Summary
Analysis of the complete flagellin glycosylation locus of Campylobacter jejuni strain 81–176 revealed a less complex genomic organization than the corresponding region in the genome strain, C. jejuni NCTC 11168. Twenty-four of the 45 genes found between Cj1293 and Cj1337 in NCTC 11168 are missing in 81–176. Mutation of 6 new genes, in addition to three previously reported, resulted in a non-motile phenotype, consistent with a role in synthesis of pseudaminic acid (PseAc) or transfer of PseAc to flagellin. Mutation of Cj1316c or pseA had been shown to result in loss of the acetamidino form of pseudaminic acid (PseAm). Mutation of a second gene also resulted in loss of PseAm, as well as a minor modification that appears to be PseAm extended with N-acetyl-glutamic acid. Previously described mutants in C. jejuni 81–176 and Campylobacter coli VC167 that produced flagella lacking PseAm or PseAc failed to autoagglutinate. This suggests that interactions between modifications on adjacent flagella filaments are required for autoagglutination. 81–176 mutants defective in autoagglutination showed a modest reduction in adherence and invasion of INT407 cells. However, there was a qualitative difference in binding patterns to INT407 cells using GFP-labelled 81–176 and mutants lacking PseAm. A mutant lacking PseAm was attenuated in the ferret diarrheal disease model.
Introduction
Although originally considered a eukaryotic-specific phenomenon, there is an increased awareness of prokaryotic glycoproteins, particularly bacterial surface proteins (reviewed in Schmidt et al., 2003 and Szymanski and Wren, 2005). The most heavily glycosylated bacterial proteins described to date are the flagellins from Campylobacter jejuni and Campylobacter coli. Flagellin from C. jejuni strain 81–176 has been shown to be glycosylated at 19 serine or threonine residues, and C. coli VC167 at approximately 16 sites (Thibault et al. 2001; Logan et al., 2002). Although the glycans show variability among strains and can confer serospecificity (Logan et al., 2002), the carbohydrate modifications appear to be based on 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic acid (pseudaminic acid), a 9-carbon sugar that is structurally similar to sialic acid (NeuNAc; Thibault et al., 2001). The major modifications identified on flagellin from C. jejuni strain 81–176 were pseudaminic acid (PseAc, m/z 317) and an acetamidino form of pseudaminic acid (PseAm, m/z 316). There were also minor amounts of an acetylated form of pseudaminic acid (PseOAc, m/z 359), and a form in which the PseAm sugar had an N-acetyl glutamic acid attached (PseAmOGlnNAc; m/z 486; Schirm et al., 2005).
All genes that have been shown to be involved in flagellin glycosylation to date map near the two flagellin structural genes, flaA and flaB, in a region that is one of the more variable regions in the C. jejuni chromosome (Dorrell et al., 2001; Fouts et al., 2005; Pearson et al., 2004) and includes genes encoding enzymes that have recently been shown to be part of the PseAc biosynthetic pathway (Chou et al., 2005; Schoenhofen et al., 2006), as shown in Fig. 1. Six genes in this region in C. jejuni 81–176 have been mutated. A mutant in Cj1293 or pseB, which encodes the first step in biosynthesis of PseAc (Schoenhofen et al., 2006), was non-motile and accumulated unglycosylated flagellin intracellularly, demonstrating a requirement for glycosylation of flagellin for flagella filament biogenesis (Goon et al., 2003). Mutants in Cj1314c, Cj1315c, as well as Cj1317, which encodes the enzyme recently shown to condense a deoxyhexose precursor with PEP to produce pseudaminic acid (Chou et al., 2005), were also non-motile (Thibault et al., 2001). Karlyshev et al. 2002 have reported on a family of related genes in NCTC 11168 called maf genes (for motility accessory factor). Mutation of one of these maf genes in NCTC 11168 (maf5 or Cj1337) resulted in a non-motile phenotype, and more recently this same group has reported that the corresponding mutation in C. jejuni 81–176 also resulted in a non-motile phenotype (Karlyshev and Wren, 2005). The product of the fourth 81–176 gene, Cj1316c or pseA, appears to synthesize PseAm directly from PseAc by transfer of an acetamidino group (Thibault et al., 2001). Thus, a mutant in this gene was fully motile, but flagellin from the mutant had all PseAm modifications replaced with PseAc (Thibault et al., 2001). Other strains of campylobacter appear to have an additional, alternate pathway for synthesis of a related form of PseAm (Guerry et al., 1996; Parkhill et al., 2000; Logan et al., 2002; Fouts et al., 2005). Genetic analysis in C. coli VC167 has identified 6 genes, including additional homologs of NeuNAc biosynthetic genes, that are involved in direct synthesis of PseAm independently of PseAc synthesis, and these have been termed ptmA-F (Guerry et al., 1996; Logan et al., 2002). Thus, there appears to be variability in PseAm biosynthetic pathways among Campylobacter strains. Strains like NCTC 11168 have homologs of both the ptm genes and pseA, and would be predicted to have two distinct pathways directed towards the synthesis of PseAm. In contrast, 81–176 has a single pathway by which all PseAm is synthesized via a PseAc precursor, and C. coli VC167, which lacks a functional pseA gene, can synthesize PseAm only by the ptm pathway (Logan et al., 2002).
Fig. 1. Pathway of pseudaminic acid synthesis in C. jejuni from UDP-GlcNAc.
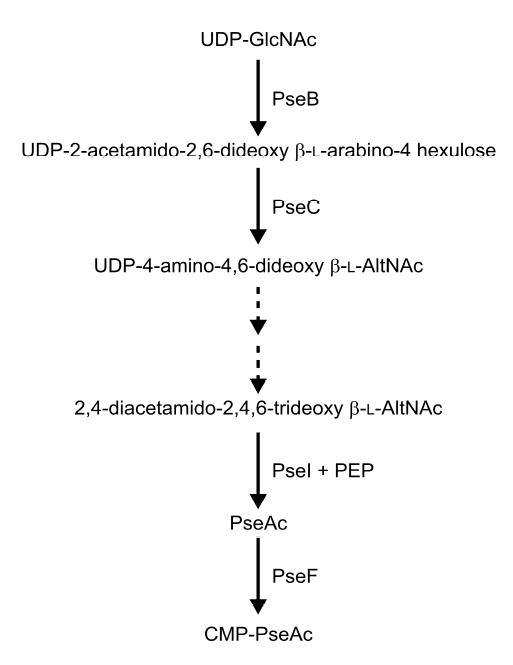
The enzymatic activity of the products of PseB (Cj1293) and PseC (Cj1294) have been described in Schoenhofen et al. 2005 and that of PseI (Cj1317c) by Chou et al., (2005). Broken arrows indicate biosynthetic steps for which the respective enzymatic activities have still to be assigned to a campylobacter gene. The presumed activity of PseF is based on homology to the corresponding enzyme in the sialic acid biosynthetic pathway, NeuA (Vann et al., 1987).
Here we characterize the complete flagellin glycosylation locus of C. jejuni strain 81–176. The simplicity of this locus in strain 81–176 has facilitated identification of the minimum number of genes required for synthesis of the two major flagellin modifications, PseAc and PseAm. Also, the glycans on flagellin have been suggested to play a role in autoagglutination and virulence (Golden and Acheson, 2002), and we have reexamined this association using a collection of defined mutants in both 81–176 and C. coli VC167 (Guerry et al., 1996; Thibault et al., 2001; Logan et al., 2002).
