Abstract
We previously identified three Avr9/Cf-9 Rapidly Elicited (ACRE) genes essential for Cf-9– and Cf-4–dependent hypersensitive response (HR) production in Nicotiana benthamiana. Two of them encode putative E3 ubiquitin ligase components. This led us to investigate other ACRE genes associated with the ubiquitination pathway. ACRE74 encodes a U-box E3 ligase homolog, highly related to parsley (Petroselinum crispum) CMPG1 and Arabidopsis thaliana PLANT U-BOX20 (PUB20) and PUB21 proteins, and was called Nt CMPG1. Transcript levels of Nt CMPG1 and the homologous tomato (Solanum lycopersicum) Cmpg1 are induced in Cf9 tobacco (Nicotiana tabacum) and Cf9 tomato after Avr9 elicitation. Tobacco CMPG1 possesses in vitro E3 ligase activity. N. benthamiana plants silenced for Nt CMPG1 show reduced HR after Cf-9/Avr9 elicitation, while overexpression of Nt CMPG1 induces a stronger HR in Cf9 tobacco plants after Avr9 infiltration. In tomato, silencing of Cmpg1 decreased resistance to Cladosporium fulvum. Overexpression of epitope-tagged tobacco CMPG1 mutated in the U-box domain confers a dominant-negative phenotype. We also show that Nt CMPG1 is involved in the Pto/AvrPto and Inf1 responses. In summary, we show that the E3 ligase Nt CMPG1 is essential for plant defense and disease resistance.
INTRODUCTION
Plants are continuously exposed to pathogen attack, but successful infection is rare. Many plant–microbe interactions can be described by the gene-for-gene model, in which a plant, via a resistance (R) gene, has the capacity to recognize a pathogen carrying the corresponding avirulence (Avr) gene (Dangl and Jones, 2001). This interaction induces a hypersensitive response (HR), comprising localized cell death around the infection site. Many R genes have been cloned from plants (Meyers et al., 2005), but the mechanisms involved in signal transduction after pathogen recognition are poorly understood.
Cladosporium fulvum is a biotrophic fungus that causes leaf mold in tomato (Solanum lycopersicum). Several resistance genes to C. fulvum have been cloned, of which Cf-9 is one of the best characterized (Rivas and Thomas, 2005). Cf genes encode receptor-like proteins, type I transmembrane protein with extracellular leucine-rich repeats, and a short cytoplasmic region (Thomas et al., 1998; Fritz-Laylin et al., 2005).
Cf-9 confers resistance to C. fulvum strains carrying the Avr9 gene, and infiltration of Avr9 peptide induces HR in tobacco (Nicotiana tabacum) and tomato plants carrying Cf-9 (Hammond-Kosack and Jones, 1997). Since the cloning of Cf-9, we have investigated how the defense response is produced after Cf-9/Avr9 recognition. Several early responses have been characterized that are activated in a Cf-9/Avr9-dependent manner, including ion fluxes (Blatt et al., 1999), production of reactive oxygen species (Piedras et al., 1998), and activation of mitogen-activated and calcium-dependent protein kinases (Romeis et al., 1999, 2000, 2001). An essential role for calcium-dependent kinases (Romeis et al., 2001) has been shown, as well as for SUPPRESOR OF G-2 ALLELE OF SKP1 (SGT1), a protein putatively required for the function of SCF (for Skp1/Cullin/F-box) ubiquitin ligases (Peart et al., 2002). More recently, Cf-9–interacting thioredoxin was identified, which negatively regulates the Cf-9–dependent responses (Rivas et al., 2004).
To get further insight into the molecular mechanism regulating Cf-9 function, many Avr9/Cf-9 Rapidly Elicited (ACRE) genes were identified that change their expression rapidly after Avr9 elicitation in Cf9 tobacco cells (Durrant et al., 2000). Most of those genes encode components of signaling pathways, including transcription factors, protein kinases, and ubiquitination pathway-related proteins. Virus-induced gene silencing (VIGS) of 42 ACRE genes identified three genes essential for Cf-9/Avr9 and Cf-4/Avr4 HR: a protein kinase–encoding gene (Avr9/Cf-9-INDUCED KINASE [ACIK1]), an F-box–encoding gene (ACRE189), and a U-box–encoding gene (ACRE276) (Rowland et al., 2005). The F-box and U-box proteins are ubiquitin ligase components (Deshaies, 1999; Hatakeyama et al., 2001), indicating a major role for this regulatory mechanism in the control of Cf-9–dependent responses.
Until recently, ubiquitination was primarily associated with proteasome-mediated protein degradation, but it is now clear that ubiquitination also regulates protein function in a proteasome-independent way. Ubiquitination alters protein localization, activity, and interactions (Schnell and Hicke, 2003). Ubiquitin is a 76–amino acid polypeptide that is attached to a protein target through an enzymatic cascade comprising a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (Glickman and Ciechanover, 2001). The E3 ligase confers the specificity of the reaction (Hershko et al., 1983; Finley et al., 2004) and can be a single protein or a protein complex. Single protein E3 ligases include HECT, RING finger, and U-box domain proteins (Moon et al., 2004). U-box domains resemble the RING finger domain (Aravind and Koonin, 2000). The relevance of the U-box domain for the ubiquitin activity of U-box proteins has been shown in different ways. The U-box domain interacts with E2 proteins (Pringa et al., 2001), and deletion of the U-box domain leads to a lack of ubiquitination activity (Ohi et al., 2003; Stone et al., 2003; Zeng et al., 2004). Arabidopsis thaliana encodes many U-box proteins compared with other organisms, and plant U-box proteins have been classified in different groups based on the presence of other domains (Azevedo et al., 2001; Mudgil et al., 2004). Few physiological roles for plant U-box proteins have been established. Plant U-box proteins are involved in hormone regulation (Amador et al., 2001), biotic and abiotic response (Yan et al., 2003; Zeng et al., 2004), self-incompatibility (Stone et al., 2003), and plant development (Kim et al., 2003).
In plants, ubiquitination modulates environmental and endogenous signals, including responses to pathogen attack (Hare et al., 2003). Despite much effort showing regulation of defense by the ubiquitination pathway, the identity of the involved E3 ligases remains almost unknown (Devoto et al., 2003). The only E3 ligases involved in plant pathogen response identified to date are the rice (Oryza sativa) U-box SPOTTED LEAF11 (SPL11; Zeng et al., 2004), the Arabidopsis RING finger proteins RPM1-INTERACTING PROTEIN2 (RIN2) and RIN3 (Kawasaki et al., 2005), and two ACRE genes identified in our laboratory (Rowland et al., 2005). In animals, ubiquitination regulates innate immune responses through Toll-Like Receptor (TLR) in different ways. Tumor necrosis factor Receptor-Associated Factor6 (TRAF6) is a RING domain–containing protein that is essential in Nuclear Factor-κB activation downstream of TLRs (Akira and Takeda, 2004). Another example is the RING finger protein Triad3A that ubiquitinates TLR4 and TLR9, controlling signaling through those receptors and avoiding a harmful effect after activation (Chuang and Ulevitch, 2004). Recent work also shows that the Shigella flexneri effector Outer Shigella protein G (OspG) controls human innate immunity through binding of ubiquitin-conjugating enzymes (Kim et al., 2005).
The discovery that two out of the three ACRE genes essential for Cf-9– and Cf-4–dependent HR encode E3 ligases led us to consider other ACRE genes encoding putative ubiquitin ligases for analysis in Cf-mediated defense mechanisms. In this work, we describe the characterization of tobacco CMPG1, a U-box protein essential for Cf-9–dependent HR production and for Cf-9–mediated resistance to C. fulvum. Tobacco CMPG1 can ubiquitinate in vitro, and mutation in the U-box domain abolishes this activity and confers a dominant-negative function on Nt CMPG1, indicating that this domain is essential for the physiological role of the protein. We also describe a role of tobacco CMPG1 in the Pto/AvrPto interaction and in the response to Inf1 elicitor.
RESULTS
ACRE74 Is a U-Box Protein with High Similarity to Parsley CMPG1 and Arabidopsis PLANT U-BOX20 and 21
We earlier reported the homologies of 178 cDNA-amplified fragment length polymorphism (AFLP) fragments, corresponding to ∼110 ACRE genes (Rowland et al., 2005). A BLAST search using one of those fragments, ACRE74, identified parsley (Petroselinum crispum) CMPG1, a rapidly induced gene after elicitation with Pep13 (Kirsch et al., 2001). We therefore refer to ACRE74 as Nt CMPG1. Tobacco CMPG1 also showed high similarity with Arabidopsis PLANT U-BOX20 (PUB20) and PUB21, both encoding U-box proteins (Azevedo et al., 2001). These sequence similarities suggest a role for Nt CMPG1 in early signaling in defense and in protein modification by ubiquitination, making tobacco CMPG1 an interesting candidate for further investigation in plant defense.
