Abstract
Transposable genetic elements are abundant in the genomes of most organisms, including humans. These endogenous mutagens can alter genes, promote genomic rearrangements, and may help to drive the speciation of organisms. In this study, we identified almost 11,000 transposon copies that are differentially present in the human and chimpanzee genomes. Most of these transposon copies were mobilized after the existence of a common ancestor of humans and chimpanzees, ∼6 million years ago. Alu, L1, and SVA insertions accounted for >95% of the insertions in both species. Our data indicate that humans have supported higher levels of transposition than have chimpanzees during the past several million years and have amplified different transposon subfamilies. In both species, ∼34% of the insertions were located within known genes. These insertions represent a form of species-specific genetic variation that may have contributed to the differential evolution of humans and chimpanzees. In addition to providing an initial overview of recently mobilized elements, our collections will be useful for assessing the impact of these insertions on their hosts and for studying the transposition mechanisms of these elements.
Transposable genetic elements collectively occupy ∼44% of the human genome (International Human Genome Sequencing Consortium 2001). Although most of these transposons lost the ability to transpose long ago, some copies have transposed in relatively recent human history (Kazazian et al. 1988; Wallace et al. 1991; Moran et al. 1996; reviewed by Ostertag and Kazazian 2001; reviewed by Batzer and Deininger 2002; Bennett et al. 2004). These recently mobilized transposons are of great interest for a number of reasons. First, recent insertions within or near genes may cause phenotypic changes in humans, including diseases (Kazazian et al. 1988; Wallace et al. 1991; reviewed by Ostertag and Kazazian 2001; reviewed by Batzer and Deininger 2002). Several dozen transposon insertions have been identified to date that cause human diseases, and human populations are likely to harbor additional transposon insertions that influence phenotypes as well. Some of these recently mobilized transposons also remain actively mobile today and continue to generate new transposition events elsewhere in the genome (Moran et al. 1996; reviewed by Ostertag and Kazazian 2001; Dewannieux et al. 2003). Active retrotransposons in particular have been observed to be the most potent endogenous mutagens in humans, and these elements continue to generate mutations and genetic variation in human populations (reviewed by Ostertag and Kazazian 2001). In some cases, transposon insertions also may go on to create genomic rearrangements by recombining with other transposon copies (reviewed by Ostertag and Kazazian 2001). Thus, recently mobilized transposons continue to restructure the human genome through a variety of mechanisms.
The completion of a draft chimpanzee genome sequence provided an opportunity to identify these recently mobilized transposons in both humans and chimpanzees (Chimpanzee Sequencing and Analysis Consortium 2005). Transposons that inserted into either of these genomes during the past ∼6 million years (i.e., since the existence of the most recent common ancestor of humans and chimpanzees) would be expected to be present in only one of the two genomes. We used a comparative genomics approach to identify these recently inserted transposon copies (fig. 1). We began by aligning the sequences of the human and chimpanzee genomes to identify all insertions and deletions (indels).
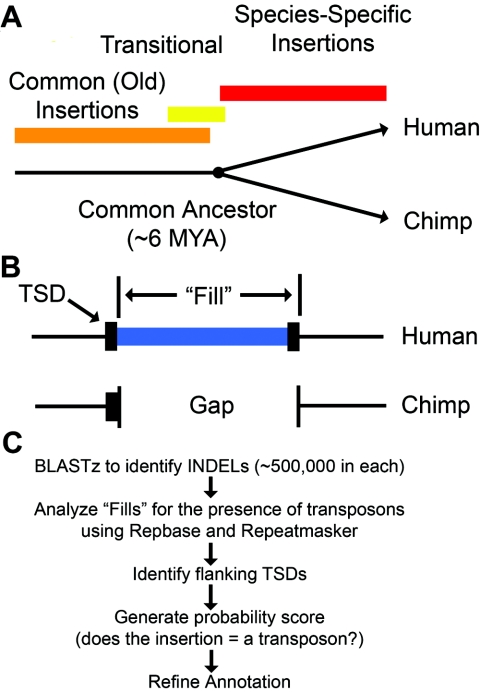
Figure 1.
