) and appropriate templates in a 20 µl reaction. PCR reactions were heated to 95°C for 3 min and then for 40 cycles with steps of 95°C for 30 s, 56°C for 30 s, 72°C for 30 s. The generation of specific PCR products was confirmed by melting curve analysis and gel electrophoresis. Samples in which the reverse transcriptase was omitted in reverse transcriptase reactions were used as negative controls. The standard curve method was employed for relative quantification and gapA was a housekeeping gene control. Genomic DNA from parent strains LO or I110 was used as PCR templates to generate a standard curve for each gene. Relative starting quantities (SQ) of mRNAs for interested genes and gapA were calculated from corresponding standard curves. Quantity of interested genes was then normalized to quantity of gapA for each condition. The results were based on the average of triplicates and standard deviation was shown as the error bar in the results.
Iron uptake assays
Cells were grown to mid-log phase in low- or high-iron media, centrifuged, washed twice and resuspended in uptake buffer to a OD540 of 0.4. Uptake buffer contained 0.2 M MOPS, 20 mM citrate and 2% (w/v) glucose, pH 6.8. Thirty millilitres of cells were placed into a 125 ml flask and preincubated at 30°C with shaking. At time zero, 59FeCl3 was added to a final concentration of 0.05 µM (1.8 µCi). One millilitre aliquots (c. 125 µg protein) were removed at various times and added to 3 ml quench buffer precooled on ice. The quench buffer contained 0.1 M Tris, 0.1 M succinate and 10 mM EDTA, pH 6.0. The quenched cells were collected on 0.45 µm filters presoaked in quench buffer containing 40 µM Fe-EDTA, and then counted on an LKB gamma-counter.
Iron content measurement using atomic absorption spectroscopy
Forty millilitres of cultures grown to mid-log phase (OD540 0.4–0.6) were harvested by centrifugation at 7000 r.p.m., 4°C for 10 min. The pellet was washed twice with 20 ml of ice-cold quench buffer (0.1 M Tris, 0.1 M succinate and 10 mM EDTA) and once with metal-free double distilled water to remove salts. To lyse cells, the pellet was resuspended in 100 µl of 70% HNO3 (J.T. Baker, AAS grade), lightly vortexed and heated at 75°C for 3–5 min until cells were completely lysed. One millilitre of metal-free double distilled water was added to the lysed cells and mixed by vortex. Samples were centrifuged at 13 000 r.p.m. for 5 min and the supernatant was analysed for Fe content. Atomic absorption was performed in the furnace mode on a Perkin Elmer Atomic Absorption Spectrometer model 1100B equipped with a model HGA 700 graphite furnace. All samples were diluted with metal-free double distilled water to contain 1% HNO3. To set up a Fe standard curve, the stock solution Fe 2 ng/10 µl in 2% HNO3 (Perkin Elmer) was diluted to 0.25 ng/10 µl, 0.5 ng/10 µl, 0.75 ng/10 µl, 1 ng/10 µl in 1% HNO3. Ten microlitres of samples were used for each measurement and 4–6 replicates were performed for each sample. The program for Fe element included five steps: 90°C for 45 s, 1400°C for 30 s, 30°C for 20 s, 2400°C for 10 s and 2650°C for 5 s. Protein concentrations were determined from another 40 ml cells by BCA assay (Pierce) using bovine serum albumin standards. The Fe content was normalized to the protein concentration.
Electrophoretic mobility shift analysis of ICE motifs
The ICE motifs were identified using the fuzznuc program (http://emboss.sourceforge.net/apps/fuzznuc.html) sing the imperfect inverted repeat sequence in the hmuR promoter as the input data (Nienaber et al., 2001) to find motifs within the B. japonicum genome (http://www.kazusa.or.jp/rhizobase/index.html). EMSAs were carried out to determine Irr binding to putative ICE motifs upstream of the blr3904 and bll5796 genes as previously described (Friedman and O’Brian, 2003). The purified recombinant Irr protein (500 nM) that was overexpressed and purified as previously described (Qi et al., 1999) and 100 pM 32P-labelled probes were used. A non-specific DNA fragment from plasmid pSK was used as a negative control. Protein–DNA complexes were resolved on 5% non-denaturing gels and visualized by autoradiography. DNA oligonucleotides used as probes are: blr3904 (5′-CAGG AATTCGATACTTTAGAACCGTTTGAAACTGACCTC-3′, 5′-GTCGAG-GTCAGTTTCAAACGGTTCTAAAGTATCGAATTC-3′); bll5796 (5′-CAGGAATTCGATA-GCTTAGAACGCTTCTA TGCCGACCTC-3′, 5′-GTCGAGGTCGGCATAGAAGCGTTC TA-AGCTATCGAATTC-3′); control (5′-CAGGAATTCGATATC AAGCTTATCGATACCGTCG-ACCTC-3′, 5′-GTCGAGGTCG ACGGTATCGATAAGCTTGATATCGAATTC-3′).