Results
DNA sequence analysis of the flagellin glycosylation locus of 81–176
A 32.4 kb contiguous region of the C. jejuni strain 81–176 chromosome that spanned Cj1291 to Cj1342 was sequenced, as shown in Table 1. Strain 81–176 is missing 25 genes that are found in NCTC 11168, namely Cj1296, Cj1297, Cj1301, Cj1304, Cj1306-Cj1310, Cj1318-Cj1332, and Cj1340. There are some genes that appear rearranged in 81–176 compared to the NCTC 11168 chromosome. Thus, the homolog of Cj1300c maps between Cj1295 and Cj1298 in 81–176, in place of Cj1296 and Cj1297. Also, the 81–176 gene that shows the highest homology to Cj1333 maps between Cj1334 and Cj1337. Of the seven maf gene homologs described by Karlyshev et al. 2002 in NCTC 11168, only five, Cj1333 (maf2), Cj1334 (maf3), Cj1337 (maf5), Cj1341c (maf6) and Cj1342c (maf7), were found in 81–176.
Table 1.
Genes in the flagellin glycosylation locus of C. jejuni 81–176.
| NCTC homolog1 | Gene/annotation2 | Homology to 111683 | length bp | Position of insertion4 | Phenotype of mutant | Reference |
|---|---|---|---|---|---|---|
| Cj1292 | dcd; dCTP deaminase | 98/99 (186) | 561 | N/A | ND | N/A |
| Cj1293 | pseB | 94/98 (334) | 1005 | 395 NP | mot− | Goon et al., 2003 |
| Cj1294 | pseC | 88/91 (376) | 1131 | 654 NP | mot− | this work |
| Cj1295 | amidopeptidase | 95/97 (233)5 | 708 | 335 P | mot+; wt IEF | this work |
| Cj1300c | unknown | 50/68 (295) | 897 | 287 NP | mot+; wt IEF | this work |
| Cj1298 | unknown | 46/63 (234) | 720 | 259 NP | mot+; wt IEF | this work |
| Cj1299 | accP | 72/85 (75) | 228 | ND | NA | NA |
| None (CJB1301) | NA | none | 1359 | 807 NP/663 P | mot+; wt IEF | this work |
| Cj1302 | unknown | 95/95 (520) | 1563 | 122 P | mot+; wt IEF | this work |
| Cj1303 | fabH2 | 99/99 (353) | 1062 | 302 NP | mot+; wt IEF | this work |
| Cj1304 | accP | 95/97 (73) | 221 | 81 NP | mot+; wt IEF | this work |
| Cj1305c | unknown | 81/86 (405) | 1215 | 271 NP | mot+; wt IEF | this work |
| Cj1311 | pseF | 97/98 (232) | 699 | 304 P | mot− | this work |
| Cj1312 | pseG | 92/92 (198) | 597 | 304 P | mot− | this work |
| Cj1313 | pseH | 88/95 (127) | 384 | 246 P | mot− | this work |
| Cj1314c | probable cyclase | 93/93 (248) | 747 | 134 NP | mot+; wt IEF | Thibault et al., 2001 |
| Cj1315c | amidotransferase | 98/99 (201) | 606 | 205 NP | mot+; wt IEF | Thibault et al., 2001 |
| Cj1316c | pseA | 99/99 (378) | 1137 | NA | mot+; PseAm− | Thibault et al., 2001 |
| Cj1317 | pseI | 92/93 (343) | 1032 | NA | mot− | Thibault et al., 2001 |
| Cj1334 | unknown (maf3) | 71/84 (544) | 1638 | 817 P | mot+; wt IEF | this work |
| Cj1333 | pseD | 64/79 (653) | 1965 | 1407 NP | mot+/PseAm−; 486−; 487+ | this work |
| Cj1337 | pseE | 61/76 (653) | 1899 | 998 NP | mot− | this work |
| Cj1338c | flaB | 75/81 (576) | 1731 | NA | N/A | this work |
| Cj1339c | flaA | 75/81 (576) | 1731 | NA | N/A | NA |
| Cj1341c | unknown (maf6) | 58/72 (605) | 1827 | 1121 NP | mot+; wt IEF | this work |
| Cj1342c | unknown (maf7) | 71/80 (404) | 1251 | 1062 NP | mot+; wt IEF | this work |
The NCTC 11168 that showed the highest homology to 81–176 is listed; as stated in the text, some genes in 81–176 are in a different order than those in NCTC 11168;
Annotations are the original annotations from Parkhill et al. (2000), Karlyshev et al., (2002) or from this work based on loss of PseAm or lack of motility suggesting a role in PseAc synthesis or attachment; annotations in parentheses indicate motility associated genes in NCTC 11168 but not 81–176;
homology to NCTC11168 was done by blastp analysis at campyDB (http://campy.bham.ac.uk/) and is given at the % identity/% similarity (number of amino acids);
position of the transposon insertion was determined by sequence analysis in E. coli and is given from the ATG start in bp;
The 81–176 allele of Cj1295 was truncated due to a possible slip strand mismatch at a homopolymeric tract of As; if the slip strand was corrected, the predicted protein showed 94% identity/96% similarity to the NCTC 11168 gene over 418 amino acids.
There was one gene unique to 81–176 that showed no homology to other known campylobacter genes (Fouts et al., 2005; Parkhill et al., 2000). This gene, termed CjB1301, encodes a predicted protein of 52.2 kDa, pI 9.4. The predicted protein showed significant homology to a dehydratase/ketoreductase domain of BryA, an enzyme from a bacterial symbiont of a marine bryozoan that is involved in polyketide synthesis (Hildebrand et al., 2004).
There were three genes in this region that had homopolymeric tracts of 9 C’s or G’s within the coding region, suggesting possible slip strand mismatch repair; these were Cj1295, Cj1305c and Cj1342c. However, Cj1295 was truncated as cloned from 81–176 into two ORFs by what appears to be slip strand mismatch repair at a homopolymeric tract of As. However, none of these three genes appear to be involved in flagellin glycosylation (see below).
Mutational analysis of the flagellin glycosylation locus
As mentioned above, mutants in 6 genes in this locus (Cj1293/pseB, Cj1317, Cj1316c/pseA, Cj1315c, Cj1314c and Cj1337) in 81–176 have been reported previously (Thibault et al., 2001; Goon et al., 2003; Karlyshev and Wren, 2005). A total of 17 additional genes in this region were mutated. The two genes that were not mutated were Cj1299 and Cj1292. No transposon insertion was isolated in Cj1299; in the case of Cj1292 a transposon insertion was obtained in E. coli, but multiple attempts to electroporate the mutated allele into 81–176 were unsuccessful. All mutants were screened for motility and, if positive, flagellins were purified and characterized on IEF gels (Doig et al.1995; Thibault et al., 2001; Logan et al., 2002; Goon et al., 2003). Mutants displaying an altered IEF pattern were then examined by mass spectrometry. The data are summarized in Table 1 and those genes that appear to be involved in glycosylation have been designated “pse” (for biosynthesis or attachment of PseAc or PseAm; Thibault et al., 2001; Goon et al., 2003) as indicated in Table 1, and this nomenclature will be used throughout.
In addition to the previously described non-motile 81–176 mutants in Cj1317 or pseI (Thibault et al., 2001) and Cj1337 or pseE (Karlyshev and Wren, 2005), mutation of 3 additional genes resulted in a totally non-motile phenotype. These included the homolog of Cj1311, which encodes a protein with similarity to NeuA, the enzyme that activates NeuNAc; this gene has been reannotated as pseF. Mutation of pseF has been shown to result in a non-motile phenotype in C. jejuni strain G1, but not NCTC 11168 (Linton et al., 2000), which is likely a reflection of the presence of the alternate ptm pathway for PseAm synthesis in the latter strain (Parkhill et al., 2000). 81–176 mutants in Cj1312 and Cj1313, which were originally annotated in the NCTC 11168 genome as a putative flagellar genes based on homology to putative flagellin glycosylation genes of Caulobacter crescentus, were also non-motile; these genes have been called pseG and pseH, respectively. Mutation of Cj1294, designated pseC (Schoenhofen et al., 2006), also resulted in a non-motile phenotype, and it has been shown to be involved in synthesis of PseAc (Fig. 1; Schoenhofen et al., 2006). An 81–176 mutant in Cj1337 generated in this study was also non-motile, as previously reported for NCTC 11168 maf5 (Karlyshev et al., 2002) and more recently for 81–176 (Karlyshev and Wren, 2005); this 81–176 gene has been called pseE. Mutants in pseF, pseG, pseH and pseE lacked flagella filaments and hook structures as determined by transmission electron microscopy (data not shown). When the pseC and pseE mutants were complemented in trans using an expression plasmid (Larsen et al., 2004), motility was restored (data not shown). Although the mutations in pseG and pseH were not complemented, 20 independent mutations in pseG and 14 independent mutations in pseH were non-motile, suggesting that the phenotype was not due to phase variation at a distal locus.