Overview of our transposon insertion–discovery pipeline. A, The time line for speciation of humans and chimpanzees is compared with the time line for the generation of transposon insertions. Common insertions occurred a very long time ago and are fixed in both species. “Species-specific” insertions are differentially present in the two species and occurred mostly during the past ∼6 million years. MYA = million years ago. B, Our strategy for identifying new transposon insertions in humans and chimpanzees. Recently mobilized transposons are flanked by TSDs and are precisely absent from one of the two genomes. One of the two copies of the TSD is actually found within the indel. Thus, the transposon plus one TSD copy equals the “fill.” C, Our computational pipeline. The five sequential steps of our computational pipeline for discovering species-specific transposon insertions in humans and chimpanzees are depicted. The draft chimpanzee-genome (build panTro1) and human-genome (build hg17) sequences were obtained from the University of California Santa Cruz browser (Kent et al. 2002). BAC clone sequences for the chimpanzee genome were obtained from GenBank (National Center for Biotechnology Information [NCBI]). BLAST programs also were obtained from NCBI. Repeatmasker was obtained from Arian Smit (Institute for Systems Biology). RepBase version 10.02 and the consensus sequence for the L1-Hs element were obtained from Jurzy Jurka (Jurka 2000). Full-length consensus sequences for L1-PA2, L1-PA3, L1-PA4, and L1-PA5 were obtained from GenBank (Boissinot et al. 2000). Custom MySQL databases and PERL scripts were generated as necessary. All analysis was performed locally on SUN SunFire v40z or Dell Power Edge 2500 servers running Linux operating systems. Our computational pipeline began with identification of all indels in humans versus chimpanzees using genomic alignments that were generated with BLASTz. Next, indels containing transposons were identified using Repeatmasker (A. Smit, unpublished material) and RepBase version 10.02 (Jurka 2000). RepBase libraries for humans and chimpanzees were modified to include full-length L1-PA2, L1-PA3, L1-PA4, L1-PA5 consensus sequences (Boissinot et al. 2000). TSDs were identified using a Smith-Waterman local alignment algorithm on the regions flanking each indel junction. The algorithm was restricted to require the optimum alignment to be located within 5 bp of the indel junction. Aligned sequences smaller than 4 bp or having an identity <90% were not scored as TSDs. A probability scoring system was developed to determine the likelihood that a given indel was caused by a single transposon insertion plus its TSD. This score was obtained by adding together the fraction of the indel that was accounted for by the transposon, its TSD, and a poly (A) tail (if present). A score of 1.0 indicated that the gap was fully accounted for by the transposon and associated sequences. We empirically determined that a lower cutoff of 0.85 provided accurate results while eliminating few, if any, true positives. SVA elements initially were annotated poorly by Repeatmasker. This program often split SVA elements into 2–3 segments (and thus counted most elements more than once). We developed a new method to reassemble these segments into a single element, where appropriate.
We screened indels for the presence of transposable elements by comparing each indel to a library of known transposons (RepBase v. 10.02) (Jurka 2000). Using this approach, we initially identified a total of 14,783 transposon copies that were differentially present in the two genomes. Many of these copies appeared to be recently mobilized transposon insertions, whereas others were simply transposon copies that happened to be located within larger genomic duplications or deletions in the two genomes.
To identify all of the insertions that were caused by actual transposition events, we next screened our collections for insertions that (1) were precisely flanked by target-site duplications (TSDs) and (2) precisely accounted for a gap in one of the two genomes. Using these criteria, we identified 10,719 insertions of single transposon copies that appeared to have been caused by transposition events (see the tab-delimited ASCII files, which can be imported into spreadsheets, of data sets 1 and 2 [online only]). The remaining 4,064 examples lacked TSDs or, in general, did not precisely account for the indels, which suggests that they were caused by alternative mechanisms. Of the 10,719 transposon insertions, 7,786 (72.6%) were found in humans and only 2,933 (27.4%) were found in chimpanzees. Therefore, it appears that transposons have been significantly more active in the human genome during the parallel evolution of these organisms (see data sets 1 and 2 [online only]). The different population dynamics of these organisms during the past several million years also may have helped to shape the final patterns of transposons observed.
The most abundant classes of new transposon insertions in both chimpanzees and humans were Alu, L1, and SVA element insertions, and these three classes collectively accounted for >95% of the recently mobilized transposons in both species (table 1 and fig. 2). However, the relative abundance of these elements and their subfamilies differed between the two species (table 1; data sets 1 and 2 [online only]; fig. 2; see below). Other, less-abundant classes of transposon insertions also were identified in our study. For example, long terminal repeat (LTR) retroelement insertions were observed in both species, including insertions of human endogenous retroviruses (HERVs) and solo LTRs of these elements (data set 1 [online only]). Solo LTR insertions have been shown to influence the expression of nearby genes, which makes these insertions of particular interest (Landry and Mager 2003). Also identified were five full-length HERV-K insertions with relatively long ORFs (up to several thousand amino acids in length) that could remain capable of retrotransposition (data set 1 [online only]). Insertions of chimpanzee endogenous retroviruses (CERVs) also were identified (data set 2 [online only]) (Yohn et al. 2005). Finally, mammalian interspersed repetitive elements, copies of satellite DNA flanked by unusual TSDs, and small numbers of other interesting transposable elements were identified in the two species (data sets 1 and 2 [online only]).