An Nt CMPG1 cDNA-AFLP fragment was used as a probe to screen a cDNA library from Avr9-elicited tobacco cells (Durrant et al., 2000). Several independent cDNA clones were sequenced, and a clone containing the exact sequence of the Nt CMPG1 cDNA-AFLP fragment was selected for further studies.
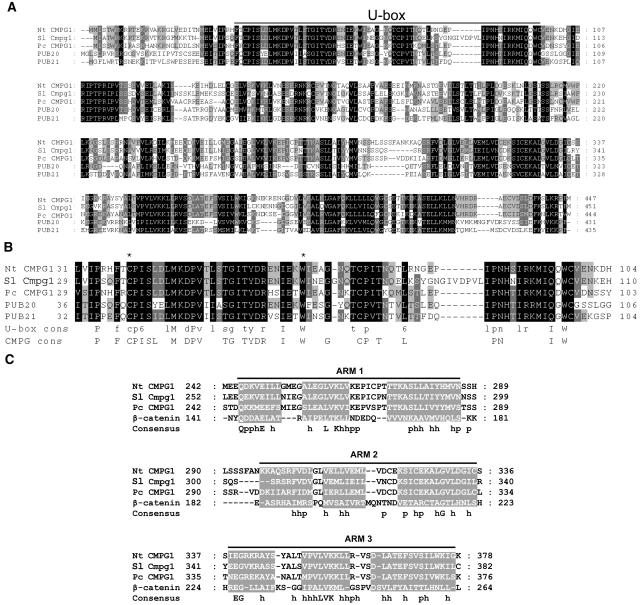
We identified in The Institute for Genomic Research (TIGR) tomato database an EST sequence with 73.6% identity to Nt CMPG1 (sequence identification TC159549) that we designated Sl Cmpg1. Primers were designed for 3′-rapid amplification of cDNA ends (RACE) of tomato Cmpg1 to obtain the full-length cDNA sequence. Three RACE fragments were identified that overlap with TC159549. Primers were designed based on the 5′ and 3′ cDNA sequences to isolate the full length of tomato Cmpg1 cDNA. Tobacco CMPG1 was predicted to encode a protein of 447 amino acids with a molecular mass of 50 kD. A SMART (Schultz et al., 1998; Letunic et al., 2004) search reveals an N-terminal U-box domain (Figure 1A) in the Nt CMPG1 protein. For tomato Cmpg1, the predicted protein had 451 amino acids with a molecular mass of 50 kD. These protein sequences share 73.8% identity.
Figure 1.
Sequence of Nt CMPG1 Protein and Alignment with Homologs.
(A) Protein sequence comparison between tobacco Nt CMPG1, tomato Sl Cmpg1, parsley Pc CMPG1, and Arabidopsis PUB20 and PUB21 was performed using ClustalW software. Position of the U-box domain is indicated. Shading represents the conservation of the residues among the sequences of the alignment: black, the residue is conserved in all the sequences; dark gray, the residue is conserved in four proteins of the alignment; light gray, the residue is conserved in three proteins of the alignment.
(B) Protein sequence alignment of the U-box domain of Nt CMPG1 homologs and comparison with U-box and CMPG consensus sequences. Asterisks show the amino acid residues mutated in this work to generate the dominant-negative versions of Nt CMPG1.
(C) Protein sequence comparison between the ARM repeats of Nt CMPG1, Sl Cmpg1, Pc CMPG1, and β-catenin (from human). The gray boxes indicate the protein region where an α-helix secondary structure was predicted. The consensus sequence is indicated, where p indicates polar residue and h indicates hydrophobic residue.
A BLAST search revealed parsley CMPG1 as the closest homolog of tobacco CMPG1 in the database. Two highly similar proteins are also found in Arabidopsis, PUB20 and PUB21. Figure 1A shows the protein alignment of Nt CMPG1 with putative orthologs in tomato (Sl Cmpg1), parsley (Pc CMPG1), and Arabidopsis (At PUB20 and At PUB21). Parsley CMPG1 shares 62.8% identity with tobacco CMPG1 protein. Of the two Arabidopsis proteins, Nt CMPG1 shows 43.1% identity to PUB20 and 36.2% identity to PUB21. The U-box domain is highly similar in all these proteins (Figures 1A and 1B). A comparison of U-box and CMPG1 consensus domains (Figure 1B) showed that most of the residues are conserved between them. An ARMADILLO (ARM) repeat domain has been identified in Pc CMPG1, At PUB20, and At PUB21. Parsley CMPG1 contains three ARM repeats from residue 255 to 375 (Zeng et al., 2004). Arabidopsis PUB20 and PUB21 were originally described as class III plant U-box proteins, which were characterized by a C-terminal leucine-rich region (Azevedo et al., 2001). However, using a sophisticated approach, Mudgil et al. (2004) identified ARM repeats in proteins of this class, including PUB20 and PUB21 where four and five repeats were found, respectively. Despite the low similarity of Sl Cmpg1 and Nt CMPG1 protein sequences with other ARM repeat–containing proteins, analysis of protein secondary structure using PSIPRED (Jones, 1999; McGuffin et al., 2000) and JPRED servers (Cuff et al., 1998) revealed a series of short α-helices consistent with the presence of three ARM repeats in both tobacco and tomato CMPG1 at the same position as for parsley CMPG1 (Figure 1C). Figure 1C shows the predicted secondary structure of the putative ARM repeat domain of tomato, tobacco, and parsley CMPG1. The same pattern of α-helices as for the β-catenin ARM repeat domain was found in the C-terminal region of the three proteins, indicating a conserved ARM repeat domain among tomato, tobacco, and parsley CMPG1.
Nt CMPG1 and Sl Cmpg1 mRNAs Are Induced after Avr9 Elicitation in Cf9 Tobacco and Tomato
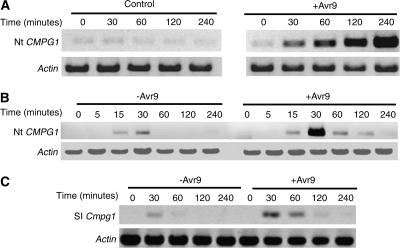
To confirm the Cf-9/Avr9-dependent induction of tobacco CMPG1 gene expression, we analyzed Nt CMPG1 transcript levels after Avr9 elicitation. Cf9 tobacco cell cultures were elicited with Avr9 peptide (+Avr9) or mock inoculated (control), and tobacco CMPG1 transcript levels were analyzed at different times after elicitation. RT-PCR was conducted with tobacco CMPG1-specific primers. Actin primers were used as a control for a constitutively expressed gene (Figure 2A). Consistent with the cDNA-AFLP data, Nt CMPG1 mRNA levels were upregulated 30 min after Avr9 elicitation, reaching a maximum level by 4 h. No transcript induction was detected in the negative control. We also observed an induction of Nt CMPG1 in Cf9 tobacco leaves after infiltration with a solution of intercellular fluid (IF) from Avr9-transgenic N. tabacum plants (+Avr9) or IF from tobacco wild-type plants (−Avr9). Elevated transcript levels were detected 15 min after infiltration with a maximum level at 30 min in both treatments (Figure 2B). However, the addition of IF(+Avr9) produced a stronger induction of tobacco CMPG1 gene expression at the 30-min time point, and the expression was maintained over 2 h of the time course. Transient upregulation of gene expression by the mechanical stress caused by infiltration has been observed for other ACRE genes (Durrant et al., 2000; Rowland et al., 2005).
Figure 2.
Expression Patterns of Nt CMPG1 and Sl Cmpg1 after Avr9 Elicitation.
Cf9 tobacco cell cultures (A) were treated with Avr9 peptide (+Avr9) or untreated (control), and samples were harvested at the time points indicated. Cf9 tobacco plants (B) or Cf9 tomato plants (C) were infiltrated with IF that contains Avr9 (+Avr9) or that does not contain Avr9 (−Avr9), and plant leaf discs were harvested at the time points indicated. Total RNA was isolated and used for RT-PCR with specific primers for Nt CMPG1, Sl Cmpg1, and Actin.
To investigate the role of tomato Cmpg1 as a putative ortholog of tobacco CMPG1, we analyzed the expression pattern of Sl Cmpg1 after Avr9 elicitation. Cf9 tomato plants were infiltrated with IF with or without Avr9, and tomato Cmpg1 transcript levels were analyzed by RT-PCR (Figure 2C). As observed for Nt CMPG1, Sl Cmpg1 transcript levels were induced after infiltration with both IF(+Avr9) and IF(−Avr9), but again, the induction was stronger and lasted longer after infiltration with IF(+Avr9). These results suggest a role for tomato Cmpg1 in the Cf-9/Avr9-dependent response and the possibility that Sl Cmpg1 is the Nt CMPG1 ortholog.
