Abstract
Stuttering is a speech disorder long recognized to have a genetic component. Recent linkage studies mapped a susceptibility locus for stuttering to chromosome 12 in 46 highly inbred families ascertained in Pakistan. We report here on linkage studies in 100 families of European descent ascertained in the United States, Sweden, and Israel. These families included 252 individuals exhibiting persistent stuttering, 45 individuals classified as recovered from stuttering, and 19 individuals too young to classify. Primary analyses identified moderate evidence for linkage of the broader diagnosis of “ever stuttered” (including both persistent and recovered stuttering) on chromosome 9 (LOD = 2.3 at 60 cM) and of the narrower diagnosis of persistent stuttering on chromosome 15 (LOD = 1.95 at 23 cM). In contrast, sex-specific evidence for linkage on chromosome 7 at 153 cM in the male-only data subset (LOD = 2.99) and on chromosome 21 at 34 cM in the female-only data subset (LOD = 4.5) met genomewide criteria for significance. Secondary analyses revealed a significant increase in the evidence for linkage on chromosome 12, conditional on the evidence for linkage at chromosome 7, with the location of the increased signal congruent with the previously reported signal in families ascertained in Pakistan. In addition, a region on chromosome 2 (193 cM) showed a significant increase in the evidence for linkage conditional on either chromosome 9 (positive) or chromosome 7 (negative); this chromosome 2 region has been implicated elsewhere in studies on autism, with increased evidence for linkage observed when the sample is restricted to those with delayed onset of phrase speech. Our results support the hypothesis that the genetic component to stuttering has significant sex effects.
Developmental stuttering is a communication disorder that begins in early childhood and is characterized by involuntary disruptions in the fluency of verbal expression. The most typical core behaviors are repetitions of sounds or syllables and articulatory fixation, resulting in blocks or prolongations of sounds. When the disorder continues into later childhood and beyond, it may cause serious interference with both personal and professional pursuits. Evidence from twin and family studies has clearly established a role for genetic factors in the development of stuttering (Howie 1981; Kidd 1984; Yairi et al. 1996). Increased concordance rates in MZ twins versus DZ twins have been consistently observed, with pairwise concordance estimates ranging from 20% to 83% for MZ twins and from 4% to 19% for DZ twins (Howie 1981; Andrews et al. 1991; Felsenfeld et al. 2000), depending on the screening method used in the study. Both Andrews et al. (1991) and Felsenfeld et al. (2000) calculated that ∼70% of the variance in liability for stuttering could be attributed to additive genetic effects, whereas the remaining 30% was due to nonshared environmental effects. Familial aggregation of stuttering has been extensively documented, with an increased incidence of ∼15% in first-degree relatives of probands, as compared with a 5% lifetime risk in the general population (Kidd 1984; Ambrose et al. 1993). Segregation analysis has suggested different genetic models for the inheritance of stuttering in families, including multifactorial/polygenic transmission (Cox et al. 1984), a major locus with multifactorial/polygenic background (Ambrose et al. 1993), and, recently, an autosomal dominant locus (Viswanath et al. 2004).
The onset of stuttering usually occurs in childhood, between the ages of 3 and 6 years, with reported rates of natural, unassisted recovery of ∼75% (Yairi and Ambrose 1999, 2005). So, whereas the lifetime incidence of stuttering is estimated at ∼4%–5%, its prevalence is lower, at 0.5%–1% (Bloodstein 1995; Felsenfeld 2002). An investigation into the relationship between persistence and recovery in stuttering, with the use of pedigree and segregation analysis, suggested that the two phenomena were not genetically independent disorders, though the persistence of stuttering may require the transmission of additional genetic factors (Ambrose et al. 1997). In contrast, previous investigations of stuttering severity (as measured by the frequency or length of disfluencies) showed no relationship between any measure of severity and genetic liability to stuttering (Kidd et al. 1980). It, therefore, seems prudent to use both broad (any individual who ever stuttered) and narrow (those with persistent stuttering) definitions of affection status in linkage studies of stuttering, although there is little justification for using measures of stuttering severity in genetic studies.
There is also a significant sex bias in the incidence of stuttering, with a male-to-female ratio of ∼2:1 during childhood increasing to 4:1 or 5:1 in adulthood (Bloodstein 1995; Yairi and Ambrose 2005). The increased polarity with age of affected males versus females suggests that recovery from stuttering is considerably more frequent in girls than in boys (Yairi and Ambrose 1999). Thus, investigations into the genetic basis underlying sex effects in stuttering are warranted.
To date, only two whole-genome scans designed to detect genetic variation related to stuttering have been published. Riaz et al. (2005) reported genomewide-significant evidence for linkage of stuttering to chromosome 12q (LOD=4.61) and suggestive evidence for linkage to chromosome 1q (LOD=2.93) in 46 highly inbred Pakistani families. Shugart et al. (2004) observed modest evidence for linkage to chromosome 18q (NPLALL=1.51) in 68 families of European ancestry, in which only individuals with persistent stuttering were considered as affected.