Table 1.
Summary of Transposon Insertions
|
No. (%) of Transposon Insertions |
||
|
Transposon Class |
Human (n=7,786) |
Chimpanzee (n=2,933) |
| Alu (All): | 5,530 (71.0%) | 1,642 (56.1%) |
| Alu S | 263 (3.3%) | 50 (1.7%) |
| Alu Ya5 | 1,709 (21.9%) | 10 (.3%) |
| Alu Yb8 | 1,290 (16.6%) | 9 (.3%) |
| Alu Y | 484 (6.2%) | 360 (12.3%) |
| Alu Yc1 | 356 (4.6%) | 979 (33.4%) |
| Alu Yg6 | 261 (3.4%) | 1 (.1%) |
| L1 (All): | 1,174 (15.1%) | 758 (25.9%) |
| L1 Hs (Ta) | 271 (3.5%) | 0 (.0%) |
| L1 Hs (Non Ta) | 252 (3.2%) | 210 (7.2%) |
| L1 PA2 | 490 (6.3%) | 476 (16.2%) |
| SVA (All) | 864 (11.1%) | 396 (13.6%) |
| Other | 219 (2.8%) | 127 (4.4%) |
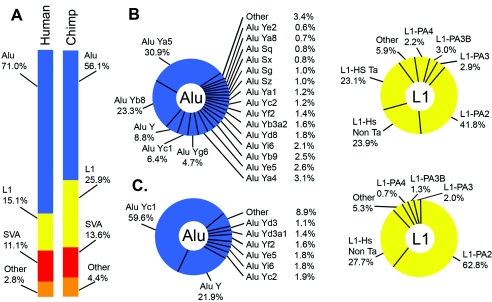
Figure 2.
Classes of species-specific transposons in humans and chimpanzees. A, The overall composition of species-specific insertions in humans and chimpanzees. Note that 97.2% of all insertions in humans and 95.6% of all insertions in chimpanzees are Alu, L1, and SVA insertions. B, The distributions of Alu and L1 subfamilies for humans. C, The distributions of Alu and L1 subfamilies for chimpanzees. Note that different Alu and L1 subfamilies were amplified in humans (B) and chimpanzees (C).
Alu insertions were by far the most abundant class of transposon insertions in both humans and chimpanzees, and these insertions collectively accounted for the bulk of transposons in our study (table 1 and fig. 2). The number of Alu insertions in humans (5,530) was 3.4-fold higher than the number observed in chimpanzees (1,642). The distributions of these elements among various Alu subfamilies also differed between the two organisms (table 1; data sets 1 and 2 [online only]; fig. 2). For example, Alu Ya5, Alu Yb8, Alu Y, and Alu Yc1 were highly abundant in humans, whereas only Alu Yc1 and Alu Y were highly abundant in chimpanzees. Our data indicate that Alu S elements, which have been presumed to have been inactive for the past 35 million years (Johanning et al. 2003), apparently have been active in humans and less active in chimpanzees during the past ∼6 million years (table 1). It is possible that some of these older Alu S “insertions” were caused by the precise deletion of Alu S elements from one of the two genomes (van de Lagemaat et al. 2005) or by gene-conversion events (reviewed by Batzer and Deininger 2002). However, these results also are in agreement with recent data from our laboratory, which indicates that a small number of younger Alu S elements are polymorphic in humans and appear to have transposed more recently than the bulk of Alu S elements (Bennett et al. 2004). Overall, our results indicate that the human genome has supported higher levels of Alu retrotransposition and has amplified a different set of Alu elements than has the chimpanzee genome (table 1 and fig. 2). These results confirm and extend previous classifications of Alu elements of chimpanzee chromosome 22 (Hedges et al. 2004; Watanabe et al. 2004).