Tobacco CMPG1 Has in Vitro Ubiquitination Activity
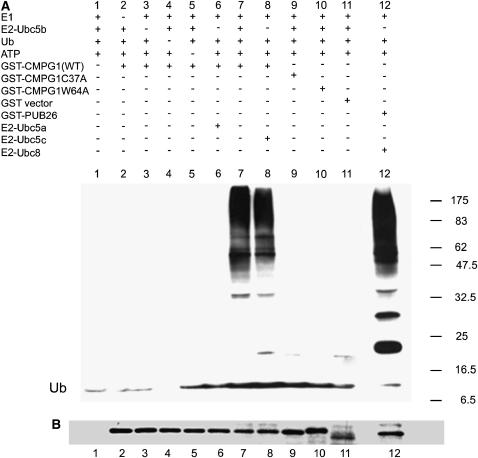
We wanted to determine if tobacco CMPG1 has in vitro ubiquitination activity. A full-length clone of Nt CMPG1 was expressed in Escherichia coli as a C-terminal fusion with glutathione S-transferase (GST) and purified by affinity chromatography. Ubiquitination assays were performed using yeast E1 and three different E2 enzymes (UbcH5a, UbcH5b, and UbcH5c). As a positive control, the Arabidopsis U-box protein PUB26 (Azevedo et al., 2001) fused to GST was assayed. The reactions were separated on an SDS gel and transferred to nitrocellulose membranes. Ubiquitinated proteins were detected using anti-ubiquitin antibody, and CMPG1 was detected using anti-GST antibody (Figures 3A and 3B). Tobacco CMPG1-GST displayed E3 ligase activity in the presence of UbcH5b and UbcH5c but not with UbcH5a (Figure 3A, lines 6 to 8). E3 ligase activity was not observed in the absence of any of the essential components of the reaction (E1, E2, E3, ubiquitin, or ATP; Figure 3, lines 1 to 5, respectively) or using GST alone as ubiquitin ligase (Figure 3, line 11). Thus, tobacco CMPG1 has in vitro ubiquitination activity.
Figure 3.
Nt CMPG1 Is an E3 Ligase, and Mutations in Its U-Box Domain Abolish Its Activity.
GST-Nt CMPG1, GST-Nt CMPG1C37A, and GST-Nt CMPG1W64A proteins were expressed and purified from E. coli and tested for E3 ligase activity using yeast E1 and three differents E2s. GST-PUB26 with the E2 Ubc8 was used as a positive control (line 12). Lines 1 to 5 and 11 are negative controls. The reactions were analyzed by protein gel blots using anti-ubiquitin antibody (A) or anti-GST antibody (B).
We then investigated if, as for other U-box proteins described up to date, the U-box domain is essential for the E3 ligase activity of CMPG1. The U-box domain of E3 ligases is predicted to be involved in the interaction with the E2 ubiquitin-conjugating enzyme (Pringa et al., 2001), and several of its amino acids have been described to be essential for this interaction (Pringa et al., 2001; Ohi et al., 2003). The putatively E2-interacting amino acid Trp-64 and the highly conserved Cys-37 were mutated to Ala in the tobacco CMPG1 protein (Figure 1B). Both mutated proteins were expressed as GST fusions, and the E3 ligase activity was assayed as for the wild type. Figure 3 shows that neither of the CMPG1 mutants displayed ubiquitination activity (lines 9 and 10). Therefore, an intact U-box domain is essential for Nt CMPG1 E3 ligase activity.
Tobacco CMPG1 Is Required for Cf-9/Avr9-Dependent HR in Nicotiana benthamiana Plants
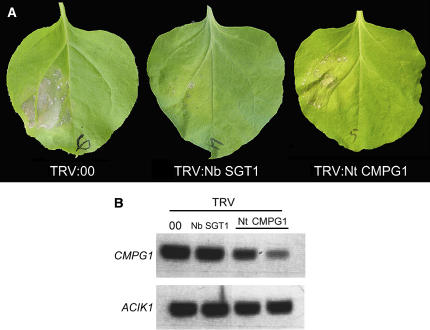
To investigate if CMPG1 protein is essential for Cf-9/Avr9-dependent HR, we performed VIGS in N. benthamiana plants using tobacco rattle virus (TRV; Ratcliff et al., 2001) as a viral vector. A fragment of 374 bp from the 3′ end of tobacco CMPG1 was cloned in a TRV vector (TRV:Nt CMPG1). TRV empty vector (TRV:00) was used as a negative control. No differences in morphology were observed between CMPG1-silenced plants and those silenced with the empty vector. SGT1 has been previously shown to be essential in Cf-9/Avr9-dependent HR production (Peart et al., 2002); therefore, the TRV:Nb SGT1 construct (Peart et al., 2002) was used as a positive control. N. benthamiana seedlings were inoculated via Agrobacterium tumefaciens with TRV:00, TRV:Nb SGT1, or TRV:Nt CMPG1. Three weeks later, agrobacteria expressing Cf-9 and Avr9 were delivered into the silenced plants, and HR was assessed after 5 d. No HR was observed in plants silenced for SGT1, consistent with previous results (Figure 4A, Table 1). We observed strong HR in 36 out of 43 TRV:00-silenced plants. The number of CMPG1-silenced plants that developed HR was much lower than in the TRV:00-silenced plants (Figure 4A, Table 1). Figure 4B shows that CMPG1 transcript accumulation is reduced in two different plants carrying the TRV:Nt CMPG1 construct compared with control. This experiment was repeated three times, and in each repetition, lower levels of tobacco CMPG1 were observed in at least three plants carrying TRV:Nt CMPG1 and showing low HR symptoms. These data indicate that CMPG1 is essential for efficient production of Cf-9/Avr9-mediated HR in N. benthamiana plants.
Figure 4.
Nt CMPG1 Is Required for Efficient HR Mediated by Cf-9 in N. benthamiana.
(A) VIGS in N. benthamiana using TRV. Seedlings were inoculated with TRV:Nt CMPG1, TRV:Nb SGT1, or empty vector as a control (TRV:00). Three weeks after inoculation, leaves were infiltrated with agrobacteria delivering Cf-9 and Avr9. Pictures were taken 4 d after agrobacteria infiltration.
(B) RT-PCR from silenced plants. N. benthamiana leaves silenced with the indicated TRV construct were infiltrated with water to induce Nb CMPG1 expression. Samples were collected 30 min after infiltration, and RNA was isolated and used for RT-PCR. Nb ACIK1 was used as a control for induction by flooding (bottom panel).
Table 1.
Number of TRV-Silenced Leaves of N. benthamiana Showing HR after Cf-9/Avr9 Elicitation
| Silencing Construct | Leaves Producing HR | Total Leaves Infiltrated | Leaves Producing HR (%) |
|---|---|---|---|
| TRV:00 | 36 | 43 | 83.7 |
| TRV:Nb SGT1 | 2 | 43 | 4.6 |
| TRV:Nt CMPG1 | 13 | 40 | 32.5 |
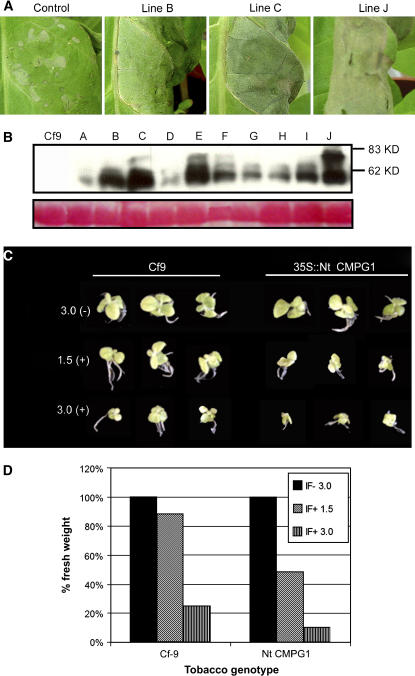
Overexpression of Nt CMPG1 Increases the HR in Cf9 Tobacco Plants after Avr9 Elicitation
To confirm the role of tobacco CMPG1 in HR, we overexpressed Nt CMPG1 in tobacco plants. We engineered a construct to produce tandem affinity purification (TAP)–tagged (Rigaut et al., 1999) CMPG1 protein under the control of the 35S promoter, and stable transformants in Cf9 tobacco were generated. Ten independent transgenic lines were analyzed for HR and for CMPG1:TAP protein accumulation. Tobacco leaves from transgenic and nontransgenic (control) plants were infiltrated with 1/16 dilution of IF(+Avr9) and IF(−Avr9) (Figure 5A). A weak HR was produced at this low concentration of Avr9 in control plants (Figure 5A, left panel), while 3 out of the 10 transgenic tobacco lines developed strong HR symptoms (Figure 5A, lines B, C, and J). CMPG1:TAP protein levels were analyzed by immunoblots in the same tobacco leaves that were infiltrated for HR production (Figure 5B). We observed that the three tobacco lines that responded with a strong HR accumulate high levels of CMPG1 protein. A higher molecular mass (∼75 kD) second band can be detected in some of the tobacco transgenic lines. This band was found in different lines in different experiments, and it does not correlate with the strong HR phenotype. We speculate that this higher molecular form of tobacco CMPG1 might be due to a posttranslational modification that has not been further investigated. We chose the line with the strongest HR phenotype (line J) to analyze the response to Avr9 using a different approach. Ten-day-old seedlings from tobacco CMPG1:TAP line J and control were transferred to Murashige and Skoog (MS) medium containing different concentrations of IF(+Avr9) or IF(−Avr9) (Figures 5C and 5D). The presence of Avr9 in the medium elicited a response in Cf9 seedlings that is visualized by a growth inhibition effect in an Avr9 concentration-dependent manner (Figure 5C, left panel). Plants overexpressing CMPG1 are more sensitive to Cf-9/Avr9-dependent growth inhibition at two different Avr9 concentrations (Figures 5C and 5D). In conclusion, by two different approaches, we have observed a stronger Avr9-dependent HR in tobacco plants overexpressing tobacco CMPG1. These results suggest that Nt CMPG1 is a rate-limiting protein in HR production triggered by Cf-9.