Both aforementioned studies used microsatellite markers at an average spacing of ∼10 cM. However, with the advent of high-throughput genotyping methods that allow tens to hundreds of thousands of SNPs to be genotyped at a relatively low cost, using high-density SNP maps to detect linkage seems an attractive alternative to microsatellite mapping. In fact, there is growing evidence that, compared with using traditional microsatellite maps, using high-density SNP maps lends greater power to detect linkage (Kruglyak 1997; Evans and Cardon 2004) and improved definition of underlying disease/trait loci (John et al. 2004). For example, Evans and Cardon (2004) showed that, when parental genotypes were available, a 2-cM–dense SNP map was sufficient to extract almost 100% of the inheritance information, whereas a 10-cM–dense microsatellite map could extract only ∼70%.
Here, we present the results of a genomewide scan with >10,000 SNPs genotyped in 100 families that have at least two relatives affected with stuttering. We have considered both susceptibility to stuttering and persistence of stuttering in our analyses, by using two phenotype definitions that distinguish between individuals who have ever stuttered (broad) and those with persistent stuttering (narrow). The notable male bias in the prevalence of stuttering led us to investigate whether there are sex-specific genes that underlie susceptibility to the disorder, as has been suggested for autism, which also shows a profound difference in risk by sex (Veenstra-VanderWeele and Cook 2004). These studies led us to consider criteria for assessing genomewide significance of sex-specific analyses. Finally, because stuttering is a complex disorder with multiple interacting genetic and environmental factors for susceptibility, we performed secondary analyses that allowed us to assess statistical interactions of loci with the most significant effects in primary analyses.
Material and Methods
Families
The sample comprised 110 families, each with at least two non–parent/offspring relatives affected with stuttering. The families were collected from the University of Illinois at Urbana-Champaign (nf=76); the Tel Aviv University School of Medicine, Israel (nf=16); and the Helsingborg Hospital, Sweden (nf=18). All samples were collected with the approval of local institutional review boards and with informed consent. The primary group of American families were identified (1) from pedigrees of 180 children previously evaluated for stuttering through a longitudinal study (the Illinois Stuttering Research Program), (2) through a well-established, nationwide network of speech-language pathologists specializing in stuttering, and (3) through publications, announcements, and fliers at meetings of organizations geared to provide support and information for people who stutter. Families from the Swedish and Israeli centers were identified and recruited by specialists in stuttering. Since the families were referred through a broad network of pediatricians, day care centers, and speech-language pathologists and by word of mouth, they were considerably more representative of the population than a typical clinical sample. Blood samples from a total of 585 individuals were collected. There were 362 males, 233 adult and 129 minor, and 223 females, 182 adult and 41 minor. A total of 365 affected-relative pairs were identified.
Diagnosis
Individuals were categorized as having ever stuttered (currently or in the past) if they met one or more of the following criteria: (1) direct observation of stuttering by one of the speech investigators, (2) diagnosis and/or treatment by speech-language pathologists, or (3) detailed description by a participant of stuttering behaviors, judged sufficient to positively identify the disorder by a speech-language pathologist with expertise in stuttering. The stuttering was classified as persistent if the individual stuttered for a minimum of 4 years and continued to stutter into later childhood and adulthood. Particular care was taken to verify any cases of naturally recovered stuttering (see Yairi and Ambrose [1999] for full treatment of this issue). If evidence was vague or conflicting, the stuttering was classified as “status unknown.” Those reported as having ever stuttered also filled out a questionnaire regarding their history of stuttering. An earlier version of this questionnaire has been used in past research (Ambrose et al. 1997). Stuttering was classified as recovered in 45 individuals. Of the remainder, 252 exhibited persistent stuttering, and 19 were too young to determine if they would recover or become persistent.
Genotyping
Ten milliliters of blood was drawn from an antecubital vein into two 5-ml plastic vacutainer tubes containing ethylenediaminetetraacetic acid. Half of each blood sample was stored at −70°C as a backup, and the other half was used for DNA extraction and genotyping. DNA was extracted from 3 ml of blood by use of the PureGene DNA extraction kit (Gentra), and the remainder of the blood was frozen. The Affymetrix Mapping Array 10K set was used for genotyping as per the standard protocol described elsewhere (Matsuzaki et al. 2004). Three pairs of MZ twins available in the data set were used to assess the discordance of genotype calls, with only one from each of the twin sets used in subsequent linkage analyses. The observed level of genotype disagreement was low, at 0.00012–0.00037.
Quality Control (QC)
Of the 11,561 total markers, those that were not placed on the deCODE map or that had a missing rate >10% were discarded from analysis. Extensive relationship checking was performed using PREST (Sun et al. 2002) to assess the evidence for the specified within-family relationships and using RELPAIR (Epstein et al. 2000) to check across families for unrecognized relationships and/or switched or duplicated samples. Three families were dropped because of unresolvable errors in pedigree specification. Mendelian incompatibilities were identified using PEDCHECK (O’Connell and Weeks 1998). Any markers that were associated with Mendelian errors in more than two families were dropped from subsequent analyses. All remaining incompatibilities were coded as “missing.”