L1 insertions also were abundant in both organisms. In humans, almost 1,200 recently mobilized L1 insertions with TSDs were identified that precisely accounted for gaps in the chimpanzees genome (data set 1 [online only] and table 1). These human L1 elements predominantly included members of the L1-Hs and L1-PA2 families (data set 1 [online only]; table 1; fig. 2) (Boissinot et al. 2000; Brouha et al. 2003). The human L1-Hs elements included members of the pre-Ta, Ta0, and Ta1 subfamilies (data set 1 [online only]; grouped together as “L1-Hs Ta” in table 1 and fig. 2), which are known to be highly active in humans (Brouha et al. 2003). Also identified in humans were additional L1-Hs and L1-PA2 subfamilies that had unique base combinations at the nine key positions described elsewhere (data set 1 [online only]; grouped together as “L1-Hs non-Ta” or “L1-PA2” in table 1 and fig. 2) (Boissinot et al. 2000; Brouha et al. 2003). These novel subfamilies contained 3–65 copies (data set 1 [online only]). The remaining L1 insertions in humans belonged to older L1-PA2, L1-PA3, and L1-PA4 groups (data set 1 [online only] and fig. 2).
The L1 insertions identified in chimpanzees, in contrast, were notably different from those outlined above for humans (table 1; data set 2 [online only]; fig. 2). For example, fewer recently mobilized L1 insertions were identified in chimpanzees than in humans (758 in chimpanzees vs. 1,174 in humans). Only 4 of the chimpanzee L1 insertions were full-length (compared with >200 new full-length insertions in humans), and none of the chimpanzee L1 insertions had intact ORFs (data set 2 [online only]). The initial draft sequence of the chimpanzee genome is likely to contain assembly errors that may account for at least some of these observed differences. However, we also observed differences in the L1 subfamilies of these organisms that are unrelated to genome assembly issues. For example, proportionally more L1-PA2 insertions and fewer L1-Hs insertions were observed in chimpanzees than in humans (data set 2 [online only] and fig. 2). Initially, we were surprised to find L1-Hs elements in chimpanzees at all, since these elements were expected to be found only in humans. However, further analysis revealed that most of the L1-Hs elements in chimpanzees actually were “intermediate” elements that matched L1-Hs overall but had ORF1 sequences that were more similar to L1-PA2 elements (data set 2 [online only]). Therefore, the L1-Hs family of elements includes subfamilies that are truly human specific as well as other L1-Hs–like elements that are not human specific. We also aligned and analyzed all of our chimpanzee L1 insertions, using ClustalW and PAUP, to determine whether any new L1 subfamilies (equivalent to L1-Ta elements in humans) were present in chimpanzees. In addition, we classified all of our chimpanzee L1 insertions, using the nine key positions that have been used elsewhere to classify human L1 elements (Boissinot et al. 2000; Brouha et al. 2003). In both cases, we failed to identify any new extended subfamilies of L1 elements within our collection of chimpanzee insertions. Therefore, a dominant class of new offspring elements analogous to L1-Ta elements in humans does not appear to have been produced in recent chimpanzee history (data set 2 [online only]).
We next examined all of the existing L1 ORFs in the human and chimpanzee genomes to further characterize possible differences between the L1 elements of these species. We screened the human and chimpanzee genomes for ORFs in all nearly full-length elements (>5,500 bp) and identified 633 L1 elements with intact ORF1 sequences in the human genome but only 39 elements with intact ORF1 sequences in the draft chimpanzee sequence (see the tab-delimited ASCII files, which can be imported into spreadsheets, of data sets 3 and 4 [online only]). Moreover, we identified 205 L1 elements with intact ORF2 sequences in the human genome (table 2) but failed to detect intact ORF2 sequences in the draft chimpanzee sequence (data set 3 [online only]). These results suggested that functional L1 elements were likely to be rare in chimpanzees. As outlined above, however, it also was possible that the quality of the chimpanzee draft sequence affected our ability to detect ORFs accurately. We determined that the sequence quality of the >5,500 bp L1 elements in chimpanzees had average scores that generally were high (>40 Phred scores) (Ewing et al. 1998; Kent et al. 2002; data set 3 [online only]). However, single bases of low quality (<10 Phred scores) (Ewing et al. 1998) also were distributed throughout the draft sequence at sporadic intervals (Kent et al. 2002). These single bases, although rare, often resulted in frameshifts. Therefore, it was possible that these sporadic low-quality bases were interfering with our ability to detect ORFs accurately.
Table 2.