Figure 5.
Overexpression of Nt CMPG1 Induces a Stronger HR Response to Avr9 in Cf9 Tobacco Plants.
(A) HR in N. tabacum. Cf9 tobacco plants were transformed with 35S:Nt CMPG1:TAP and analyzed for Avr9-dependent HR production. Tobacco leaves from three independent transgenic lines and a control were infiltrated with a 1/16 dilution of IF(+Avr9). Pictures were taken 3 d after infiltration.
(B) Nt CMPG1 protein levels were analyzed by protein gel blots. Leaf discs were harvested from different Cf9 tobacco lines expressing 35S:Nt CMPG1:TAP or control leaves. Total proteins were separated in SDS gels and analyzed by inmunoblotting using PAP antibody for Nt CMPG1:TAP detection. Bottom panel shows Ponceau S Red staining of ribulose-1,5-biphosphate carboxylase/oxygenase for confirmation of equal loading.
(C) Growth inhibition assay. Cf9 tobacco seedlings expressing Nt CMPG1:TAP (line J) and control were transferred to MS medium containing the indicated amount of IF(+Avr9) (+) or IF(−Avr9) (−) (μL per mL of MS medium). Pictures were taken 10 d after transfer to MS medium.
(D) Growth inhibition assay. Cf9 tobacco seedlings were transferred to MS medium containing IF(−Avr9) or IF(+Avr9) as described in (C). Ten days after the transfer, the same number of seedlings was weighted for each treatment. The percentage was calculated based on the weight of seedling growth in IF(−Avr9) containing MS medium.
Mutations in Essential Amino Acids of Nt CMPG1 U-Box Domain Confer a Dominant-Negative Effect in Cf9 Tobacco Plants
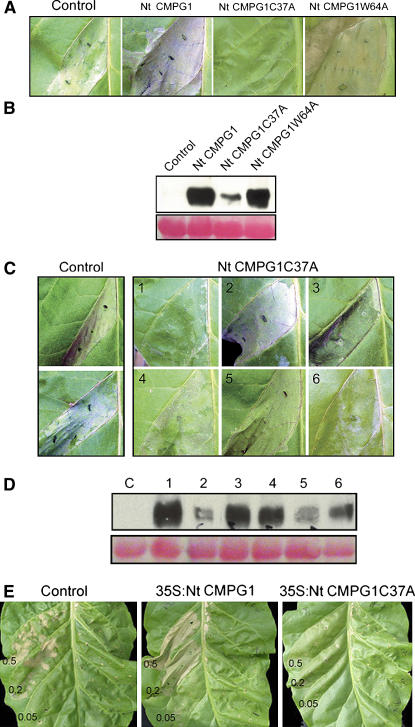
We wanted to investigate if the role of tobacco CMPG1 in defense is dependent on its E3 ligase activity. For this purpose, we compared Cf-9–dependent HR production in tobacco plants overexpressing tobacco CMPG1 wild type and the two mutants (CMPG1W64A and CMPG1C37A) that did not show activity in the ubiquitination experiments (Figure 3).
We generated transgenic plants carrying 35S promoter fusions for overexpression in Cf9 tobacco of TAP-tagged CMPG1W64A (11 plants) and CMPG1C37A (eight plants). Untransformed (control), wild-type Nt CMPG1-transformed, and mutated Nt CMPG1-transformed tobacco leaves were infiltrated with IF(+Avr9), and HR was observed 3 d after infiltration. We observed a reduced HR compared with controls after Avr9 elicitation in 2/8 primary transformants carrying CMPG1C37A (Figure 6A), suggesting a dominant-negative effect of this mutation. By contrast, none of the CMPG1W64A T1 transgenic lines showed a clear HR reduction compared with untransformed controls (Figure 6A). Immunoblot analysis indicated the presence of the protein in the leaves tested for HR. The differences observed in protein level accumulation are correlated with the locus number of the transgene (based on segregation analysis) (Figure 6B; see Supplemental Table 1 online). Segregating T2 tobacco plants carrying either tobacco CMPG1 mutant were analyzed for HR and protein levels (Figures 6C and 6D). We observed no HR in the tobacco plants that accumulated high levels of CMPG1C37A protein. Similar results were found in the analysis of CMPG1W64A lines B and D, where six segregating T2 tobacco plants were analyzed for protein accumulation and HR production. High levels of CMPG1W64A protein were correlated with a reduced HR (see Supplemental Table 1 and Figure 1 online) in both lines. Supplemental Figure 1 online shows the HR production and CMPG1W64A protein accumulation of the line D. Similar results were found for line B. Only two of the lines that carried more than one T-DNA locus (based on segregation results; data not shown) showed a dominant-negative phenotype in the T2 for the Nt CMPG1W64A mutant (see Supplemental Table 1 online).
Figure 6.
Overexpression of Nt CMPG1 Dominant-Negative Mutants.
(A) Leaf panels from Cf9 tobacco control plants (control) and Cf9 tobacco plants transgenic for Nt CMPG1 wild type and mutants (C37A line I and W64A line B) were infiltrated with IF(+Avr9) for HR induction. Pictures were taken 3 d after infiltration.
(B) Tobacco leaf discs were harvested from the same leaves shown in (A), and Nt CMPG1:TAP protein levels were determined as described in Figure 5B.
(C) The presence of high levels of Nt CMPG1C37A correlates with a reduced HR production. Two leaves of Cf9 tobacco control plants (left panels) and of six different plants of the segregating population of transgenic Nt CMPG1C37A tobacco (right panels) were infiltrated with IF(+Avr9) for HR induction. Pictures were taken 3 d after infiltration.
(D) Nt CMPG1C37A protein levels were analyzed by protein gel blots. Tobacco leaf discs were harvested from the same leaves shown in (C), and Nt CMPG1C37A-TAP protein levels were determined as described in Figure 5B. Numbers on top of the lanes correspond to the numbers of the leaves shown in (C). C, control leaf.
(E) Leaf panels of Cf9 tobacco control and Cf9 tobacco overexpressing Nt CMPG1 wild type or Nt CMPG1C37A were infiltrated with agrobacteria delivering Pto and AvrPto at the indicated cell densities. Pictures were taken 4 d after infiltration.
Tobacco CMPG1 Is Involved in Pto/AvrPto and Inf1-Mediated Cell Death
To investigate if tobacco CMPG1 function is specific to Cf-9–dependent responses or if it is involved in different defense mechanisms, we analyzed the Pto/AvrPto-dependent (gene-for-gene system) and Inf1-dependent (general elicitor) HR in tobacco plants overexpressing CMPG1 and CMPG1C37A. Pto is a tomato R protein that confers resistance against Pseudomonas syringae carrying AvrPto (Pedley and Martin, 2003). By contrast, Inf1 is a secreted protein from Phytophthora infestans that induces HR in Nicotiana spp (Kamoun et al., 1998).
Tobacco leaves were infiltrated with different densities of agrobacteria that confer transient expression of Pto and AvrPto (Figure 6E) or Inf1 (see Supplemental Figure 2A online) and HR assessed. Pictures were taken 4 d after infiltration. Tobacco CMPG1 proteins levels are homogeneous among the individual plants in each line. Interestingly, Figure 6E showed that after Pto/AvrPto infiltration, HR was stronger in tobacco leaves overexpressing CMPG1 compared with nontransgenic control, while no HR was observed in tobacco plants expressing CMPG1C37A. A weaker but consistent effect on HR production was observed in similar experiments with Inf1 elicitor (see Supplemental Figure 2A online). To confirm the involvement of tobacco CMPG1 in Inf1-dependent HR production, stable tobacco lines carrying a hairpin tobacco CMPG1 fragment were engineered. Ten independent tobacco Cf9 transformants were generated, and two lines showing low Cf-9–dependent HR production were selected (see Supplemental Figure 2B online). Inf1 elicitor was infiltrated via agrobacteria in the same tobacco lines carrying CMPG1 hairpin and untransformed controls. Supplemental Figure 2C online shows that the HR production is clearly reduced in the two lines silenced for tobacco CMPG1. RT-PCR experiments showed that the accumulation of CMPG1 transcript was clearly reduced in both lines (see Supplemental Figure 2D online). These results suggest that tobacco CMPG1 is involved in plant defense mechanisms triggered by various elicitors.