In addition to the previously reported Cj1314c and Cj1315c (Thibault et al., 2001), mutation of 11 other genes resulted in a fully motile phenotype with no discernible changes in flagellin glycosylation based on IEF analysis. These genes, most of which encode proteins of unknown function, were Cj1295, Cj1300c, Cj1298, CjB1301, Cj1302, Cj1303, Cj1304, Cj1305c, Cj1334, Cj1341, and Cj1342. Mutation of Cj1333 in 81–176 is discussed below.
Characterization of a mutant in Cj1333
Although Cj1333 was designated as maf2 based on homology to other maf genes (Karlyshev et al, 2002), a mutant in 81–176 was fully motile. IEF analysis of the flagellin from a Cj1333 mutant was intermediate between wildtype and a pseA mutant as shown in Fig. 2A. The molecular mass profile obtained from the nanoelectrospray mass spectrometry analysis of C. jejuni Cj1333 mutant flagellin is presented in Figure 3A, and shows a distinct pattern to that of the wild type strain of C. jejuni or the Cj1316c mutant (Thibault et al., 2001). The MS-MS analysis of the multiply-charged flagellin precursor ions at m/z 985.5 revealed the presence of PseAc residues (m/z 317) with no detectable PseAm (Figure 3B). The modified residue previously observed in 81–176 flagellin at m/z 486 is now observed at m/z 487 suggesting a conversion of PseAm to PseAc. Confirmation of this was obtained from the second generation product ion of m/z 487 produced from in-source fragmentation of the flagellin multiply-charged ions (Figure 3C). The MS-MS spectrum of m/z 487 clearly shows a fragment ion at m/z 171 for the acylium ion of the GlnAc residue together with fragment ions at m/z 317, 299 and 281 corresponding to the oxonium ion of PseAc and consecutive H2O losses. This top-down analysis indicated that Cj1333 mutant strain has lost the ability to decorate flagellin with PseAm (m/z 316) and PseAmOGlnAc (m/z 486) residues and can now only glycosylate the flagellin monomer with N-acetylated monosaccharide residues (PseAc, m/z 317) which at certain sites are further substituted with OGlnAc (m/z 487). Complementation of the mutant in trans on a shuttle plasmid resulted in a restoration of the wildtype IEF pattern, as shown in Fig. 2A. Intact mass analysis of purified flagellin from the complemented mutant showed a return to the more heterogeneous wildtype profile (Fig. 4A). The MS/MS fragmentation pattern of the precursor ion m/z 486 from the complemented mutant (Fig. 4B) is consistent with that obtained from the precursor ion m/z 486 from 81–176 (see Schirm et al., 2005). These data confirmed restoration of both the m/z 316 and 486 groups on flagellin, although the amounts of these glycans were reduced compared to wildtype, suggesting possible instability of the complementing plasmid. The data suggest that the product of Cj1333 is involved in attachment of PseAm and the gene has been annotated as pseD.
Fig. 2. Isoelectric focusing pattern of flagellins.
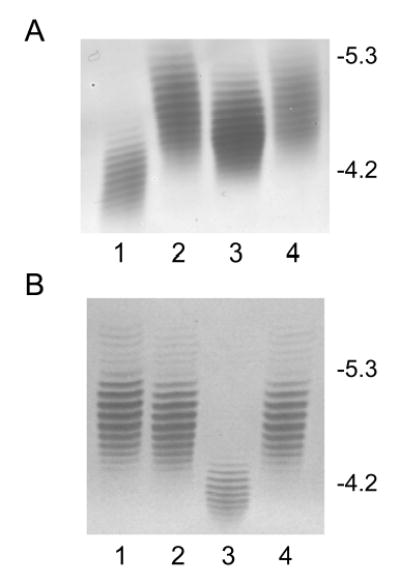
Purified flagellins from 81–176 and various mutants were separated on an ampholyte mixture of pH 4–6 and stained with Gel Code Blue. A. Characterization of flagellin from the pseD mutant. Lane 1, 81–176 pseA::cat (Thibault et al., 2001); lane 2, wildtype 81–176; lane 3, 81–176 pseD::cat; lane 4, 81–176 pseD::cat (pRY107/28+pseD). B. Complementation of pseA by insertion into atsA. Lane 1, 81–176 atsA::aphA3; lane 2, wildtype 81–176; lane 3, 81–176 pseA::cat; lane 4, 81–176 pseA::cat, atsA::pseA+aphA3. The position of IEF markers is indicated on the right.
Fig. 3. Structural analysis of flagellin from a mutant in Cj1333.
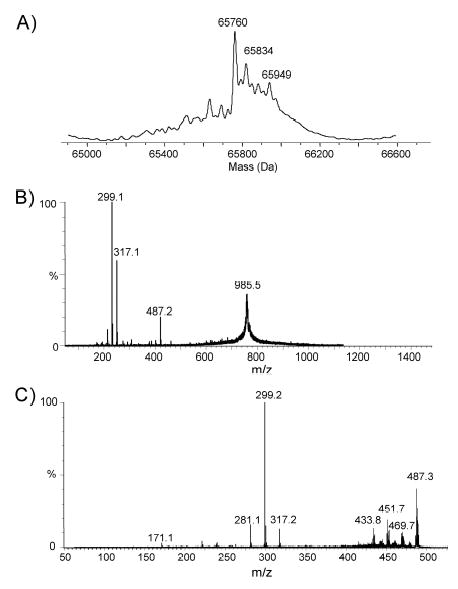
A. Molecular mass determination of intact flagellin from the mutant in pseD. B. Tandem mass spectrometry analysis of the multiply charged ion at m/z 985 from flagellin from the pseD mutant. C. Second generation product ion spectra of oxonium ion m/z 487 obtained from in source dissociation of multiply charged flagellin ion. Conditions: In-source fragmentation was obtained by increasing the RF Lens 1 from 50 to 125 V to produce oxonium fragment ions (m/z 487) from the native flagellin in the orifice/skimmer region. Argon was used as target gas at collision energy of 30 V.
Fig. 4. Structural analysis of flagellin from 81–176 pseD (pRY107/pseD).
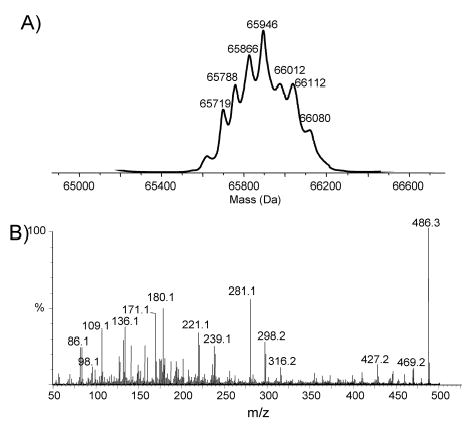
A. Molecular mass determination of intact flagellin from the complemented strain.
B. Second generation product ion spectra of oxonium ion m/z 486 obtained from in source dissociation of a multiply charged flagellin ion. The presence of m/z 316 product ion confirms the restoration of both groups on this flagellin (see Schirm et al., 2005). Conditions used for in source fragmentation are as described in figure 3.