Analysis of L1 ORFs
|
No. of Elements |
|||
| Chimpanzee |
|||
| ORF | Full Human Genome (version hg17) |
BACs (260 Mb) |
Full Genome (Extrapolation) |
| L1 >5,500 bp | 8,483 | 702 | 8,100 |
| Intact ORF1 (1,017 bp) | 633 | 20 | 230 |
| Intact ORF2 (3,828 bp) | 205 | 4 | 46 |
| Intact ORF1 and ORF2 | 126 | 2 | 23 |
To independently examine the frequency of intact L1 ORFs in chimpanzees, we analyzed all L1 elements that were present in finished BAC sequences that had been generated for the chimpanzee genome project (see the tab-delimited ASCII file, which can be imported into a spreadsheet, of data set 5 [online only]). Approximately 260 Mb of finished sequence was available in GenBank from chimpanzee BACs, and the quality of these sequences was identical to that of the finished human genome sequence. We identified a total of two L1 elements in these BACs that were >5,500 bp in length and also had intact ORFs (data set 5 [online only] and table 2). Neither of these two elements was present in the draft sequence, so it is unclear whether the quality of the draft affected our ability to detect these ORFs. Nevertheless, extrapolation of these results to the whole genome (3,000 Mb) predicts that chimpanzees harbor ∼23 full-length L1s with intact ORFs (compared with 126 in humans; table 2). Thus, the chimpanzee draft sequence indicates that L1 elements with intact ORFs are up to 20-fold less abundant in chimpanzees than in humans, whereas the BACs indicate that such elements are ∼5.5-fold less abundant (table 2). Since the draft chimpanzee sequence contains some sporadic low-quality bases, the BAC estimate is likely to be more accurate. Both of these estimates indicate that functional L1 elements are less abundant in chimpanzees than in humans.
We next examined the L1 ORF1 and ORF2 coding regions from chimpanzees to determine whether the encoded proteins are likely to remain active today. Brouha et al. (2003) showed elsewhere that human L1 elements that differ by an average of only 21 nucleotide changes from an active human L1 consensus were inactive. Thus, even elements that were >99% identical to this consensus could be inactive, and no human elements that were <99% identical were found to be active. We determined that the human genome contains at least 119 elements with >99% nucleotide identity to the active human L1 consensus (Brouha et al. 2003) within the regions encoding ORF1 and ORF2 (data set 4 [online only]). In contrast, no L1 ORFs in the chimpanzee genome or BACs had >99% identity to this active human L1 consensus (data set 3 [online only]). We cannot rule out the possibility that some of the chimpanzee L1 elements that are <99% identical to the human L1 consensus could remain active. Other “hot L1” elements might have evolved separately in chimpanzees that are >1% variant from the most-active human elements. However, since we failed to detect extended subfamilies of L1-PA2 or L1-Hs elements within our collection of chimpanzee insertions (analogous to the L1-Ta subfamily in humans), such elements generally would be present at low copy numbers in the chimpanzee genome. Thus, the landscape of potentially functional L1 elements in chimpanzees appears to be quite different from the landscape of active L1 elements in humans.
To verify these ORF results, we used an ORF1-trapping method to recover full-length ORF1 sequences from L1 elements in humans and chimpanzees (Ivics and Izsvak 1997). We recovered and sequenced 41 intact ORF1 sequences from humans and 51 intact ORF1 sequences from chimpanzees (see the tab-delimited ASCII files, which can be imported into spreadsheets, of data sets 6 and 7 [online only]) and observed results that were very similar to those obtained through the computational methods described above. None of the intact chimpanzee ORF1 sequences recovered through ORF1 trapping were >99% identical to the active human L1 consensus, whereas 17 (41%) of the 41 intact human ORF1 sequences were >99% identical to the active human L1 consensus (data sets 6 and 7 [online only]). Almost all of the intact ORF1 sequences trapped from chimpanzees (48/51; 94%) were L1-PA2 ORF1 sequences (data set 6 [online only]). In contrast, only 21 (51%) of the 41 intact ORF1 sequences recovered from humans were L1-PA2 ORF1 sequences and 18 (44%) were L1-Hs sequences (data set 7 [online only]). Thus, our ORF1-trapping experiments confirmed that the most recently active elements in chimpanzees (i.e., those with intact ORFs) contained ORF1 sequences that were divergent from the active L1 consensus in humans (Brouha et al. 2003).