Cmpg1 Is Involved in Resistance to C. fulvum in Tomato Cf9 Plants
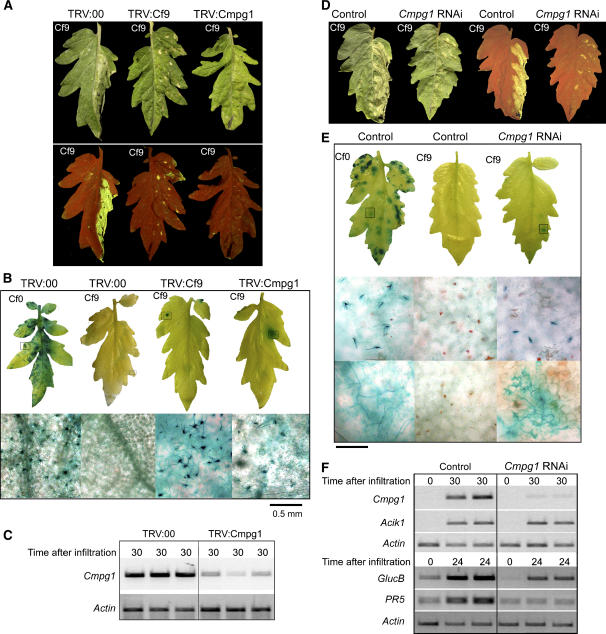
To investigate if CMPG1 is involved in resistance of tomato to C. fulvum, in addition to HR production, we silenced Sl Cmpg1 in tomato plants using two different approaches, VIGS and double-stranded RNA hairpin-induced silencing. Similar to CMPG1-silenced tobacco plants, no differences in morphology were observed when tomato Cmpg1 was silenced with any of the approaches. For the first approach, the TRV-based vector described by Liu et al. (2002a) was used. A 200-bp fragment of the 5′ coding sequence of tomato Cmpg1 was cloned in the TRV vector (TRV:Cmpg1). As controls, TRV empty vector (TRV:00) and TRV carrying a Cf-9 fragment previously described (TRV:Cf9; Rowland et al., 2005) were used. TRV constructs were inoculated via agrobacteria in cotyledons of either Cf0 tomato seedlings or tomato lines transgenic for the Cf-9 gene. Three weeks after inoculation, tomato plants were analyzed for HR, C. fulvum infection, and tomato Cmpg1 transcript accumulation. Three independent tomato plants were silenced with each construct in each experiment. Tomato-silenced plants were infiltrated with IF(+Avr9) or IF(−Avr9) (Figure 7A, right and left halves of the leaf, respectively) to confirm the HR phenotype observed in N. benthamiana plants silenced for tobacco CMPG1 (Figure 4A). Figure 7A shows HR production in Cf9 tomato plants silenced with the different TRV constructs. As expected, HR was highly reduced in tomato plants silenced for Cmpg1 and Cf-9, but strong HR was observed in tomato plants silenced with the empty vector (Table 2). These data confirm those obtained using VIGS in N. benthamiana.
Figure 7.
Tomato Cmpg1 Is Required for Full Cf-9–Mediated Resistance to C. fulvum Expressing Avr9.
(A) VIGS in tomato. Transgenic Cf9 tomato cotyledons were inoculated with agrobacteria carrying the indicated TRV construct. Three weeks after infiltration, the right half of the tomato leaf was infiltrated with IF(+Avr9), and pictures were taken 3 d after infiltration under daylight (top panel) or UV light (bottom panel).
(B) Transgenic Cf9 or Cf0 tomato seedlings were silenced with the indicated TRV construct as described in (A). Three weeks later, plants were infected with C. fulvum race 4 GUS. Leaves were stained with X-gluc 3 weeks after C. fulvum inoculation, when pictures were taken (top panel). The fungal cycle was complete, as indicated by the conidiophores extruding from the stomata observed under the microscope (bottom panel).
(C) Tomato Cmpg1 transcript levels were analyzed by RT-PCR. Leaf discs from the indicated silenced tomato plants were harvested 30 min after infiltration with water. Total RNA was extracted and used for RT-PCR analysis with Sl Cmpg1-specific primers. Equal amounts of cDNA were used, as shown by the amplification with the constitutively expressed Actin gene.
(D) RNAi in tomato. Cf9 tomato plants were transformed with pHellsgate containing a tomato Cmpg1 hairpin. One leaf half from the transgenic (Cmpg1 RNAi) and untransformed (control) Cf9 tomato plants was infiltrated with IF(+Avr9) for HR induction. Pictures were taken 3 d after infiltration under daylight (left leaves) and UV light (right leaves).
(E) Cf0 and Cf9 untransformed (control) and Cf9 transgenic (Cmpg1 RNAi) tomato plants were infected with C. fulvum race 4 GUS. Leaves were stained with X-gluc 3 weeks after infection (top panel). Fungal cycle was complete, as indicated by the conidiophores extruding from the stomata observed under the microscope (middle panel). Hyphal growth inside the leaves was confirmed under the microscope (bottom panel).
(F) Tomato Cmpg1 mRNA levels were analyzed by RT-PCR. Leaf discs from the indicated tomato lines were harvested 30 min after IF(+Avr9) infiltration and processed as described in (C). Additionally, equal induction treatment is shown by the amplification of Sl Acik1. From the same leaves, samples were harvested after 24 h, and transcript levels for the defense-related genes GlucB and PR5 were determined using gene-specific primers.
Table 2.
Proportion of VIGS-Silenced Tomato Leaves Developing HR or Fungal Infection
| Silencing Construct | Tomato Genotype | HR | C. fulvum Growth |
|---|---|---|---|
| TRV:00 | Cf0 | NTa | 35/35 |
| TRV:00 | Cf-9 | 25/32 | 0/33 |
| TRV:Cf-9 | Cf-9 | 11/30 | 16/42 |
| TRV:Cmpg1 | Cf-9 | 4/29 | 18/50 |
NT, not tested.
To study the role of Cmpg1 in resistance to C. fulvum, tomato plants were infected with C. fulvum race 4 β-glucuronidase (GUS) 3 weeks after silencing. The expression of the GUS gene in the fungal race allows the detection of the fungus after staining with 5-bromo-4-chloro-3-indolyl-β-glucuronide (X-gluc). Three weeks after inoculation, at least 12 leaves from three different plants were stained with X-gluc for each TRV construct (Figure 7B, top panel). We observed blue staining in the leaves of Cf0 tomato plants, indicating fungal infection. We examined the blue spots under the microscope, where we observed the conidiophores emerging from the stomata, indicating that the fungus completed its cycle (Figure 7B, bottom panel). Interestingly, we also observed X-gluc staining in the Cf9 tomato leaves from plants silenced for Cf-9 or tomato Cmpg1. The same reproductive structures as in Cf0 plants were observed under the microscope. This result indicates that Cmpg1 is essential for full tomato resistance to C. fulvum. The number of infection patches was much lower in Cf9 plants silenced for Cf-9 or Cmpg1 in comparison with Cf0 plants. However, this phenotype was consistent, and for tomato Cmpg1-silenced plants, 18 out of 50 leaves showed at least one blue spot (Table 2), while in Cf-9–silenced plants, 16 out of 42 leaves presented at least one blue spot. No blue staining was detected in Cf9 tomato leaves silenced with the empty vector, indicating that the phenotype observed is not due to the virus. Finally, we confirmed that Cmpg1 transcript levels were reduced in the silenced plants. mRNA levels were determined by RT-PCR using tomato Cmpg1- specific primers and Actin primers as a control, in both empty vector and Cmpg1-silenced plants (Figure 7C). We could observe highly reduced levels of transcript accumulation in comparison with the empty vector in three independent silenced plants showing low HR accumulation.
To confirm VIGS results, RNA interference (RNAi)-induced silencing was performed. Cf9 tomato plants carrying Cf-9 and -4 linked paralogs (Parniske et al., 1997) were transformed with the pHellsgate8 vector (Wesley et al., 2001; Helliwell and Waterhouse, 2003) carrying a tomato Cmpg1 hairpin fragment. Eight independent transformant plants were obtained. Six leaves from each line were infiltrated with IF(+Avr9), and HR was observed. Six out of eight transformants showed clear reduction in HR in the six infiltrated leaves, while the other two lines showed similar HR to the nontransformed Cf9 tomato plants. Three lines were further analyzed, and we could correlate the levels of tomato Cmpg1 mRNA with PATHOGENESIS-RELATED5 (PR5) mRNA levels, HR development, and C. fulvum infection symptoms (see Supplemental Figure 3 online). C. fulvum infection was only observed in the line showing the lowest Cmpg1 and PR5 mRNA accumulation. The result obtained with the best line is shown in Figure 7. RT-PCR was conducted on RNA from untransformed (control) and transgenic (Cmpg1 RNAi) Cf9 plants after elicitation with flooding (Figure 7F, top panel). Tomato Acik1 (Rowland et al., 2005) and Actin primers were used as a control for an inducible and a constitutively expressed gene, respectively. Tomato Cmpg1 transcript levels were highly reduced in Cf9 plants carrying the hairpin construct. Untransformed (control) and transgenic (Cmpg1 RNAi) Cf9 tomato plants were also infiltrated with IF(+Avr9) or IF(−Avr9) (Figure 7D, right and left half of the leaf, respectively). Three days later, HR was examined under daylight (left leaves) and UV light (right leaves). Sixteen out of 18 control leaves showed a strong HR as in the picture, while only 6 out of 18 Cmpg1 RNAi leaves showed HR (Figure 7D, Table 3). Cf9 control, Cf9 Cmpg1 RNAi, and Cf0 control plants were infected with C. fulvum race 4 GUS as before. Three weeks after infection, leaves were stained with X-gluc to score for fungal growth. Again, we could observe strong X-gluc staining in Cf0 tomato plants, where the presence of conidiophores was confirmed under the microscope (Figure 7E). We also detected patches of fungal growth in Cmpg1 RNAi Cf9 tomato. As before, the infection in Cmpg1-silenced Cf9 plants was much less severe than in the Cf0 controls. We could observe fungal hyphae growing inside the leaves in all the blue patches of the transgenic plants (Figure 7E, bottom panel, Table 3), but we only detected reproductive structures in 10 out of the 19 leaves with blue patches (Figure 7E, middle panel, Table 3).