Changes in flagellin modifications affect autoagglutination
Autoagglutination (AAG) of C. jejuni has been shown to be associated with flagellin (Misawa and Blaser, 2000), and possibly flagellin glycans (Golden and Acheson, 2002), and to be dependent on quorum sensing (Jeon et al., 2003). We examined this further using a set of mutants described in Table 2. As shown in Fig. 5A, a mutant of 81–176 in flaA (Yao et al., 1994) failed to autoagglutinate (column 2), similar to results reported by Misawa and Blaser (2000) for a flaA flaB mutant of 81–176. A luxS mutant of another C. jejuni strain has been reported to be reduced in both AAG and motility (Jeon et al., 2003). However, an 81–176 luxS mutant was fully motile and flagellin purified from the strain showed no change in IEF pattern (data not shown). The luxS mutant (column 6) showed an intermediate level of AAG between the levels of flaA (column 2) and wildtype 81–176 (column 1). A paralyzed mutant of 81–176 that synthesizes a filament, but is non-motile (Yao et al., 1994) autoagglutinated as well as wildtype (column 3), indicating that flagella structure rather than motility is required for AAG. We also examined previously described mutants of 81–176 in two other surface structures, kpsM, lacking capsular polysaccharide (Bacon et al., 2001), and neuC, which has an altered LOS core lacking NeuNAc (Guerry et al., 2001). The neuC mutant (column 5) showed an AAG level comparable to luxS, whereas the capsule mutant was only slightly less proficient in AGG (column 4) than wildtype.
Table 2.
Other mutants used in this study.
| Parent Strain | Genotype | Phenotype | Reference |
|---|---|---|---|
| C. jejuni 81–176 | |||
| pseA::cat | lacks PseAm on flagellin | Thibault et al. (2001) | |
| pseA::cat, astA::pseA, aph3 | cis complement of pseA::cat; AstA- | This study | |
| astA::aph3 | AstA- | This study | |
| pseD::cat | lacks PseAm + 486 Da group on flagellin | This study | |
| pseD::cat (pRY107/pseD) | in trans complement of pseD::cat | This study | |
| flaA:: aph3 | non-motile; non-flagellated | Yao et al. (1994) | |
| pflA::aph3 | paralyzed flagella; non-motile | Yao et al. (1994) | |
| kpsM::aph3 | non-encapsulated | Bacon et al. (2001) | |
| neuC::cat | lacks sialic acid on LOS | Guerry et al. (2001) | |
| luxS::cat | quorum sensing mutant | This study | |
| C. coli VC167 T2 | |||
| ptmD::aph3 | lacks PseAm on flagellin | Logan et al. (2002) | |
| ptmD::aph3 (pRY111/ptmD) | in trans complement of ptmD::aph3 | Logan et al. (2002) | |
| pseB::cat | lacks PseAc on flagellin | Goon et al. (2003) | |
| pseB::cat (pRY107/pseB) | in trans complement of pseB::cat | Goon et al. (2003) | |
Fig. 5. Autoagglutination of Campylobacters.
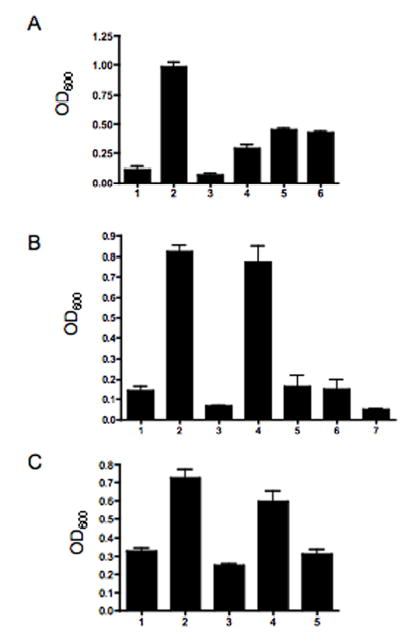
Suspensions of bacteria were set to an OD600 of 1.0 and incubated without shaking. After 18 h incubation at room temperature, the OD600 of the top 1 ml of the tube determined. Mutants used in this study are listed in Table 2. A. Comparison of wildtype 81–176 with isogenic mutants. Column 1, wild-type 81–176; column 2, 81–176 flaA::aph3; column 3, 81–176, pflA::aph3; column 4, 81–176 kpsM::aph3; column 5, 81–176 neuC1::cat; column 6, 81–176 luxS::cat. B. Flagellin glycosylation mutants of C. jejuni 81–176. Column 1, wild-type 81–176; column 2, 81–176 pseD::cat; column 3, 81–176 pseD::cat (pRY107/pseD); column 4, 81–176 pseA::cat; column 5, 81–176 pseA::cat, astA::pseA, aph3; column 6, 81–176 astA::aph3; column 7, 81–176 CjB1301::cat. C. Flagellin glycosylation mutants of C. coli VC167 T2. Column 1, wild-type VC167 T2; column 2, VC167 T2 ptmD::aph3; column 3, VC167 T1 ptmD::aph3 (pRY111/ptmD); column 4, VC167 T2 pseB::cat; column 5, VC167 T2 pseB::cat (pRY107/pseB). The graphs show the mean and standard deviations of 2–8 independent determinations.
Fig. 5B shows AGG using 81–176 glycosylation mutants. Mutants in pseA and pseD both lacked PseAm on flagellin and both failed to autoagglutinate (columns 2 and 4). AAG was restored when the mutations were complemented either in trans for pseD (column 3) or by insertion of a wildtype allele into the arylsulfatase (astA) gene for pseA (column 5). A control for the atsA insertion is also shown (atsA::aph3; column 6). The mutant in CjB1301 (column 7), which showed no detectable flagellin changes by IEF (Table 1), agglutinated as well as wildtype (column 1).
VC167 has the genes encoding the ptm pathway for synthesis of PseAm independent of PseAc synthesis, and the pseA gene, which converts PseAc to PseAm in C. jejuni 81–176 (Goon et al., 2003), is a pseudogene in VC167 (Logan et al., 2002). Thus, mutants have been described in C. coli VC167 that lack either all PseAm or lack all PseAc (Guerry et al., 1996; Logan et al., 2002). We used these mutants in AAG assays to confirm and extend the observations made with C. jejuni 81–176 and the results are shown in Fig. 5C. The VC167 ptmD mutant, that has flagellin substituted with PseAc in place of PseAm, failed to autoagglutinate (column 2), but AAG was restored by complementation with the ptmD gene in trans (column 3). A mutant in pseA, which has PseAm moieties in sites normally occupied by PseAc (Goon et al., 2003), also failed to autoagglutinate (column 4); the ability of this mutant to autoagglutinate could also be complemented in trans (column 5).
Effect of flagellin modification/autoagglutination on adherence and invasion of intestinal epithelial cells
Since the ability of C. jejuni to autoagglutinate has been associated with changes in adherence and/or invasion (Misawa and Blaser, 2000; Golden and Acheson, 2002), the ability of wildtype 81–176 and the pseA mutant to adhere to and invade intestinal epithelial cells was compared. The results, shown in Fig 6A, indicated that the pseA mutant (column 2) adhered to INT407 cells at about 20% the level of wildtype (column 1) and that the defect was restored in the complemented strain (column 3). A control, in which only a kanamycin resistance gene was inserted into atsA adhered at levels comparable to wildtype (column 4). The effect of mutation of pseA on invasion of INT407 cells was more modest (Fig. 6B); the pseA mutant (column 2) invaded at about 42% the level of wildtype (column 1), although invasion levels were increased in the complemented strain (column 3).
Fig. 6. The effect of loss of PseAm on adherence and invasion of INT407 cells.