Since linkage studies are sensitive to the misspecification of marker-allele frequencies, we compared the marker-allele frequencies of the collected families on the basis of ethnicity. We observed that the seven non-European families had allele frequencies for the included SNPs sufficiently different from those in the European families that results could be compromised, and we, therefore, excluded them from these analyses. SNPs were also checked for departures from Hardy-Weinberg equilibrium (HWE). Significant departures from HWE were observed in only 0.171% of the markers (P<.00001) and showed no clustering; therefore, all markers meeting the above-described QC standards were kept for our analyses. We note, however, that the stringent criteria we used for completion of genotyping (we dropped markers with >10% missing data) had largely eliminated the markers with highly significant departures from HWE.
It has been shown elsewhere that failure to account for intermarker linkage disequilibrium (LD), especially when parental genotypes are missing, can lead to inflated linkage scores (Huang et al. 2004). The effect of intermarker LD was examined by removing markers that were in high LD (r2⩾0.8) and conducting linkage analyses on the modified data set. The average difference in LOD scores observed was low, at 0.025. This is consistent with the previous reports, in that most families in our sample contain parental data, and the potential inflation in linkage evidence is more marked when parental data are unavailable. When we eliminated the parental genotypes in our data set, we did detect differences in the evidence for linkage across the genome, depending on whether markers in LD were included or excluded in the analysis. Given that there appeared to be no detectable effect in our data of including markers in some LD (because of the presence of parental data), we report here the results of linkage analyses using all markers passing the QC standards.
Although the genotyping accuracy of SNPs is extremely high (Matsuzaki et al. 2004), the fact that they encode binary information makes it harder to detect erroneous genotypes just by checking for Mendelian incompatibilities (Douglas et al. 2002). MERLIN (Abecasis et al. 2002) was, therefore, used to identify any unlikely recombination events that were present in the data set. The baseline event rate was determined by running MERLIN on five sets of simulated data that were known to have no errors. We observed a two- to threefold increase in the number of events relative to the baseline, with the exception of a few chromosomes (5, 18, 20, and 22) for which the number of observed events was similar to the baseline. All unlikely recombination events that were flagged by MERLIN were coded as missing, and linkage analyses were conducted on the modified data set to determine the effect of genotyping error on evidence for linkage across the genome. The mean difference in LOD scores was negligible, at 0.048 over the genome. We, therefore, report here on analyses using all markers passing QC standards. A total of 9,144 markers were used in the final analysis, with an average genomewide density of 0.3 cM.
Analysis
Linkage analysis was performed using ALLEGRO (Gudbjartsson et al. 2000) for both broad (nf=100) and narrow (nf=86) phenotype definitions. The exponential model was used to obtain nonparametric LOD scores (Kong and Cox 1997), and a total LOD score for families under heterogeneity (HLOD) was also calculated using a parametric model derived from previous segregation analyses of stuttering (Ambrose et al. 1993).
The sex bias observed in stuttering led us to investigate whether there was sex-specific evidence for linkage in these families. The larger ever-stuttered data set was stratified on the basis of sex, with the male data set generated by setting the affection status of all affected females to “unknown,” and vice versa for the female data set. The new male (nm=74) and female (nf=9) data sets were used in linkage analyses as described above.
Simulations consisting of 1,000 replicates of the overall data set, as well as the male-only and female-only data subsets, were generated using MERLIN to determine the genomewide significance of the linkage results, including the sex-specific findings. In assessing the genomewide significance of sex-specific signals, we tallied signals for the sex-specific subsets of replicates at least as large as those observed in the real sex-specific data that also had a sex-specific LOD difference, ΔLOD (ΔLOD = maximum local LOD score in sex-specific subset − maximum local LOD score in overall data), at least as large as that observed for the actual data. These criteria explicitly acknowledge that signals will be considered sex-specific only when the LOD score in a sex-specific data subset is larger than that in the overall sample and that the significance of signals with the same LOD score for sex-specific data subsets will differ depending on the magnitude of the signal in the overall sample. Because the SNP maps are relatively dense, we expected a 5-cM window for calculating ΔLOD to be sufficient but examined the robustness of results when using a 5-cM window (LOD for the overall data is the maximum LOD score within 5 cM on either side of the location of the maximum LOD score for the largest sex-specific LOD score in that region) and a 10-cM window.
To assess potential interactions between linkage signals of interest and the rest of the genome, conditional analyses were performed. Three weighting functions (described below) were used to generate sets of family-specific weights based on the largest of the primary signals obtained for the ever-stuttered and persistent-stutter diagnoses, as well as for the largest sex-specific signal in males (chromosome 9, 60 cM; chromosome 13, 14 cM; chromosome 15, 19 cM; chromosome 7, 153 cM). Weighted linkage analyses were then performed on the remainder of the genome, as described above. Weighting functions were as follows:
-
1.
Weight0–1: Families with a negative NPL score at the location under investigation were assigned a weight of 0 (i.e., they were excluded from the subsequent weighted analysis), whereas families with a positive NPL score were assigned a weight of 1 (included in weighted analysis).
-
2.
Weight0–NPL: Families with a negative NPL score received a weight of 0, whereas families with a positive NPL were assigned a weight equivalent to the value of the NPL score.
-
3.