Table 3.
Proportion of Hairpin-Silenced Tomato Leaves Developing HR or Fungal Infection
| Transgene | Tomato Genotype | HR | C. fulvum Infection/Growth |
|---|---|---|---|
| – | Cf0 | NTa | 43/43 |
| – | Cf9 | 16/18 | 0/55 |
| Cmpg1 RNAi | Cf9 | 6/18 | 19/60 |
NT, not tested.
To further characterize the Cf-9–dependent defense response in tomato Cmpg1-silenced plants, the induction of defense-related genes was analyzed by RT-PCR. Cmpg1 RNAi and control tomato plants were infiltrated with IF(+Avr9), and samples were taken at 0 and 24 h after infiltration. Transcript levels of two defense-related genes, PR5 (Cornelissen et al., 1986; Hejgaard et al., 1991) and basic β-1, 3-glucanase (GlucB; Niki et al., 1998), were analyzed (Figure 7F, bottom panel). Cmpg1 RNAi plants showed a reduced induction of both genes at 24 h after infiltration in two independent samples.
DISCUSSION
Tobacco CMPG1 Encodes a U-Box E3 Ligase and Is Rapidly Upregulated after Stress Stimuli
We sought to identify genes upregulated during the Cf-9/Avr9 defense response to gain further insight into the mechanisms involved. This generated a list of upregulated genes that was scrutinized for potential signaling components and assessed for whether they are required for the Cf-9–mediated HR. Previously, we reported that two of the three ACRE genes identified as essential for Cf-9–mediated HR are predicted to encode components of the ubiquitylation machinery (Rowland et al., 2005). Further analysis of the previously published ACRE74 cDNA fragment revealed homology to parsley CMPG1, a rapidly elicited gene after elicitor treatment (Kirsch et al., 2001). A full-length ACRE74 cDNA was named Nt CMPG1. It shows strong similarity to Arabidopsis PUB20 and PUB21 and encodes a protein with an N-terminal U-box domain. Tobacco CMPG1 displays E3 ligase activity that is dependent on an intact U-box domain. Three highly similar E2 proteins (UbcH5a, UbcH5b, and UbcH5c; Kraft et al., 2005) were used in our experiments. We showed that Nt CMPG1 ubiquitinates with two E2 proteins (UbcH5b and UbcH5c), while no activity was observed with UbcH5a. These data indicate that the E2 specificity is a determining factor in the activity of tobacco CMPG1.
Tobacco and tomato CMPG1 gene expression are induced after elicitor and wounding treatments (Figure 2), a typical ACRE gene expression pattern (Durrant et al., 2000; Rowland et al., 2005). Parsley CMPG1 gene expression is induced 5 min after elicitation in parsley cell cultures (Kirsch et al., 2001), reaching maximum levels 1 h after elicitor treatment. Expression of the Arabidopsis CMPG1 homologs is also elevated after pathogen treatment (Heise et al., 2002; Navarro et al., 2004). In a search for Arabidopsis orthologs of parsley CMPG1, Heise et al. (2002) identified and classified several At PUB proteins as At CMPG-like. In this classification, At PUB20 corresponds to At CMPG1, and At PUB21 to At CMPG5. Arabidopsis PUB20 expression is induced after treatment with the pathogen elicitors Pmg and flg22 and after infection with P. syringae pv tomato strain DC3000, reaching maximum expression between 30 and 60 min after elicitation (Heise et al., 2002; Navarro et al., 2004). These data suggest a similar role for tobacco, tomato, Arabidopsis, and parsley CMPG1 in plant defense signaling.
An alignment of Nt CMPG1 with Sl Cmpg1, Pc CMPG1, At PUB20, and At PUB21 revealed high similarity in the U-box domain and also in the C-terminal region of the proteins (Figure 1A). The presence of ARM repeats has been described for Pc CMPG1, PUB20, and PUB21. ARM repeats are highly divergent in class III U-box proteins to which tomato and tobacco CMPG1 belong (Mudgil et al., 2004). Based purely on sequence homology, ARM repeats were not found in Nt CMPG1 and Sl Cmpg1 proteins but were revealed by analysis of secondary structure. The putative ARM repeats are conserved among tobacco, tomato, and parsley CMPG1 proteins (Figures 1A and 1C; Zeng et al., 2004), suggesting that this domain is essential for the function of CMPG1-like proteins.
Tobacco CMPG1 Is a Positive Regulator of Plant Defense Mechanisms
Using VIGS, we showed that tobacco and tomato CMPG1 are essential for efficient HR activated by Cf-9 (Figures 4 and 7). Also, overexpression of tobacco CMPG1 enhances the Cf-9–mediated HR in tobacco, consistent with the idea that CMPG1 positively contributes to the HR (Figure 5). Because a loss of HR is not always correlated with a loss of resistance (Bendahmane et al., 1999; Sharma et al., 2003), we also analyzed the response of Sl Cmpg1-silenced Cf-9–containing tomato plants to C. fulvum. Using two different silencing methods, we showed that Cf9 tomato plants silenced for Cmpg1 allowed C. fulvum growth (Figures 7B and 7E). C. fulvum infection symptoms were weaker in tomato Cmpg1-silenced plants using hairpin-induced silencing in comparison with VIGS. This difference might be due to the degree of silencing obtained with each method or because VIGS was performed on plants into which Cf-9 was introduced by transformation, and the hairpin silencing was performed in Cf9 plants that carried other Cf homologs, including Cf-9B (Panter et al., 2002).
Although with both silencing techniques the fungal colonization was weaker in Cf9 plants silenced for Cmpg1 than in Cf0 control plants, in our VIGS experiments the results from silencing tomato Cmpg1 and Cf-9 were comparable. Similar experiments showed that silencing of R genes does not lead to complete elimination of mRNA or resistance (Liu et al., 2002b; Brigneti et al., 2004). This suggests that tomato Cmpg1 has a rate-limiting role in Cf-9–mediated resistance to C. fulvum. Finally, we showed that tomato Cmpg1 regulates the expression of two different PR genes, GlucB and PR5 (Figure 7F), identifying one of the defense pathways through which Cmpg1 activates Cf-9–mediated resistance.
VIGS and hairpin-induced silencing of tomato Cmpg1 were performed using a 200- and 400-bp fragment, respectively. Searches in the tomato TIGR database reveal no sequences with matches of 25 nucleotides or more with the fragments of Cmpg1 used for silencing, indicating that the phenotype we observe is likely due to the specific silencing of Cmpg1 and not to cross-silencing of other genes. Sequence analysis of the closest Arabidopsis homologs (PUB20 and PUB21) showed only one stretch with 25 identical nucleotides, which is not enough to permit PUB20 to silence PUB21 or vice versa. However, since the tomato and tobacco EST databases are not complete, we cannot exclude the possibility that other closely related genes might be targeted in our silencing experiments.
While rapid induction after elicitation has been previously shown for tobacco CMPG1 homologs in parsley and Arabidopsis (Kirsch et al., 2001; Heise et al., 2002; Navarro et al., 2004), our data provide a functional assessment of this class of protein for defense, both for Cf-9– and Pto-mediated resistance and Inf1-triggered elicitation. The importance of the ubiquitylation pathway in plant defense mechanisms has been previously indicated (Devoto et al., 2003). The first evidence was obtained for SGT1 (Azevedo et al., 2002; Peart et al., 2002). The exact role of this protein is not clear yet, but yeast SGT1 interacts with the SCF complex, a ubiquitin E3 ligase (Kitagawa et al., 1999). Silencing of SGT1 compromises gene-for-gene and general defense mechanisms (Peart et al., 2002). Other reports suggest that the SCF complex is involved in regulation of plant defense mechanisms (Liu et al., 2002b; Xu et al., 2002). SPL11 is a U-box and ARM repeat domain–containing protein. Rice spl11 mutants display spontaneous necrosis and enhanced resistance to fungal and bacterial pathogens, which surprisingly and paradoxically, is the opposite effect to the one observed for loss of CMPG1 function (Zeng et al., 2004). SPL11 could be involved in the activation of a negative regulator of defense, and its disruption would induce defense activation. This would reflect how the multiple regulators of plant defense need to be tightly coordinated in time and space and how this coordination has to be performed before (like SPL11, activating negative regulators) and after (like CMPG1, activating positive regulators; see below) pathogen elicitation.