Our method for trapping ORF1 in humans and chimpanzees employed the pβFUS plasmid (Ivics and Izsvak 1997). Briefly, full-length ORF1 sequences were amplified from human and chimpanzee genomic DNA (NA1MR91 and NA03448A, respectively [Coriell Cell Repository]) using PCR. The PCR primers were identical for humans and chimpanzees, and had the following sequences 5′-CCTGATCTGCGGCCGCATGGGGAAAAAACAGAACAGAAAAACTGG-3′ and 5′-CGTCCGAACGATATCCATTTTGGCATGATTTTGCAGCGGCTGG-3′. We used a combination of human and chimpanzee ORF1 sequences to design these primers. The ORF1 sequences identified in our chimpanzee BAC experiments were aligned to generate a consensus sequence using ClustalW. This sequence was compared to the human L1 consensus, and we determined that the primers chosen were conserved in human L1 sequences. Finally, we compared the candidate primer regions with L1-PA2, L1-PA3, and L1-PA4 elements and determined that the primer sequences also were completely conserved in these elements. Thus, the primers chosen were capable of amplifying a wide spectrum of ORF1 sequences in both humans and chimpanzees (including L1-Hs, L1-PA2, L1-PA3, and L1-PA4 elements). The NotI and EcoRV restriction sites that were introduced for cloning purposes are underlined. PCR products were cut with these enzymes and ligated to NotI/SmaI-digested pßFUS, such that the complete ORF1 sequence would be inframe with the AUG-less lacZ in the plasmid. Recombinants were identified on LB medium containing X-gal (recombinants with ORFs were blue, whereas those without ORFs and the empty vector alone were white). DNA was prepared and sequenced at Agencourt Biosciences using the primers 5′-CCAGTCACGTTGTAAAACGAC-3′ and 5′-CTAGGCCTGTACGGAAGTGTTAC-3′. High-quality sequences were analyzed and assembled using Sequencher version 4.1.2.
In addition to Alu and L1 insertions, we also found that SVA elements have been highly active in humans and chimpanzees (table 1). In fact, SVA insertions were almost as abundant as L1 insertions in humans during the past ∼6 million years (table 1). SVA is an unusual composite element that contains four components: (1) a tandem repeat of TCTCCC(n), (2) an unusual Alu element in reverse orientation, (3) a central variable-number-of-tandem-repeat (VNTR) region that is rich in CpG sequences, and (4) a SINE-R sequence that was derived from an LTR element (Shen et al. 1994). SVA ends with a poly (A) tail and is flanked by TSDs that closely resemble the TSDs of Alu and L1 elements (Ostertag et al. 2003; Bennett et al. 2004). SVA recently was found to be highly polymorphic among humans (Bennett et al. 2004), and a few instances have been reported of SVA insertions causing diseases (Kobayashi et al. 1998; Ostertag et al. 2003). Our study now provides further evidence that SVA has been actively mobile in relatively recent primate history and may remain active today.
The ORF1 and ORF2 proteins of L1 elements perform a specific retrotransposition mechanism known as “target-primed reverse transcription” (TPRT) (Luan et al. 1993), in which L1 mRNAs are copied into cDNAs and integrated into the genome (reviewed by Ostertag and Kazazian 2001). Alu RNAs (and other cellular RNAs) can compete for the L1 machinery during the TPRT process, which leads to the retrotransposition of these alternative RNAs instead of the normal L1 mRNAs (Esnault et al. 2000; Wei et al. 2001; Dewannieux et al. 2003). This “trans” mechanism of retrotransposition is thought to have led to the massive expansion of Alu (Dewannieux et al. 2003) and SVA (Ostertag et al. 2003; Bennett et al. 2004) elements in the human genome. Therefore, if L1 elements are indeed less functional in chimpanzees, as predicted above (table 2), we likewise might expect to see fewer Alu and SVA insertions in the chimpanzee genome. Table 1 and figure 3 show that this is, in fact, the case. Since other factors also influence the amplification rates of Alu (and probably SVA) elements, these differences may not be totally caused by lower levels of L1 activity in chimpanzees. It is possible, for example, that humans had a larger number of potentially active Alu and SVA source elements than did chimpanzees in recent history. However, when combined with our other data demonstrating that chimpanzees (1) have fewer full-length L1 insertions than humans, (2) have fewer L1 elements with intact ORFs than humans, (3) have ORF sequences that are divergent from active human L1 elements, and (4) lack extended subfamilies of new insertions, these data collectively indicate that chimpanzees are likely to have supported lower levels of L1 activity in recent history compared with humans.