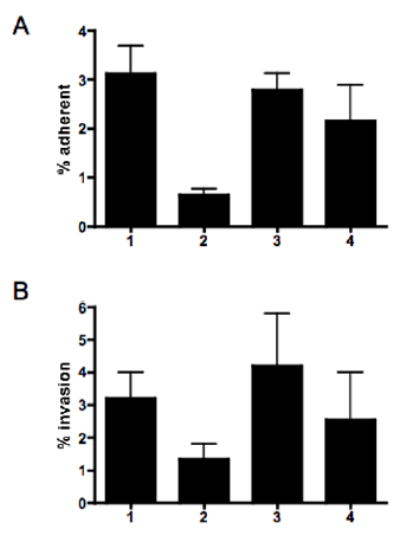
A. Adherence. B. Invasion. Column 1, wild-type 81–176; column 2, 81–176 pseA::cat; column 3, 81–176 pseA::cat, astA::pseA+aphA3; column 4, 81–176 atsA::aphA3.
Comparison of patterns of adherence of GFP tagged bacteria to INT407 cells
Since autoagglutination is often associated with microcolony formation in other pathogens (Skurnik et al., 1984; Nataro et al., 1987; Blake et al., 1995; Chiang et al., 1995; Roggenkamp et al., 1995; Bieber et al., 1998; Knutton et al., 1999; Frick et al., 2000; Kirn et al., 2000; Schembri et al., 2001; Sherlock et al., 2005), the patterns of adherence of wildtype 81–176 and the pseA mutant, each of which was carrying pCE111/28/GFP (Hickey et al., 2005), to INT407 cells were compared. Examination of INT407 cells infected with either strain at 6 h showed single fluorescent bacteria (data not shown). However, after 18 h of incubation, distinct patterns could be observed in the wildtype 81–176 but not the pseA mutant (Fig. 7A). The wildtype strain often formed apparent chains, which were also visible in the phase contrast images, while the mutant adhered as single cells. By 24 h (Fig. 7B and 7C), the wildtype strain had formed apparent microcolonies and chains of bacteria were often visible protruding from the microcolony. The amount of binding of the pseA mutant at 24 h appeared to increase, but the pattern of mutant binding remained more diffuse than that of wildtype (Fig. 7B). The pattern of binding of a GFP-tagged version of the pseD mutant was indistinguishable from that of the pseA mutant (data not shown).
Fig. 7. Comparison of adherence patterns to INT407 cells of GFP tagged wildtype 81–176 with GFP tagged 1906 mutant.
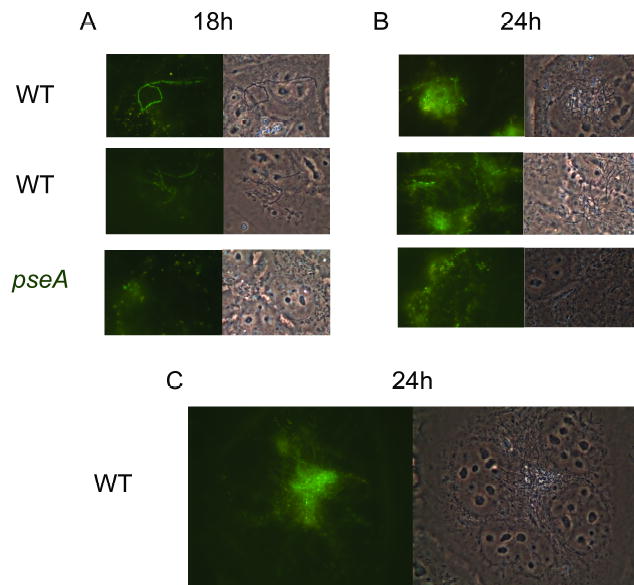
Panel A shows the adherence patterns of wildtype 81–176 and the pseA mutant following 18 h incubation. Panels B and C show adherence patterns following 24 h incubation. Fluorescence images are on the left and phase images are on the right. Final magnification of panels A and B, 1000X; final magnification of panel C, 1500X.
Effect of loss of PseAm on virulence in vivo
To determine if these differences in adherence patterns corresponded to decreased virulence in vivo, the pseA mutant and the complemented strain were compared to wildtype 81–176 in the ferret diarrheal disease model. A total of 16 animals were fed 3.8–4.5 x 1010 CFU of each strain and the numbers of animals developing diarrhea over the next 4 days was monitored. The results are summarized in Fig. 8. A total of 10/16 animals fed wildtype 81–176 and the complemented mutant developed diarrhea compared to only 3/16 animals fed the pseA mutant (P=0.029).
Fig. 8. Virulence in ferrets.
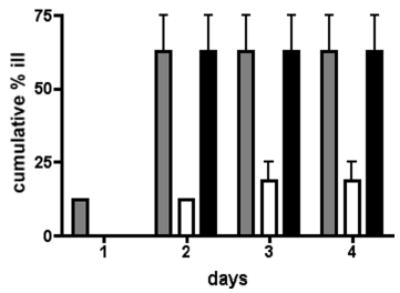
Animals were fed C. jejuni at doses ranging from 3.8 x 1010–4.5 x 1010 and monitored for diarrheal disease for 4 days. The data are presented as the cumulative % animals that developed diarrhea over time. Grey bars, 81–176; white bars, 81–176 pseA::cat; black bars, 81–176 pseA::cat, astA::pseA + aph3. The data represent the two separate experiments with 8 animals per group or a total number of 16 animals per strain.
Discussion
The flagellin glycosylation locus of C. jejuni 81–176 is much simpler than the corresponding locus in C. jejuni NCTC 11168, having approximately half the number of genes and lacking the so-called “second set” of sialic acid genes or ptm genes that are directly involved in synthesis of PseAm (Guerry et al., 1996; Logan et al., 2002). Thus, the region corresponding to the 45 genes spanning Cj1293 to Cj1337 in NCTC 11168 contains 21 genes in 81–176, and mutation of only 9 of these resulted in a phenotype affecting modification of flagellin with either PseAc or PseAm. Mutation of a total of seven genes resulted in a non-motile phenotype. This suggests that these genes are involved in either synthesis of PseAc, through which PseAm is synthesized in this strain, or attachment of both PseAc and PseAm to flagellin. As shown in Fig. 1, four of these genes, pseB, pseC, pseF and pseI, encode enzymes in the PseAc pathway. The roles of PseG, PseH, and PseE remain to be elucidated.
The loss of PseAm in mutants of pseA and pseD suggests that both genes encode proteins that are part of a pathway in biosynthesis of PseAm from PseAc or transfer of PseAm to flagellin. While flagellin from the pseD mutant had lost both PseAm and the novel group of m/z 486, this mutant now appeared capable of transfer of OGlnAc to PseAc (m/z 487). In contrast, the pseA mutant, while losing both m/z 316 and m/z 486 was unable to transfer OGlnAc to PseAc for reasons that remain to be determined. The enzyme or enzymes responsible for attachment of this novel OGlnAc group remain unknown, although the data suggest that PseA may influence this process. Since the predicted phenotype of a mutant lacking this m/z 486 group would be motile with no change in IEF pattern, there are numerous candidate genes within the glycosylation locus, although identification of this gene will require MS analysis of flagellins from each mutant.
All five maf gene homologs were mutated in 81–176 but mutants in only two, pseD and pseE, showed an observable change in flagellin. As reported here and previously by another group (Karlyshev and Wren, 2005), the 81–176 pseE mutant is non-motile, which is the predicted phenotype of maf mutants in NCTC 11168 (Karlyshev et al., 2002). Colegio et al. 2001 reported that a transposon insertion into Cj1318, one of the maf genes, of 81–176 was non-motile, but the data presented here indicate that there is no copy of Cj1318 in the flagellin glycosylation locus of 81–176. Either there is a Cj1318 copy elsewhere on the chromosome, or the insertion was actually in pseE, which shows the highest homology (39% identity/53% similarity) to Cj1318. However, the motile phenotype and the observed changes in glycosylation of the pseE mutant are different than what is predicted for a maf gene phenotype.