Finally, a role in HR production has been shown for the two RING finger E3 ligases RIN2 and RIN3. The double mutant rin2 rin3 develops a weaker RPM1- and RPS2-dependent HR, but it does not affect the growth of the pathogen in the plant (Kawasaki et al., 2005). As with CMPG1, these two proteins are positive regulators of HR, but distinct from CMPG1, they are not involved in restricting pathogen proliferation. This indicates the presence of common and independent signaling pathways between HR production and resistance, both controlled by E3 ligases.
It is interesting to note that all the E3 ligases identified in defense seem to play different roles in the signaling pathway. This variety of function is consistent with the presence of a high number of E3 ligases in plants (Downes and Vierstra, 2005), since the E3 ligase confers the specificity of the ubiquitination reaction (Hershko et al., 1983; Finley et al., 2004). The mode of action of the previously described E3 ligases is not known in any case. Only the targets of RIN2 and RIN3 have been identified, but the mechanism by which they positively regulate HR production is not understood (Kawasaki et al., 2005).
Mutations in the Tobacco CMPG1 U-Box Domain Confer a Dominant-Negative Phenotype
Tobacco CMPG1 displayed E3 in vitro ligase activity (Figure 3). To investigate if this activity is essential in the role of CMPG1 in plant defense mechanisms, we analyzed the defense response in Cf9 tobacco plants expressing the Nt CMPG1:TAP protein mutated in the U-box domain. We predicted that if CMPG1 with a mutated U-box binds to its target, it might be unable to complete the next step, resulting in a dominant-negative allele. We mutated the Trp in position 64 and the Cys in position 37 (of tobacco CMPG1). Several reports suggest that Trp is involved in the interaction with the E2 ubiquitin-conjugating enzyme in RING finger and U-box proteins (Joazeiro et al., 1999; Zheng et al., 2000; Andersen et al., 2004), and mutations in the Cys confer a dominant-negative phenotype to the RING finger E3 ligase c-Cbl (Waterman et al., 1999). Mutation in either of these amino acids abolished the E3 ligase activity of tobacco CMPG1 (Figure 3). We also observed less Cf-9/Avr9-dependent HR when we overexpressed Nt CMPG1W64A and Nt CMPG1C37A (Figure 6; see Supplemental Table 1 online). In U-box domains, the Cys-37 residue is highly conserved and is adjacent to the E2-interacting amino acid Ile. In Arabidopsis PUB14, the Cys-37 corresponding residue is involved in the first hydrogen bond network of the U-box domain, and this network participates in the correct orientation of E2 binding residues (Andersen et al., 2004). Mutation of this Cys will prevent E2 interaction, which could explain the dominant-negative phenotype. The contrasting effect of overexpression of wild-type and mutant CMPG1 in tobacco suggests that the U-box domain is essential for CMPG1 function.
Model of the Role of CMPG1 in Plant Resistance
How might CMPG1 contribute to Cf-9–triggered defense mechanisms? It could activate a positive regulator. There are several examples in the literature of activation of transcription factors through ubiquitination (Muratani and Tansey, 2003). Therefore, the positive regulator could be a transcription factor that activates different defense responses. Ubiquitination also regulates transcription by modifying histones and, therefore, chromatin (Bray et al., 2005). In animals, transcriptional activation of developmentally controlled genes is associated with the ubiquitination of histones (Bray et al., 2005). Protein kinases can also be activated through ubiquitination. The regulation of kinases by monoubiquitination has been recently shown in mammals (Aebersold et al., 2004), and it opens new possibilities of signaling control through the ubiquitination pathway. Another possible scenario would be that CMPG1 activates defense by directing the degradation of negative regulators.
The fact that three E3 ligases (and two of them U-boxes) have been identified as positive regulators of Cf-9–dependent plant defense (Rowland et al., 2005) suggests that ubiquitination is a major regulator of this mechanism. This is consistent with the existence of a high number of U-box proteins in plants in comparison with other organisms (Azevedo et al., 2001), indicating that these proteins play multiple roles in plants. The U-box ACRE276 (Yang et al., 2006) regulates both HR production and pathogen arrest, while in the case of the F-box protein ACRE189, only a role in HR production has been found, and its involvement in inhibiting the pathogen growth has still to be shown (Rowland et al., 2005). How can three different ligases regulate the same defense pathway? The potential targets of ubiquitination in plant defense mechanisms are multiple; therefore, each E3 ligase could specifically modify one or several signaling components after Cf-9 response activation (either activation of positive regulators or degradation of negative regulators). Also, any of the ligases could modulate activity of the receptor after the response has been elicited.
The data presented in this article do not discriminate between these possibilities. The identification of the target protein(s) of CMPG1 and the two other E3 ligases is essential to understand the specific role of these proteins in defense mechanisms. The use of tobacco overexpressing a dominant-negative form of Nt CMPG1 could be useful for this purpose. If the target protein is ubiquitinated for degradation, the overexpression of the dominant-negative CMPG1 could result in its stabilization, facilitating its purification and identification. The dominant-negative form could facilitate the isolation of tobacco CMPG1 protein targets, which will help us to understand how CMPG1 controls Cf-9–mediated resistance in tomato.
METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana, Solanum lycopersicum, and Nicotiana tabacum plants were grown as previously described (Rowland et al., 2005). For tomato VIGS experiments, S. lycopersicum cv Moneymaker, carrying no known genes for resistance against C. fulvum (Cf-0), and the Cf-9 transgenic tomato cv Moneymaker (Hammond-Kosack et al., 1998) were used. For pHellsgate transformation, S. lycopersicum cv Moneymaker containing an introgression segment that carries the Cf-9 locus was used. For tobacco CMPG1 TAP-tagged overexpression studies, the Cf-9 transgenic N. tabacum cv Petite Havana line 8808J was used (Hammond-Kosack et al., 1998). Suspension cultures of Cf9 tobacco cells derived from line 34.1B were subcultured as previously described (Piedras et al., 1998).
Isolation of Full-Length Tobacco CMPG1 cDNA by Library Screening
The cDNA-AFLP fragment for ACRE74 was used as a probe to screen a cDNA library established from elicited tobacco cells (Durrant et al., 2000). Several clones were sequenced, and the clone containing the exact sequence of the ACRE74 cDNA-AFLP fragment was selected for further study.
Tomato Cmpg1 3′-RACE and Full-Length cDNA Cloning
The EST sequence TC159549 from the tomato TIGR gene index shared the highest identity with tobacco CMPG1 along the 950 bp of the 5′ end of Nt CMPG1 cDNA and was named Sl Cmpg1. For tomato Cmpg1 3′ cDNA cloning, total RNA was extracted from Cf9 tomato plants after 30 min of infiltration with IF(+Avr9). cDNA was synthesized from 2 μg of total RNA using Superscript II reverse transcriptase (Invitrogen). Tomato Cmpg1 3′ sequence was amplified with the gene-specific primer LeCMPG1F1 (5′-GAGGAAGTGATCTTGTCAACTCT-3′) and oligo(dT) primer (5′-TTTTTTTTTTTTTTTTTTTA/C/G-3′). Nested PCR was performed with the specific primer LeCMPG1F2 (5′-AAGTAGGAGGAATGCAGTTGTTGTC-3′) and the oligo(dT) primer. PCR products were cloned into the pGem-T Easy plasmid (Promega), and independent clones were sequenced. A cDNA fragment that matched the sequence of the 5′ end of the tomato EST TC159549 was selected. Primers LeCMPGF4 (5′-CTTCGTTAGCTGATATTTGTTTAG-3′) and LeCMPG1R1 (5′-AATTATACTAGATACTAGATACTAAACTC-3′) were used for PCR amplification of tomato Cmpg1 full-length cDNA. The PCR product was cloned into the pGem-T Easy plasmid, and several independent clones were sequenced. A clone that matched the sequence of the original 3′ and 5′ Sl Cmpg1 fragments was selected.
Sequence Analysis
Protein alignment was done using ClustalW (Thompson et al., 1994) and edited using the GeneDoc program (K.B. Nicolas and H.B. Nicolas Jr., GeneDoc: a tool for editing and annotating multiple sequence alignment; http://www.psc.edu/biomed/genedoc). The protein domains were identified using the PRODOM (Servant et al., 2002) and SMART (Schultz et al., 1998; Letunic et al., 2004) programs. Protein secondary structure predictions were done using the programs JPRED (Cuff et al., 1998) and PSIPRED (Jones, 1999; McGuffin et al., 2000).