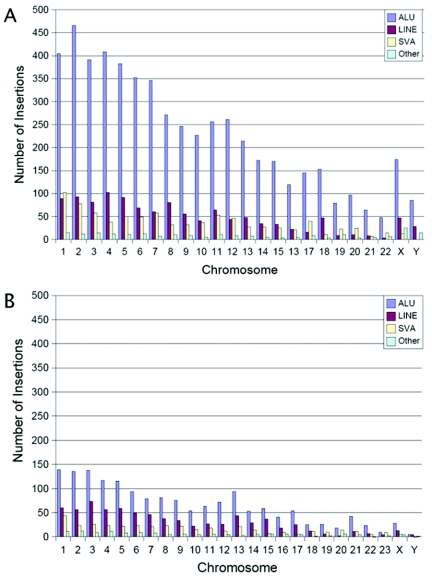
Figure 3.
Genomic distributions of transposon insertions. A, Genomic distribution of Alu, L1, SVA, and other elements in the human genome. B, Genomic distribution of Alu, L1, SVA and other elements in the chimpanzee genome. For both genomes, the number of insertions in each chromosome is generally proportional to the amount of DNA present. Note that the Y-axis is the same for both charts. Thus, many more transposon insertions are present throughout the human genome than the chimpanzee genome (compare the number of insertions depicted in panels A and B).
We next examined the genomic distributions of our recently mobilized transposon insertions. In both humans and chimpanzees, these insertions generally were distributed according to the amount of DNA that was present on each chromosome (fig. 3). We also examined the distributions of new insertions relative to genes (table 3 and tab-delimited ASCII files, which can be imported into spreadsheets, of data sets 8 and 9 [online only]). Approximately 34% of the new insertions in both genomes were located within known genes (defined as 3 kb upstream to 0.5 kb downstream of a RefSeq gene) (table 3 and data sets 8 and 9 [online only]). Using the same criteria, we determined that genes occupy ∼34% of the human and chimpanzee genomes (33.5% and 34.8%, respectively). Therefore, the fraction of insertions in genes was very close to that expected if integration (and mechanisms that subsequently remove insertions) had occurred randomly during the past ∼6 million years.
Table 3.
Transposon Insertions within Genes
| Insertions in Genes | Human | Chimpanzee |
| Total no. insertions in genes | 2,642 | 990 |
| No. of unique genes hit | 1,891 | 828 |
| No. of promoters | 50 | 13 |
| No. of exons | 7 | 4 |
| No. of introns | 2,478 | 973 |
| No. of terminators | 17 | 4 |
| No. unclassified | 90 | 0 |
| No. of insertions per gene: | ||
| 0 | 16,901 | 19,328 |
| 1 | 1,457 | 704 |
| 2 | 265 | 97 |
| 3 | 99 | 22 |
| 4 | 37 | 3 |
| 5 | 13 | 1 |
| 6 | 9 | 0 |
| 7 | 4 | 0 |
| 8 | 1 | 0 |
| 9 | 5 | 1 |
| 10 | 1 | 0 |
However, further analysis of these patterns revealed that they were not, in fact, random. Although we identified insertions in only ∼14% of all human genes, many of these genes had more than one insertion (table 3 and data set 9 [online only]). Overall, about a third of the human genes with insertions contained multiple insertions. Similar results were observed in the chimpanzee genome (16.5% of the genes with insertions had multiple insertions). We performed one-sample Z tests with our insertions and determined that the observed patterns were not consistent with a random integration model. For example, we observed 16,901 human genes that lacked new transposon insertions from our collections (table 3). The chance of observing this many human genes without such insertions is zero (P=0) with a random integration model. Similar results were observed with Z tests for the remaining integration classes listed in table 3 (data not shown). Therefore, our statistical tests allowed us to reject the hypothesis of random integration with a very high degree of confidence. On the basis of this analysis, it appears that a large fraction of the new transposon insertions in humans and chimpanzees (the majority of which were Alu, L1, and SVA elements) were targeted preferentially to specific genes. It is also possible that negative selection eliminated insertions from a larger initial collection over time, and this led to the appearance of nonrandom integration. Although targeted integration of L1 has not been observed previously in biochemical or cell culture experiments, previous studies indicate that transposons are eliminated through negative selection (Boissinot et al. 2001). Thus, negative selection is likely to have played a role in dictating the final patterns of transposons observed. Our data also may reflect an integration targeting mechanism that is not functional in cell-culture systems but is active in the germline of whole organisms, where all of our insertions occurred.
Our study indicates that a relatively large number of insertions occurred within genes during the evolution of humans and chimpanzees (2,642 in humans and 990 in chimpanzees) (table 3). It is likely that at least some of these insertions altered the expression of the target genes, perhaps to the extent that mutant phenotypes emerged. Thus, at least some of the insertions might have had an impact on the differential speciation of humans and chimpanzees by influencing the expression of nearby genes. Since humans received at least 4,853 additional transposon insertions compared with chimpanzees, the impact of transposon mutagenesis was likely to be greatest in humans during the past several million years.