Supplemental material
The following supplementary material is available for this article online:
Comparison of parent strain LO grown in iron-limited or iron replete media.
Comparison of Irr mutant strain LODTM5 to parent strain LO grown in iron-limited media.
This material is available as part of the online article from www.blackwell-synergy.com
Acknowledgments
We thank Gesine Rudolph for her help with many aspects of the work, and Andrea Patrignani, Ralph Schlapbach and Ulrich Wagner (Functional Genomics Center Zürich) for their help in the microarray experiments. This work was supported by National Institutes of Health Grant R01 GM067966 to M.R.O’B. and a grant from the Swiss National Foundation for Scientific Research to H.-M.F.
Note added in proof
The microarray data have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible throught GEO Series accession number GSE4156.
References
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, et al. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna G, Ebeling S, Hennecke H. Cloning, sequencing, and mutational analysis of the Bradyrhizobium japonicum fumC-like gene: evidence for the existence of two different fumarases. J Gen Microbiol. 1991;137:991–1000. doi: 10.1099/00221287-137-4-991. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- Chao TC, Becker A, Buhrmester J, Pühler A, Weidner S. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn (II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol. 2004;186:3609–3620. doi: 10.1128/JB.186.11.3609-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao TC, Buhrmester J, Hansmeier N, Pühler A, Weidner S. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl Environ Microbiol. 2005;71:5969–5982. doi: 10.1128/AEM.71.10.5969-5982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp RJ, Pollington A, Galea C, Jaron S, Yamaguchi-Iwai Y, Kaplan J. Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J Biol Chem. 2003;278:45499–45506. doi: 10.1074/jbc.M307229200. [DOI] [PubMed] [Google Scholar]
- Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology. 2004;150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Guest JR. Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing enzyme. Biochemistry. 1992;31:10331–10337. doi: 10.1021/bi00157a022. [DOI] [PubMed] [Google Scholar]
- Friedman YE, O'Brian MR. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J Biol Chem. 2003;278:38395–38401. doi: 10.1074/jbc.M306710200. [DOI] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O'Brian MR. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, et al. Identification of iron-activated and – repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc Natl Acad Sci USA. 2003;100:9542–9547. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I, Chauhan S, Hassett R, O'Brian MR. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J Biol Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- Hamza I, Hassett R, O'Brian MR. Identification of a functional fur gene in Bradyrhizobium japonicum. J Bacteriol. 1999;181:5843–5846. doi: 10.1128/jb.181.18.5843-5846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- Martinez M, Ugalde RA, Almiron M. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology. 2005;151:3427–3433. doi: 10.1099/mic.0.28213-0. [DOI] [PubMed] [Google Scholar]
- Nap J-P, Bisseling T. Developmental biology of a plant-prokaryote symbiosis: the legume root nodule. Science. 1990;250:948–954. doi: 10.1126/science.250.4983.948. [DOI] [PubMed] [Google Scholar]
- Nienaber A, Hennecke H, Fischer HM. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol Microbiol. 2001;41:787–800. doi: 10.1046/j.1365-2958.2001.02555.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Vasil ML. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Vasil AI, Vasil ML. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- Platero R, Peixoto L, O'Brian MR, Fabiano E. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol. 2004;70:4349–4355. doi: 10.1128/AEM.70.7.4349-4355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Artymiuk PJ, Guest JR. The aconitase of Escherichia coli. Nucleotide sequence of the aconitase gene and amino acid sequence similarity with mitochondrial aconitases, the iron-responsive-element-binding protein and isopropylmalate isomerases. Eur J Biochem. 1992;204:599–609. doi: 10.1111/j.1432-1033.1992.tb16673.x. [DOI] [PubMed] [Google Scholar]
- Qi Z, O'Brian MR. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell. 2002;9:155–162. doi: 10.1016/s1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- Qi Z, Hamza I, O'Brian MR. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc Natl Acad Sci USA. 1999;96:13056–13061. doi: 10.1073/pnas.96.23.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol. 2006;188:733–744. doi: 10.1128/JB.188.2.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AW. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology. 2002;148:4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Yumoto N, Tokushige M, Fukui K, Ohya-Nishiguchi H. Purification and characterization of two types of fumarase from Escherichiay coli. J Biochem (Tokyo) 1991;109:728–733. doi: 10.1093/oxfordjournals.jbchem.a123448. [DOI] [PubMed] [Google Scholar]
- Vanoni MA, Curti B. Glutamate synthase: a complex iron-sulfur flavoprotein. Cell Mol Life Sci. 1999;55:617–638. doi: 10.1007/s000180050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguier CPOC, Clarke P, O'Connell M. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol Lett. 2005;246:235–242. doi: 10.1016/j.femsle.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Yang J, Ishimori K, O'Brian MR. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J Biol Chem. 2005;280:7671–7676. doi: 10.1074/jbc.M411664200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of parent strain LO grown in iron-limited or iron replete media.
Comparison of Irr mutant strain LODTM5 to parent strain LO grown in iron-limited media.