Spontaneous autoagglutination of C. jejuni strains has long been recognized as a problem in serotyping schemes (Lior et al., 1982; Penner and Hennessy, 1980; Wong et al., 1985). The phenomenon was shown to be associated with flagella by Misawa and Blaser (2000) and shown in the same study to correlate with cell surface hydrophobicity. A kpsM mutant of 81–176 showed reduced hydrophobicity (Bacon et al., 2001), a result that is consistent with a reduction in AAG for this mutant. The involvement of flagella in AAG was confirmed in a random transposon mutagenesis screen of C. jejuni strain 480 (Golden and Acheson, 2002). In that study Golden and Acheson also found non-agglutinating mutants in genes in the glycosylation locus, although the genes were not further characterized and they first reported reduced AAG in ptmA and ptmB mutants of VC167. Here, we have demonstrated that all mutants in 81–176 and C. coli VC167 that result in loss of PseAm residues from flagellin fail to autoagglutinate. Since PseAm is made via PseAc in 81–176, it is not possible to construct an 81–176 mutant that can decorate flagellin with only PseAm. However, we have previously described such a mutant in C. coli VC167 defective in pseB (Goon et al., 2003), and that mutant also showed reduced AAG. Flagellin from the VC167 mutant in pseB also had an unusual phenotype in that it was more sensitive to dissociation in 1% SDS, which is used during the flagellin purification scheme (Goon et al., 2003), and suggests that interactions among residues of PseAc and PseAm on flagellin monomers within the filament enhances the physical strength of the flagellum. The requirement for both PseAm and PseAc in the AAG process also suggests that interactions of residues across flagella filaments may be critical for the aggregative interaction. The slight reduction in AAG in a NeuNAc mutant was unexpected and requires additional investigation, but it may be that there are heterophilic interactions between PseAm residues on flagellin and the structurally related NeuNAc on LOS, similar to interactions seen between opacity proteins and sialylated LOS in Neisseria (Blake et al., 1995). This is the first report, to our knowledge, of the role of a glycan on a glycoprotein in bacterial autoagglutination. Although the glycoprotein adhesin TibA from enterotoxigenic E. coli mediates autoagglutination, the glycan is required for attachment to epithelial cells, but not for agglutination (Benz and Schmidt, 2001; Sherlock et al., 2005).
AAG has been demonstrated to be critical for virulence for a variety of pathogens, and can play a role in adherence, microcolony formation, biofilm formation, and resistance to acid and phagocytosis (Skurnik et al., 1984; Galdiero et al., 1988; Chiang et al., 1995; Frick et al., 2000; Sherlock et al., 2005). In two previous studies on AAG of C. jejuni (Misawa and Blaser, 2000; Golden and Acheson, 2002), there appeared to be an association with adherence or invasion of intestinal epithelial cells. The quantitative defects in adherence and invasion of intestinal epithelial cells of the pseA mutant of 81–176 in a standard 2 hour assay were relatively minor. However, wildtype 81–176 appears able to form distinct microcolonies on INT407 cells upon longer incubation, while mutants lacking PseAm cannot. This distinction in vitro correlates well with the observed attenuation of the pseA mutant in vivo in the ferret model. This suggests that the changes in glycosylation/AAG relate more to microcolony formation during intestinal colonization rather than to primary adherence, a distinction that has been made for several other pathogens (Bieber et al., 1998; Blake et al., 1995; Chiang et al., 1995; Frick et al., 2000; Kirn et al., 2000; Kirn and Taylor, 2005; Knutton et al., 1999). The precise role of flagellin glycosylation in the biology and pathogenesis of C. jejuni will require additional investigation, but the data presented here suggest that the glycans decorating the surface of the flagella filament play a key role in virulence.
Experimental Procedures
Strains and growth conditions
C. jejuni 81–176 has been described (Black et al., 1988). Campylobacter strains were grown routinely in Mueller Hinton agar under microaerobic conditions. Media were supplemented with chloramphenicol (10 μg/ml), trimethoprim (10 μg/ml) or kanamycin (25 μg/ml) when appropriate. Escherichia coli XL-1 Blue was the host for λZAP Express cloning experiments and DH5α was the host for subcloning and routine DNA manipulation. Shuttle plasmids were mobilized from E. coli DH5α using RK212.2 into C. jejuni, as previously described (Guerry et al., 1994). Wildtype 81–176 and mutants were tagged with GFP by conjugal transfer of shuttle plasmid pCPE111/28/GFP (Hickey et al., 2005).
DNA cloning and generation of mutants
Genes were identified from partial Sau3A libraries of C. jejuni 81–176 constructed in λZAP Express (Stratagene, La Jolla, CA). Cloned DNAs were subjected to sequence analyses using an Applied Biosystems Model 3100 DNA sequencer. Custom sequencing primers were synthesized on a Perkin Elmer Applied Biosystems Model 294 DNA synthesizer. Additional DNA sequence was obtained following transposition into some clones using an in vitro Tn5-based tranposition system (Epicentre, Madison, WI) with a campylobacter chloramphenicol resistance (cat) cassette and primers within the cassette, as previously described (Guerry et al., 2000). Gaps in the region were closed by polymerase chain reaction (PCR) using the proof-reading polymerase PfuI (Stratagene, LaJolla, CA). Transposon insertions identified during the sequencing phase were used to generate knockout mutations in C. jejuni 81–176 by electroporation. All mutants were confirmed to have undergone a double crossover by PCR with primers that bracketed the insertion point of the transposon. The position of transposon insertions in genes in the flagellin glycosylation locus are listed in Table 1. A non-polar transposon insertion that mapped 37 bp from the translational start of luxS was used to generate a luxS::cat mutant in C. jejuni 81–176.
Purification of flagellin
Flagellins were purified as described previously (Power et al. 1994).
Complementation of mutants in trans
Three mutants were complemented by expressing the wildtype alleles in a kanamycin resistant σ28 expression vector, pRY107/28 (Larsen et al., 2004). Each final construction was mobilized from E. coli DH5α into the respective mutant by RK212.1 with selection on trimethoprim, kanamycin and chloramphenicol (Labigne-Roussel et al., 1987). The pseD gene was PCR amplified with PfuI high fidelity polymerase (Stratagene) using the following primers: Cj1333F: 5’-CGGGATCCATGAAATTTAATTTAAATCAAAAAGAGC-3’ and Cj1333R: 5’-CCGCTCGAGTTCATTTGTTTGCATTTTTTATCC-3’. The primers introduced BamHI and XhoI sites, respectively. Following digestion with these restriction enzymes (New England Biolabs, Beverly, MA) the 1.9 bp amplicon was cloned into pCE107/28 (Larsen et al., 2004) that had also been cut with BamHI and XhoI.
The pseE gene was PCR amplified using Advantage HF2 high fidelity polymerase (BD Sciences, Clontech, San Jose, CA) with the following primers that introduced BamHI and EcoR1 sites: Cj1337F: 5’-CGGGATCCCGATGCAAACAAATGAAATTTTTAAAAAAAATTTAG-3’ and Cj1337R: 5’-GGAATTCCTTAGATTAAGCTTCTTTTTTCTAGCTCATCC-3’. The resulting 1.9 kb amplicon was digested with BamHI and EcoR1 and cloned into pCE107/28 (Larsen et al, 2004).
The pseC gene was PCR amplified using Advantage HF2 high fidelity polymerase with the following primers that introduced BamHI and EcoR1 sites, respectively: Cj1294F: 5’-CGGGATCCCGATGATTACTTATTCTCATCAAAATAATGATC-3’ and Cj1294R: 5’-GGAATTCCTTATCCACAATATCCCTTTTTAACTTTTTC-3’. The resulting 1131 bp product was digested with BamHI and EcoR1 and cloned into pCE107/28.