Weight1–0: Families with a negative NPL score were assigned a weight of 1, whereas families with a positive NPL received a weight of 0.
Statistical significance was assessed by permuting the original weights files (1,000 total permutations) and by conducting linkage analyses on the actual data set with use of each of the 1,000 permuted weight files.
The family-based association test (FBAT) (v.1.5.5) (Rabinowitz and Laird 2000) was used to test the null hypothesis of no association and no linkage for each marker with the persistent-stuttering phenotype under an additive genetic model. Genomewide significance was assessed by comparing the number of signals exceeding a range of P value thresholds in the actual data set with the 1,000 null replicates used in the linkage study. For associated markers showing strong LD (r2⩾0.8), only one marker was counted in the actual data set, since linkage equilibrium was assumed under the null hypothesis. Overlapping signals for FBAT and linkage (nonparametric LOD score) were considered if a marker exceeded the specified significance thresholds for both analyses. Genomewide significance for the overlapping signals was assessed by comparing the actual and the 1,000 null replicates.
Results
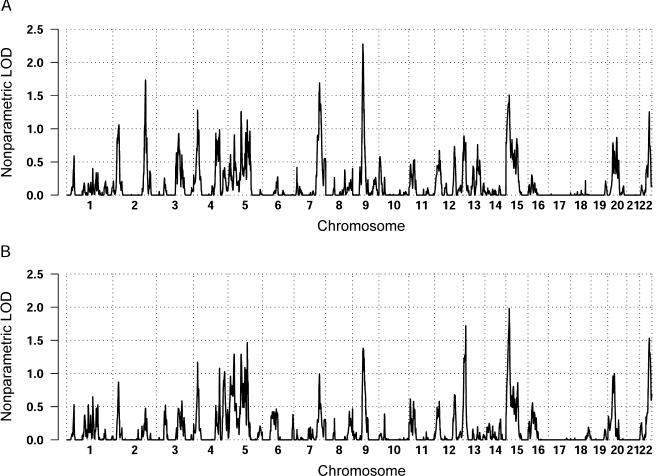
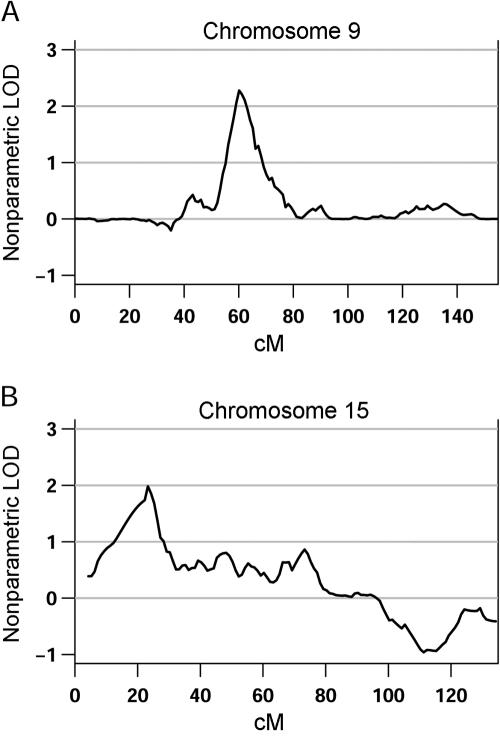
The genomewide nonparametric LOD scores (Kong and Cox 1997) for the broad and narrow phenotype definitions are shown in figure 1. The HLOD results tracked remarkably well with the nonparametric LOD scores, with a correlation over the genome of 0.784, and are, thus, not shown separately. For the ever-stuttered phenotype, a total of 100 families were included in the analysis, and the largest score was observed on chromosome 9 (60 cM), with a LOD of 2.3 (fig. 2a). Chromosomes 2 and 7 also showed nominal evidence for linkage, with LOD scores of 1.72 and 1.69, respectively. For the persistent-stuttering phenotype, a total of 86 families were included in the analysis, and positive evidence for linkage spanning a large region on chromosome 15 (5–96 cM) and peaking at 23 cM, with a LOD score of 1.95, was observed (fig. 2b). Modest evidence for linkage to chromosome 13 (14 cM), with a LOD score of 1.72, was also seen.
Figure 1.
Nonparametric LOD scores obtained across the genome with use of the broad-affection classification of “ever stuttered” (A) and the narrow-affection classification of “persistent stuttering” (B).
Figure 2.
Close-up of the chromosomes with the highest evidence for linkage in the primary analysis for each phenotype definition. Chromosome 9, with a LOD score of 2.3 at 60 cM (A), and chromosome 15, with a LOD score of 1.95 at 23 cM (B), showed the highest evidence for linkage by use of the broad and narrow phenotype definitions, respectively.