In conclusion, we have determined that the original set of transposons in the common ancestor of humans and chimpanzees behaved differently during the subsequent evolution of these organisms. More than 95% of the new transposon insertions in both organisms were Alu, L1, and SVA insertions. However, our data indicate that humans and chimpanzees have amplified very different subfamilies of these elements. Our combined data also indicate that chimpanzees have supported lower levels of L1 activity than have humans during the past several million years, and this has led to decreased levels of Alu, L1, and SVA transposition in chimpanzees. Other factors, such as differences in population sizes and differences in population bottlenecks, also are likely to have influenced the final patterns of transposon insertions observed in these organisms. In some cases, apparent “insertions” may have been caused by the precise deletion of transposon copies through homologous recombination at the TSDs flanking these elements (van de Lagemaat et al. 2005). A fraction of our insertions also may have been older polymorphisms that were subject to lineage sorting. Thus, the final patterns of transposons in these genomes are likely to have been shaped not only by integration and excision mechanisms but also by the population dynamics of these organisms during the past several million years.
Supplementary Material
Acknowledgments
We thank the Washington University Genome Sequencing Center and the Whitehead Genome Center for their chimpanzee draft sequence data. We thank Zoltan Ivics for the pβFUS plasmid. We thank Shari Corin for critical review of the manuscript and for helpful advice. We thank Haiyan Wu for help with statistical analysis. This work was supported by National Institutes of Health training grant 2T32GM00849 (to E.A.B. and R.C.I.), a grant from SUN Microsystems (to W.S.P. and S.E.D.), and National Institutes of Health grant 1R01HG002898 (to S.E.D.).
References
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE (2004) Natural genetic variation caused by transposable elements in humans. Genetics 168:933–951 10.1534/genetics.104.031757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano A (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18:926–935 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schstak J, Badge RM, Lutz-Prigg S, Farbey AH, Moran JV, Kazazian HH (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 100:5280–5285 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87 10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35:41–48 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T (2000) Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24:363–367 10.1038/74184 [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA (2004) Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res 14:1068–1075 10.1101/gr.2530404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z (1997) Family of plasmid vectors for the expression of β-galactosidase fusion proteins in eukaryotic cells. Biotechniques 22:254–256 [DOI] [PubMed] [Google Scholar]
- Johanning K, Stevenson CA, Oyeniran OO, Gozal YM, Roy-Engel AM, Jurka J, Deininger PL (2003) Potential for retroposition by old Alu subfamilies. J Mol Evol 56:658–664 10.1007/s00239-002-2433-y [DOI] [PubMed] [Google Scholar]
- Jurka J (2000) Repbase update a database and an electronic journal of repetitive elements. Trends Genet 16:418–420 10.1016/S0168-9525(00)02093-X [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Hausler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006 10.1101/gr.229102. Article published online before print in May 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-lida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, Hamano K, Sakakihara Y, Nonaka I, Nakagome Y, Kanazawa I, Nakamura Y, Tokunaga K, Toda T (1998) An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394:388–392 10.1038/28653 [DOI] [PubMed] [Google Scholar]
- Landry JR, Mager DL (2003) Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J Virol 77:7459–466 10.1128/JVI.77.13.7459-7466.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 10.1016/0092-8674(93)90078-5 [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Kazazian HH (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH Jr (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73:1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH (2001) Biology of mammalian L1 retrotransposons. Annu Rev Genet 35:501–538 10.1146/annurev.genet.35.102401.091032 [DOI] [PubMed] [Google Scholar]
- Shen L, Wu L, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY (1994) Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and C4B genes in the HLA class III region. J Biol Chem 269:8466–8476 [PubMed] [Google Scholar]
- van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL (2005) Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res 15:1243–1249 10.1101/gr.3910705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS (1991) A de novo Alu insertion results in neurofibromatosis type 1. Nature 353:864–866 10.1038/353864a0 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Fujiyama A, Hattori M, Taylor TD, Toyoda A, Kuroki Y, Noguchi H, et al (2004) DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429:382–388 10.1038/nature02564 [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21:1429–1439 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CT, Jiang Z, McGrath SD, Hayden KE, Khaitovich P, Johnson ME, Eichler MY, McPherson JD, Zhao S, Pääbo S, Eichler EE (2005) Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol 3:e110 10.1371/journal.pbio.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.