Complementation of Cj1316c by insertion into atsA
Although trans complementation of pseA has been described previously (Thibault et al., 2001), a strain was constructed in which a wildtype allele of pseA was inserted into the arylsulfatase gene (atsA; Yao and Guerry, 1996) of the 81–176 pseA mutant. This was done because some plasmid constructions are lost at relatively high frequency without antibiotic selection during adherence and invasion assays (Guerry, unpublished observations). The pseA gene was amplified by PCR from 81–176 using the following primers: 5’-TGTATGGAATAATCCTTAGGTGGTG-3’ and 5’-AGTTTTACCCGCTTTGCTTGC-3’. The PCR reaction was done for 30 cycles of 95C/1 min; 52C/1 min; 72C/1min using PfuI, and the 517 bp product was cloned into PCR-Script (Stratagene, La Jolla, CA). Clones were sequenced with the appropriate forward and reverse sequencing primers to determine the orientation of the pseA gene and to verify the sequence of the PCR amplicon. A clone containing pseA oriented such that the 3’ end was adjacent to the PstI site was selected for further manipulation. A kanamycin resistance cassette (aph3) from pILL600 (Labigne-Roussel et al., 1987) was subsequently cloned into the PstI site of this plasmid. The orientation of this cassette was determined to be in the same orientation as the pseA gene by sequence analysis. A NotI-XhoI fragment containing both pseA and Kmr was purified, made blunt-ended with Klenow I, and cloned into the unique EcoRV site within the atsA gene on pYG660 (Yao and Guerry, 1996). This plasmid was used to electroporate 81–176 pseA::cat (Thibault et al, 2001) to kanamycin resistance. Kmr colonies were screened for loss of arylsulfatase activity by growth on MH agar containing a chromogenic substrate (X-S; Sigma), as previously described (Yao and Guerry, 1996). Kmr Colonies that were phenotypically Ats-were confirmed to have the predicted genotype by PCR. One such mutant was further characterized. A control strain was also generated in which the Kmr cassette was inserted into atsA of 81–176.
Motility testing
Motility of mutants was compared with that of wildtype on semi-solid (0.4%) MH agar plates as previously described (Guerry et al., 1991).
IEF gels
Isoelectric focusing gels were prepared as previously described (Doig et al., 1995; Goon et al., 2003; Logan et al., 2002; Thibault et al., 2001).
Mass spectroscopy
Flagellin extracts were dialyzed in water (0.2% formic acid, FA) using Centricon YM-30 membrane filters (Millipore) to a final concentration of 0.2 mg/ml prior to intact mass analyses on a Waters Q-TOF™ Ultima mass spectrometer. This solution was infused into the mass spectrometer at a flow rate of 0.5 ul/min. External calibration of the TOF was performed using a 150 fmoles/ml solution of Glu-Fibrinopeptide B (50% aqueous methanol, 0.2% formic acid) and provided mass accuracy typically within ± 0.07 m/z unit across the acquisition range (m/=50–1400). Molecular mass profiles were obtained through spectal deconvolution using MaxEnt (MassLynz softwar, Waters). Collision-induced dissocation (CID) experiments were performed using argon as collision gas with differential voltage values of 10–25 V between the collision cell and the incoming ions. Second generation fragment ion spectra were obtained by increasing the RF lens 1 voltage from 50 to 125 V, thereby forming fragment ions in the high-pressure region of the skimmer/cone region of the mass spectrometer.
Autoagglutination assays
AAG assays were done as described by Misawa and Blaser (2000). Basically, bacterial growth from a MH plate was resuspended in PBS, pH 7.4 to an OD600 1.0. Two ml of each suspension was transferred to 12 x 75 mm polypropylene tubes, and incubated at room temperature for 24 h. One ml from the top of each tube was carefully removed and the OD600 measured. The drop in OD600 from the initial setting of 1.0 reflected the degree of autoagglutination.
Adherence and invasion assays
Adherence and invasion assays to INT407 cells were done at an MOI of approximately 20, as previously described (Oelschlaeger et al., 1993; Yao et al., 1997). For adherence assays, bacteria were incubated with the monolayer for 2 h at 37°C in 5% CO2/95% air. The cells were then washed four times with Hank’s balanced salt solution with strong agitation for 2 min prior to lysing the monolayer with 0.01% Triton X-100 and enumeration of total bacteria by plate count. For determination of invasion, the monolayer was washed twice with Hank’s balanced salt solution after the 2 h incubation, and fresh, pre-warmed medium containing gentamicin at 100 ug/ml was added to kill extracellular bacteria. After 2 h incubation, the monolayer was washed twice with Hank’s balanced salt solution, and lysed with 0.01% Triton X-100. The released intracellular bacteria were enumerated by plate count. For adherence assays using GFP labeled bacteria, INT407 cells were seeded onto cover slips. After infection with GFP labeled bacteria, the assays were incubated at 37°C for 6, 18 and 24 h and washed four times with Hank’s Balanced Salt Solution prior to examination in a Nikon Eclipse E400 fluorescence microscope. Images were photographed with an Olympus camera using MagnaFire software and enhanced with Adobe Photoshop 7.0.
Electron microscopy
Mutants were examined by transmission electron microscopy following negative staining with uranyl acetate to determine the presence or absence of a flagella filament.
Ferret diarrheal model
Campylobacter-free, six-week old female ferrets were purchased from Triple F Farms (Sayre, PA). After one week acclimation, animals were infected with 1.4–2.7 x 1010 bacteria by oral gavage as previously described (1) and observed for signs of diarrheal illness for four days. Experiments were done in duplicate with eight animals per group or a total of 16 animals per strain. The experiments were conducted in compliance with the Animal Welfare Act and according to the principles set forth in the “Guide for the Care and Use of Laboratory Animals,” Institute of Laboratory Animals Resources, National Research Council, National Academy Press, 1996. The P value was calculated by Fishers Exact Test using GraphPad Instat software (GraphPad, San Diego, CA).
Accession number
The complete flagellin glycosylation locus of C. jejuni 81–176 has been deposited in Genbank as AY102622.
Acknowledgments
This work was supported by NIAID grant RO1 AI43559 (to PG) and Navy Work unit no. 6000.RAD1.DA3.A0308 from the Military Infectious Diseases Program, and the Genomics Health Initiative at NRC (SML and JFK). We thank Robert Williams of NMRC for electron microscopy. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the US government.