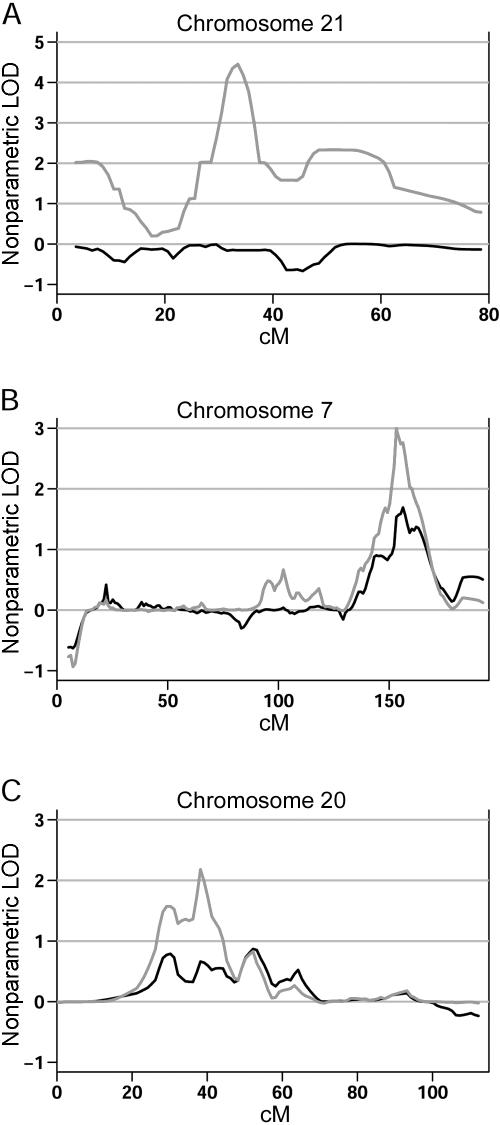
Sex-specific analyses resulted in increased evidence for linkage in three regions across the genome. Genomewide-significant evidence for linkage on chromosome 21 (LOD score of 4.5; ΔLOD = 4.46) was observed in the female-only families (fig. 3a). Within the 1,000 female-only simulated data sets, there was a peak that exceeded the 4.5 LOD threshold and the 4.46 ΔLOD threshold (with use of either a 5-cM or a 10-cM window) only five times (genomewide P=.005). Analysis of the male data set showed increased evidence for linkage on chromosome 7 at 153 cM (fig. 3b) and chromosome 20 at 38 cM (fig. 3c), with LOD scores of 2.99 and 2.18, respectively, and ΔLOD values of 1.30 (for both window sizes) and 1.52 (for 10-cM window; ΔLOD = 1.39 for 5-cM window), respectively. The genomewide significance of the chromosome 7 linkage signal was estimated as P=.04 (for both window sizes), whereas that for chromosome 20 was P=.096 for a 5-cM window and .115 for a 10-cM window.
Figure 3.
Sex-specific evidence for linkage. A, Analysis of the female-only data set (gray line) compared with the complete data set (black line) on chromosome 21, showing significant increase in the LOD score when only females are classified as affected. B, Similar significant increase in the evidence for linkage on chromosome 7 when the male-only data set (gray line) is compared with the complete data set (black line). C, Increased evidence for linkage also observed in the male-only data set (gray line) on chromosome 20.
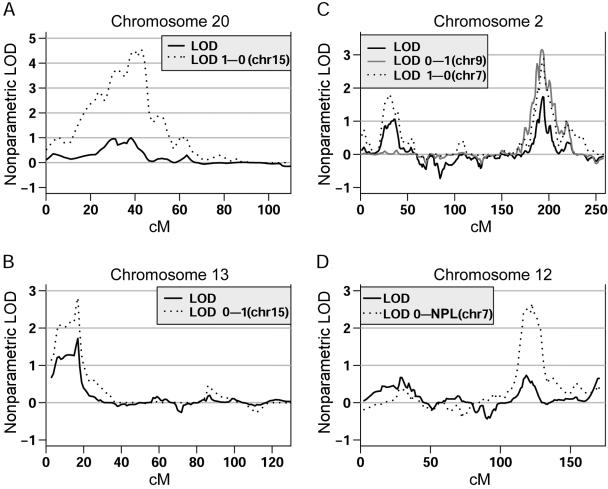
Conditional analyses identified four regions across the genome that showed a notable increase in evidence for linkage, with LOD scores >2.5. The weighting functions enable us to detect both positive (weight0–1 and weight0–NPL) and negative (weight1–0) interactions, since families are assigned weights on the basis of the presence or absence of linkage, respectively, to the signal of interest. When families were weighted on the basis of their evidence for linkage at chromosome 15 (weight0–1), a positive interaction was observed with chromosome 13, for which the evidence for linkage increased from a LOD score of 1.78 to 2.8 (P=.013) (fig. 4b). Conversely, a negative interaction was observed with chromosome 20 (weight1–0) at 40 cM, with a LOD score of 4.5 (fig. 4a). In the 1,000 simulated data sets that were generated, a peak crossing that threshold was not observed (P=.0), making this interaction highly significant. In addition, the location of the peak overlapped the male-specific linkage signal previously detected. When families were weighted on the basis of their evidence for linkage at chromosome 9, increased evidence for linkage on chromosome 2 at 193 cM was observed using the weight0–1 function, with the original LOD score of 1.63 increasing to 3.16 (P=.01). Interestingly, when families were weighted on the basis of their evidence for linkage at chromosome 7 (153 cM), analysis with the weight1–0 function led to the observation of a negative interaction in the same location on chromosome 2 (P=.013) (fig. 4c). Finally, a strong positive interaction (P=.01) with the chromosome 7 linkage signal and chromosome 12 at 120 cM was observed, with the LOD score increasing from 0.57 to 2.66 (fig. 4d).
Figure 4.
Results of conditional analyses that showed a notable increase in evidence for linkage, with LOD scores >2.5. The X-axis corresponds to the distance in cM from the P terminus of the respective chromosome. The Y-axis corresponds to the nonparametric LOD score. For all conditional analyses, the weighting function used is indicated in each legend, along with the primary signal (in parentheses) from which the weights were generated.
FBAT did not reveal any marker showing genomewide-significant association for persistent stuttering. Moreover, there were no more overlapping signals between FBAT and linkage analyses than expected by chance alone (data not shown).
Discussion
Genomewide linkage analysis of 100 families segregating for stuttering resulted in suggestive evidence for linkage on chromosome 9, by use of the broad phenotype definition, with a LOD score of 2.3 at 60 cM. Modest evidence for linkage with the persistent-stuttering diagnosis was detected on chromosomes 15 and 13, with LOD scores of 1.95 (23 cM) and 1.72 (14 cM), respectively. However, primary linkage mapping of the overall data did not yield any LOD scores in the range of criteria established for genomewide significance (Lander and Kruglyak 1995). These results are not surprising, since a large proportion of linkage studies designed to identify the underlying genetic variation of complex diseases have met with limited success. Altmuller et al. (2001) analyzed >100 published whole-genome scans of complex human diseases and found that only 32% reported significant linkage (LOD >3.6) and that only 2% of the studies showed replicated evidence for linkage. These results can be attributed both to methodological limitations, such as small sample size, genotyping errors, and/or clinical heterogeneity, and to the underlying complexity of genetic models for common diseases arising from the actions and interactions of genetic and nongenetic factors.
Recently, several studies have advocated investigating the sex-specific genetic architecture of complex disorders, especially when the disorder exhibits a skewed sex ratio (Stone et al. 2004; Weiss et al. 2005). The search for potential sex-related loci in our stuttering cohort was quite successful, leading to the identification of several regions that merit further investigation. We observed genomewide-significant evidence for linkage (genomewide P<.04) of stuttering in the male-only data set on chromosome 7 (153 cM). Evidence for linkage to this region is supported by replication of the finding (including the sex-specific effect) in the Hutterite stuttering cohort (J. K. Wittke-Thompson, unpublished data). Modest evidence for linkage to chromosome 7 (160 cM) was also observed in families ascertained for specific language impairment (SLI), a disorder characterized by impairments restricted to the domain of language skills and in the absence of other factors such as mental retardation and speech-motor or sensory deficits (Bartlett et al. 2004). This region was also identified as harboring a language-related QTL for autism (Alarcon et al. 2005), a disorder characterized by disturbances in social, communicative, and behavioral functioning and that also shows a striking male bias in prevalence, with consistent estimates of 4:1 affected males versus females (Veenstra-VanderWeele and Cook 2004). A greater-than-expected prevalence of SLI in families with autistic disorder and vice versa has been observed, as has the existence of overlapping phenotypes for the two disorders (Folstein and Mankoski 2000). It is, therefore, tempting to speculate that there are one or more genes in this region of chromosome 7 that help to shape dimensions of speech and language development and that may differ (e.g., in timing) between males and females, with genetic variation at the gene(s) affecting susceptibility to SLI, autism, and stuttering. We note, however, that this region does not encompass the FOXP2 gene identified elsewhere as the causal gene for a rare speech and language disorder segregating in a large pedigree (Lai et al. 2001).
We also obtained genomewide-significant evidence for linkage of stuttering in the female-only subgroup on chromosome 21 (33.5 cM), with a LOD score of 4.5. Although the LOD score is likely to be poorly estimated in a sample this small, simulation studies confirm that a signal of this magnitude is very unlikely by chance (genomewide P=.005). Moreover, there is some support for this finding, including the sex-specific nature of the result, in an independent stuttering data set (J. K. Wittke-Thompson, unpublished data).
Replication of evidence for linkage in complex disorders is limited, as previously mentioned. Two whole-genome scans for stuttering have been published so far, with only one significant result on chromosome 12 at 115 cM, with a LOD score of 4.61, observed to date (Riaz et al. 2005). Conditional analysis using families weighted on the basis of their evidence for linkage to chromosome 7 (153 cM), a genomewide-significant linkage signal in our data, showed a marked increase in the evidence for linkage to chromosome 12, with a resulting LOD score of 2.66 (increased from a baseline LOD of 0.57). The location of the chromosome 12 signal in our stuttering cohort overlaps with that reported by Riaz et al. (2005), providing support for the existence of a locus affecting susceptibility to stuttering on chromosome 12q23-24 in our sample, which is quite historically distinct from the Pakistani population studied in Riaz et al. (2005).