References
- Bacon DJ, Szymanski CM, Burr DH, RP Silver RP, Alm RA, Guerry P. A phase variable capsule is involved in virulence of Campylobacter jejuni 81–176. Mol Microbiol. 2001;40:769–777. doi: 10.1046/j.1365-2958.2001.02431.x. [DOI] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Glycoylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA adhesin. Mol Microbiol. 2001;40:1403–1413. doi: 10.1046/j.1365-2958.2001.02487.x. [DOI] [PubMed] [Google Scholar]
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- Blake MS, Blake CM, Apicella MA, Mandrell RE. Gonococcal opacity: lectin-like interactions between Opa proteins and lipooligosaccharide. Infect Immun. 1995;63:1434–1439. doi: 10.1128/iai.63.4.1434-1439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Taylor RK, Koomey M, Mekalanos JJ. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination and serum resistance. Mol Microbiol. 1995;17:1131–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- Chou WK, Dick S, Wararchuk WW, Tanner ME. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem. 2005;280:35922–35928. doi: 10.1074/jbc.M507483200. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Griffin IV TJ, Grindley NDF, Galan JE. In vitro transposition system for efficient generation of random mutants of Campylobacter jejuni. J Bacteriol. 2001;183:2384–2388. doi: 10.1128/JB.183.7.2384-2388.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig P, Kinsella N, Guerry P, Trust TJ. Characterization of a posttranslational modification of Campylobacter flagellin: identification of a sero-specific moiety. Mol Microbiol. 1995;19:379–387. doi: 10.1046/j.1365-2958.1996.370890.x. [DOI] [PubMed] [Google Scholar]
- Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, Barrell BG, Parkhill J, Stoker NG, Karylshev AV, Butcher PD, Wren BW. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers KT, Park SF. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signaling molecule. Microbiol. 2002;148:1475–1481. doi: 10.1099/00221287-148-5-1475. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Dougherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLOS Biology. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick IM, Morgelin M, Bjorck L. Virulent aggregates of Streptococcus pyogenes are generated by homophilic protein-protein interactions. Mol Microbiol. 2000;37:1232–1247. doi: 10.1046/j.1365-2958.2000.02084.x. [DOI] [PubMed] [Google Scholar]
- Galdiero F, Romanao-Carratelli C, Nuzzo I, Bentivoglio C, Galdiero M. Phagocytosis of bacterial aggregates by granulocytes. Eur J Epidemiol. 1988;4:456–460. doi: 10.1007/BF00146398. [DOI] [PubMed] [Google Scholar]
- Golden NJ, Acheson DWK. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect Immun. 2002;70:1761–1771. doi: 10.1128/IAI.70.4.1761-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- Guerry P, Alm RA, Power ME, Logan SM, Trust TJ. Role of two flagellin genes in Campylobacter motility. J Bacteriol. 1991;173:4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P, Doig P, Alm RA, Burr DH, Kinsella N, Trust TJ. Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol Microbiol. 1996;19:369–378. doi: 10.1046/j.1365-2958.1996.369895.x. [DOI] [PubMed] [Google Scholar]
- Guerry P, Ewing CP, Hickey TE, Prendergast MM, Moran AP. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect Immun. 2000;68:6656–6662. doi: 10.1128/iai.68.12.6656-6662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P, Szymanski CM, Burr DH, Prendergast M, Ewing CP, Hickey TE, Pattarini D, Moran AP. Phase variation of Campylobacter jejuni 81–176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect Immun. 2001;70:787–793. doi: 10.1128/iai.70.2.787-793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry, P., Yao, R., Alm, R. A., Burr, D. H., and Trust, T. J. (1994) Systems of experimental genetics for Campylobacter sp. In Methods in Enzymol.235: Bacterial pathogenesis, part A, Identification and regulation of virulence factors. Academic Press, San Diego, California. [DOI] [PubMed]
- Hickey TE, Majam G, Guerry P. Intracellular survival of Campylobacter jejuni in human monocytic cells and induction of apoptotic death by cytolethal distending toxin. Infect Immun. 2005;73:5194–5197. doi: 10.1128/IAI.73.8.5194-5197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M, Waggoner LE, Liu H, Sudek S, Allen S, Anderson C, Sherman DH, Haygood H. An unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of themarine Bryozoan Bugula neritina. Chem Biol. 2004;11:1543–1552. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Jeon B, Itoh K, Misawa N, Ryu S. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol Immunol. 2003;47:833–839. doi: 10.1111/j.1348-0421.2003.tb03449.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Linton D, Gregson NA, Wren BW. A novel paralogous gene family involved in phase variable flagella-mediated motility in Campylobacter jejuni. Microbiol. 2002;148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71:4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn TJ, Lafferty MJ, Sandoe CMP, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- Kirn TJ, Taylor RK. TcpF is a soluble colonization factor and protective antigen secreted by El Tor and Classical O1 and O139 Vibrio cholerae serogroups. Infect Immun. 2005;73:4461–4470. doi: 10.1128/IAI.73.8.4461-4470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Shaw RK, Anantha RP, Donnenberg MS, Zorgani AA. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;31:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A, Harel J, Tompkins L. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J Bacteriol. 1987;169:5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JC, Szymanski C, Guerry P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J Bacteriol. 2004;186:6508–6514. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H, Woodward DL, Edgar JA, Larouche LJ, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D, Karlyshev AV, Hitchen PG, Morris HR, Dell A, Gregson NA, Wren BW. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipooligosaccharide. Mol Microbiol. 2000;35:1120–1134. doi: 10.1046/j.1365-2958.2000.01780.x. [DOI] [PubMed] [Google Scholar]
- Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. Structural heterogeneity of carbohydrate modifications affects serospecificity of campylobacter flagellins. Mol Microbiol. 2002;46:587–597. doi: 10.1046/j.1365-2958.2002.03185.x. [DOI] [PubMed] [Google Scholar]
- Misawa N, Blaser MJ. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect Immun. 2000;68:6168–6175. doi: 10.1128/iai.68.11.6168-6175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. Patterns of adherence of diarrheagenic Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1987;60:2297–2304. [Google Scholar]
- Oelschlaeger TA, Guerry P, Kopecko DJ. Novel microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc Natl Acad Sci USA. 1993;90:6884–6888. doi: 10.1073/pnas.90.14.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–8. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pearson BM, Pin C, Wright J, I'Anson K, Humphrey T, Wells JM. Comparative genome analysis of Campylobacter jejuni using whole genome microarrays. FEBS Lett. 2004;554:224–230. doi: 10.1016/s0014-5793(03)01164-5. [DOI] [PubMed] [Google Scholar]
- Penner JL, Hennessy JN. Passive hemaglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power M, Guerry P, McCubbin WD, Kay CM, Trust TJ. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J Bacteriol. 1994;176:3303–3313. doi: 10.1128/jb.176.11.3303-3313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenkamp A, Neuberger HR, Flugel A, Schmoll T, Heesemann J. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol Microbiol. 1995;16:1207–1219. doi: 10.1111/j.1365-2958.1995.tb02343.x. [DOI] [PubMed] [Google Scholar]
- Schembri MA, Christiansen G, Klemm P. FimH-mediated autoaggregation of Escherichia coli. Mol Microbiol. 2001;41:1419–1430. doi: 10.1046/j.1365-2958.2001.02613.x. [DOI] [PubMed] [Google Scholar]
- Schirm M, Schoenhofen I, Logan SM, Waldron K, Thibault P. Identification of unusual bacterial glycosylation by tandem mass spectrometry analysis of intact proteins. Anal Chem. 2005;77:7774–7782. doi: 10.1021/ac051316y. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Riley L, Benz I. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 2003;45:267–561. doi: 10.1016/j.tim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young M, Dick S, Wararchuk WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self-recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73:1954–1963. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M, Bolin I, Heikkinen H, Piha S, Wolf-Watz H. Virulence plasmid-associated autoaglutination in Yersinia spp. J Bacteriol. 1984;158:1031–1036. doi: 10.1128/jb.158.3.1033-1036.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo EC, Aubry AJ, Logan SM, Guerry P, Kelly JF, Thibault P. The selective detection and identificationof sugar-nucleotides by CE-ESMS and its application to bacterial metabolomics. Anal Chem. 2004;76:619–626. doi: 10.1021/ac034875i. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, Trust TJ, Guerry P. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 2001;276:34862–34870. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- Vann WF, Silver RP, Abeijon C, Chang K, Aaronson W, Sutton A, Finn CW, Lindner W, Kotsatos M. Purification, properties, and genetic location of Escherichia coli cytidine 5’-monophosphate N-acetylneuraminic synthetase. J Biol Chem. 1987;262:17556–17562. [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Wong KH, Skelton SK, Patton CM, Feeley JC, Morris G. Typing of heat-stable and heat-labile antigens of Campylobacter jejuni and Campylobacter coli by coagglutination. J Clin Microbiol. 1985;21:702–707. doi: 10.1128/jcm.21.5.702-707.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Yao R, Burr DH, Doig P, Trust TJ, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni defective in invasion of eukaryotic cells: the role of flagella in invasion. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- Yao R, Guerry P. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J Bacteriol. 1996;178:3335–3338. doi: 10.1128/jb.178.11.3335-3338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