Chromosome 2q31-33 is a region of the genome that has been repeatedly implicated in autism, with evidence for linkage to this region substantially increasing when analyses were performed using a narrow diagnosis of autism with phrase speech delay (Buxbaum et al. 2001; Shao et al. 2002). Interestingly, this same region on chromosome 2 showed nominal evidence for linkage in the primary analysis of the overall data set with use of the ever-stuttered phenotype and increased significantly (conditional LOD=3.16; baseline LOD=1.63; P<.01) when families were weighted on the basis of their evidence for linkage to chromosome 9. A negative interaction between the chromosome 2 locus and the linkage signal on chromosome 7 was also detected.
The appropriate criteria for assessing genomewide significance in the context of sex-specific linkage analysis merit some additional discussion. Because the sex-specific data used in such studies are subsets of the overall data, evidence for linkage would not be considered sex-specific when the signal in the subsets is merely proportional to the data included. Rather, evidence for linkage has generally been interpreted as being sex-specific when the evidence for linkage in the male or female subset is at least as large as, and usually is substantially larger than, the evidence for linkage in the overall sample. The fixed structure of the sex-specific data subsets permits a straightforward estimation of the genomewide significance of sex-specific signals through the same simulation studies used to estimate genomewide significance of the results for the overall data. That is, because the male-only data subset is the same for each replicate, as is the female-only data subset, it is, therefore, possible to establish thresholds for the sex-specific signals and for the magnitude of the observed increase (ΔLOD), to estimate genomewide significance.
In contrast, estimating the genomewide significance of an observed conditional linkage signal from this experiment is difficult because most simulations from the null may not replicate the structure of the real data. In such circumstances, conditioning on the largest observed signal in each replicate will rarely generate a set of nonzero weights as large as that obtained for the real data and, thus, cannot appropriately estimate the genomewide significance of a conditional increase. Although such secondary analyses must, therefore, be considered “hypothesis generating,” they can be compelling when, as we have shown here, they highlight regions that have been convincingly implicated in independent studies of the same or related phenotypes.
Finally, our studies emphasize the importance of considering the sex-specific genetic architecture of complex phenotypes. Stuttering is one of many complex disorders showing striking sex differences in severity and/or prevalence, including inflammatory bowel disease (Fisher et al. 2002), mood disorders (Zubenko et al. 2003), and autism (Veenstra-VanderWeele and Cook 2004). Approaches considering sex as a factor that may influence disease risk in a genotype-specific manner have met with success in identifying a variety of sex-specific susceptibility loci for different complex phenotypes. In the case of autism, two independent studies showed significant male-specific evidence for linkage (maximized LOD score [MLS] >4.0) to chromosome 17q11, whereas analysis of the families with at least one affected female showed little evidence for linkage in the same region (MLS ⩽0.5) (Stone et al. 2004; Cantor et al. 2005). Recently, sex-specific heritability and genomewide linkages for a total of 17 quantitative traits associated with common human diseases, such as heart disease, hypertension, diabetes, asthma, and autoimmune disease, were evaluated in the Hutterites (Weiss et al. 2006). Ten of these traits showed sex-specific evidence for linkage, with two genomewide-significant linkages detected in the male-only subset and two genomewide-significant linkages detected in the female-only subset. Interestingly, of the 62 regions that showed suggestive evidence for linkage in either the male-only, female-only, or complete data set, only 4 regions reached suggestive significance in both the sex-stratified and complete data set, whereas none of the male-specific or female-specific signals overlapped. Notably, none of the sex-specific signals that showed genomewide significance reached suggestive significance in the complete data set. In light of these reports, as well as the presence of sex-specific loci observed in this study, it seems crucial to explore sex-specific genetic effects when conducting genomewide screens, especially in those disorders where sex differences in risk are observed.
Acknowledgments
This research was supported in part by Public Health Service grants DC-04415 and DK-55889 (to N.J.C.). We are grateful for the expert technical assistance of Laura Martinolich.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Alarcon M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH (2005) Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Mol Psychiatry 10:747–757 10.1038/sj.mp.4001666 [DOI] [PubMed] [Google Scholar]
- Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M (2001) Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 69:936–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose N, Cox NJ, Yairi E (1997) The genetic basis of persistent and recovered stuttering. J Speech Lang Hear Res 40:567–580 [DOI] [PubMed] [Google Scholar]
- Ambrose N, Yairi E, Cox NJ (1993) Genetic aspects of early childhood stuttering. J Speech Hear Res 36:701–706 [DOI] [PubMed] [Google Scholar]
- Andrews G, Morris-Yates A, Howie P, Martin N (1991) Genetic factors in stuttering confirmed. Arch Gen Psychiat 48:1034–1035 [DOI] [PubMed] [Google Scholar]
- Bartlett CW, Flax JF, Logue MW, Smith BJ, Vieland VJ, Tallal P, Brzustowicz LM (2004) Examination of potential overlap in autism and language loci on chromosomes 2, 7, and 13 in two independent samples ascertained for specific language impairment. Hum Hered 57:10–20 10.1159/000077385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodstein O (1995) A handbook on stuttering, 5th ed. National Easter Seal Society, Chicago [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Kono N, Duvall JA, Alvarez-Retuerto A, Stone JL, Alarcon M, Nelson SF, Geschwind DH (2005) Replication of autism linkage: fine-mapping peak at 17q21. Am J Hum Genet 76:1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Kramer P, Kidd K (1984) Segregation analyses of stuttering. Genet Epidemiol 1:245–253 10.1002/gepi.1370010304 [DOI] [PubMed] [Google Scholar]
- Douglas JA, Skol AD, Boehnke M (2002) Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data. Am J Hum Genet 70:487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Cardon LR (2004) Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet 75:687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld S (2002) Finding susceptibility genes for developmental disorders of speech: the long and winding road. J Commun Disord 35:329–345 10.1016/S0021-9924(02)00088-6 [DOI] [PubMed] [Google Scholar]
- Felsenfeld S, Kirk K, Zhu G, Statham M, Neale M, Martin N (2000) A study of the genetic and environmental etiology of stuttering in a selected twin sample. Behav Genet 30:359–366 10.1023/A:1002765620208 [DOI] [PubMed] [Google Scholar]
- Fisher SA, Hampe J, Macpherson AJ, Forbes A, Lennard-Jones JE, Schreiber S, Curran ME, Mathew CG, Lewis CM (2002) Sex stratification of an inflammatory bowel disease genome search shows male-specific linkage to the HLA region of chromosome 6. Eur J Hum Genet 10:259–265 10.1038/sj.ejhg.5200792 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Mankoski RE (2000) Chromosome 7q: where autism meets language disorder? Am J Hum Genet 67:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Howie PM (1981) Concordance for stuttering in monozygotic and dizygotic twin pairs. J Speech Hear Res 24:317–321 [DOI] [PubMed] [Google Scholar]
- Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W, Vasavda N, Mills T, Barton A, Hinks A, Eyre S, Jones KW, Ollier W, Silman A, Gibson N, Worthington J, Kennedy GC (2004) Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet 75:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd K (1984) Stuttering as a genetic disorder. In: Curlee R, Perkins W (eds) Nature and treatment of stuttering. College Hill, San Diego, pp 149–169 [Google Scholar]
- Kidd KK, Heimbuch RC, Records MA, Oehlert G, Webster RL (1980) Familial stuttering patterns are not related to one measure of severity. J Speech Hear Res 23:539–545 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L (1997) The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17:21–24 10.1038/ng0997-21 [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, Yang G, Kennedy GC, Webster TA, Cawley S, Walsh PS, Jones KW, Fodor SP, Mei R (2004) Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods 1:109–111 10.1038/nmeth718 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N (2000) A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 50:211–223 10.1159/000022918 [DOI] [PubMed] [Google Scholar]
- Riaz N, Steinberg S, Ahmad J, Pluzhnikov A, Riazuddin S, Cox NJ, Drayna D (2005) Genomewide significant linkage to stuttering on chromosome 12. Am J Hum Genet 76:647–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, Abramson RK, Wright HH, DeLong RG, Gilbert JR, Cuccaro ML, Pericak-Vance MA (2002) Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet 70:1058–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugart YY, Mundorff J, Kilshaw J, Doheny K, Doan B, Wanyee J, Green ED, Drayna D (2004) Results of a genome-wide linkage scan for stuttering. Am J Med Genet A 124:133–135 10.1002/ajmg.a.20347 [DOI] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Yonan AL, Gilliam TC, Geschwind DH, Nelson SF (2004) Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet 75:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wilder K, McPeek MS (2002) Enhanced pedigree error detection. Hum Hered 54:99–110 10.1159/000067666 [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Cook EH Jr (2004) Molecular genetics of autism spectrum disorder. Mol Psychiatry 9:819–832 10.1038/sj.mp.4001505 [DOI] [PubMed] [Google Scholar]
- Viswanath N, Lee HS, Chakraborty R (2004) Evidence for a major gene influence on persistent developmental stuttering. Hum Biol 76:401–412 [DOI] [PubMed] [Google Scholar]
- Weiss LA, Abney M, Parry R, Scanu AM, Cook EH Jr, Ober C (2005) Variation in ITGB3 has sex-specific associations with plasma lipoprotein(a) and whole blood serotonin levels in a population-based sample. Hum Genet 117:81–87 10.1007/s00439-004-1250-3 [DOI] [PubMed] [Google Scholar]
- Weiss LA, Pan L, Abney M, Ober C (2006) The sex-specific genetic architecture of quantitative traits in humans. Nat Genet 38:218–222 10.1038/ng1726 [DOI] [PubMed] [Google Scholar]
- Yairi E, Ambrose N (2005) Early childhood stuttering. Pro-Ed, Austin [Google Scholar]
- Yairi E, Ambrose NG (1999) Early childhood stuttering I: persistency and recovery rates. J Speech Lang Hear Res 42:1097–1112 [DOI] [PubMed] [Google Scholar]
- Yairi E, Ambrose N, Cox N (1996) Genetics of stuttering: a critical review. J Speech Hear Res 39:771–784 [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Maher B, Hughes HB 3rd, Zubenko WN, Stiffler JS, Kaplan BB, Marazita ML (2003) Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am J Med Genet B Neuropsychiatr Genet 123:1–18 10.1002/ajmg.b.20073 [DOI] [PubMed] [Google Scholar]