RT-PCR
Total RNA from leaves or from cell cultures was extracted using the Tri Reagent method according to the manufacturer's recommendations (Sigma-Aldrich). First-strand cDNA was synthesized from 2 μg of total RNA using Superscript II reverse transcriptase (Invitrogen). RT-PCR was performed as described before (Rowland et al., 2005) using the primers 74-1 (5′-GAGAAATGCTTCAACAGGGGGAAG-3′) and 74-2 (5′-CACCTAATGCCTTTTCACATATGC-3′) for amplification from tobacco; Le74F (5′-ATGATTGCAACATGGAGAAAAAAGAG-3′) and Le74R2 (5′-ACAAGCCCTTTTCACCCC-3′) primers were used for amplification of the tomato Cmpg1. For Actin control amplification, primers NtActin1 (5′-ATGGCAGACGGTGAGGATATTCA-3′) and NtActin2 (5′-GCCTTTGCAATCCACATCTGTTG-3′) were used. LeACIK-flfwd (5′-AGTATAACAGCTATGGCTACTGCGGA-3′) and LeACIK-RTPCRrev (5′-CCAAATCAAGCAATTTAACAG-3′) primers were used for LeACIK1 amplification; finally, RT-LePR5F (5′-ATGGGGTAAACCACCAAACA-3′) and RT-LePR5R (5′-AAGTGAACCAGGGCATTCAC-3′) primers were used for amplification of PR5. For amplification of the basic glucanase gene, primers were used as described before (Rivas et al., 2004).
Recombinant Protein Purification and E3 Ubiquitin Ligase Activity Assay
The creation of the GST-Nt CMPG1 fusion constructs (wild type, Nt CMPG1C37A, and Nt CMPG1W64A) was conducted as follows. The modified primers CMPG-GST-BamHI-F (5′-ATGCTGGATCCATGATGATTTCAACATGGAGGAAAAG-3′) and CMPG-GST-XhoI-R (5′-GTCAGACTCGAGTCAGAATGGCCTTTTGAGACTCTT-3′), constructed with BamHI and XhoI sites, respectively, were used to amplify the tobacco CMPG1 cDNA fragments (wild-type and dominant-negative mutants) using the proofreading DNA polymerase Pfu (Stratagene). PCR products were gel purified, digested with BamHI and XhoI, and ligated into the BamHI-XhoI–digested pGEX 4T1 (Pharmacia Biotech). The generated GST:Nt CMPG1 plasmids were transformed into Escherichia coli BL21(DE3) (Invitrogen) bacteria cells for protein expression. Overnight cultures of the bacteria were diluted 100 times in a total volume of 50 mL Luria-Bertani medium and further incubated at 28°C to reach an OD of 0.8. Induction of the GST:Nt CMPG1 proteins was performed with 0.1 mM isopropylthio-β-galactoside for 3 h at 28°C. After the induction, cells were harvested by centrifugation at 13,000 rpm for 15 min, washed with 1× PBS (140 mM NaCl, 2.7 mM KCl, 10 mM NaH2PO4·7H2O, and 1.8 mM KH2PO4), and further resuspended in 2.5 mL of 1× PBS amended with protease inhibitors (Roche). Cells were sonicated, and the supernatants were incubated for an additional 30 min at 4°C with 1% Triton X-100 to aid in solubilization of the proteins. The protein extracts were centrifuged, the supernatants were incubated with Glutathione Sepharose beads (Amersham Pharmacia), and the proteins were purified according to the manufacturer's recommendations. The fused GST:Nt CMPG1 wild-type and mutated proteins were eluted with 0.5 mM glutathione elution buffer and further used for the ubiquitination assays.
For the in vitro ubiquitination assays, each reaction (30 μL final volume) contained 40 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 50 mM KCl, 2 mM ATP, 1 mM DTT, 10% glycerol, 10 mM phosphocreatine, 3.5 units of phosphocreatine kinase (Sigma-Aldrich), 0.6 units of inorganic pyrophosphatase (Sigma-Aldrich), 0.1 μg of yeast E1 (Affiniti Research Products), 0.5 μg of E2 UbcH5a, UbcH5b, or UbcH5c (Affiniti Research Products), and ∼0.5 μg of E3s (eluted GST-Nt CMPG1 wild type or mutated proteins or GST-PUB26). The reactions were incubated at 30°C for 3 h and stopped by adding 2× SDS-PAGE sample buffer (0.5 M Tris-HCl, pH 6.8, 20% [v/v] glycerol, 10% [w/v] SDS, 100 mM DTT, and 1% bromophenol blue) at 65°C for 5 min and analyzed by SDS-PAGE electrophoresis followed by protein gel blotting using anti-ubiquitin antibody raised in mouse (Santa Cruz Biotechnology) and anti-GST antibody raised in goat (Amersham Pharmacia).
TRV-Based VIGS in N. benthamiana
A 380-bp fragment of tobacco CMPG1 was obtained by HindIII digestion and cloned into the HindIII site of pTV00 (Ratcliff et al., 2001) to create TRV:Nt CMPG1. The empty pTV00 vector (TRV:00) and the pTV00 vector containing the Nbsgt1.2 fragment (TRV:sgt1; Peart et al., 2002), were used as controls. All the plasmids were transformed in Agrobacterium tumefaciens strain GV3101.
Infection of plants by agroinfiltration was performed as previously described (Rowland et al., 2005). Three weeks after infiltration, plants were analyzed for RNA accumulation and HR production. Fourth and fifth leaves were infiltrated with agrobacteria carrying Cf-9/Avr9 (Thomas et al., 2000) for HR production.
Stable Transformation of Tobacco with Nt CMPG1:TAP Wild-Type and Mutant Overexpression Constructs
Mutations in tobacco CMPG1 were introduced by overlapping PCR. Two independent fragments were generated for each mutation. The 5′ cDNA fragments were amplified using the primers NtCMPG1F1 (5′-GTAATCGATATGATGATTTCAACATGGAGG-3′) and NtCMPG1R1 (5′-ATTCCCAGCCTCAATCGCTTTCTCAATATTCTC-3′) for the Trp-64 mutation and NtCMPG1F1 and NtCMPG1R4 (5′-GTCTAAGGAAATTGGAGCTGTGAAATGTCTAGG-3′) for the Cys-37 mutation. For the 3′ fragment amplification, the primers NtCMPG1F2 (5′-GAGAATATTGAGAAAGCGATTGAGGCTGGGAAT-3′) and NtCMPG1R2 (5′-GTAGGATCCGAATGGCCTTTTGAGACTCTT-3′) were used for the Trp-64 mutation and NtCMPG1F4 (5′-CCTAGACATTTCACAGCTTGTGAAATGTCTAGG-3′) and NtCMPG1R2 for the Cys-37 mutation. The 5′ and 3′ PCR fragments were gel purified and mixed in a 1:1 ratio for overlapping PCR with the primers NtCMPG1F1 and NtCMPG1R2 for each construct. For the NtCMPG1 wild type, cloning PCR was performed from the original cDNA clone using the primers NtCMPG1F1 and NtCMPG1R2. The restriction sites ClaI and BamHI were introduced with the primers NtCMPG1F1 and NtCMPG1R2, respectively. The amplification fragments were cloned into pGem-T Easy plasmid and digested with SalI and ClaI. The digestion products were cloned into the epiGreenB4 binary vector containing a 35S:GUS:TAP cassette at the same sites to generate C-terminal TAP tag–fused tobacco CMPG1 driven by the 35S promoter. To generate epiGreenB4 plasmid, pBIN19 h was digested with EcoRI and HindIII to obtain the expression cassette 35S:GUS:TAP:NOS, which was further cloned into a pGreen-derived vector (Hellens et al., 2000) at the same restriction sites. Each construct was cotransformed with the pSoup plasmid in A. tumefaciens strain Agl1, and tobacco stable transformants were generated as described (Hammond-Kosack et al., 1998).
Preparation of Protein Extract
Leaves from transgenic tobacco overexpressing Nt CMPG1:TAP and controls were homogenized in liquid nitrogen, thawed in 2 volumes of extraction buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, 2 μg/mL antipain, 2 μg/mL leupeptin, and 2 μg/mL aprotinin], filtered through two layers of Miracloth, and centrifuged at 1000g for 10 min at 4°C. The supernatant was collected for SDS-PAGE.
SDS-PAGE and Immunoblotting
SDS-PAGE and immunoblotting experiments were performed as previously described (Rivas et al., 2002), except that 8% SDS gels were used for separation.
Growth Inhibition Assay
Homozygous N. tabacum seeds were germinated on plates without any selection. At day 10, tobacco seedlings were transferred to MS liquid medium containing IF(−Avr9) or IF(+Avr9) previously filter-sterilized. Ten days later, the fresh weight was determined and pictures were taken.
TRV-Based VIGS in S. lycopersicum
A 150-bp fragment from tomato Cmpg1 cDNA was PCR amplified using the primers SulphurCMPGF (5′-AACATTGAGAAACTCGGTGGGAATCAAACA-3′) and Le74seqR1 (5′-TGAAATCGGGATTCTTGGAG-3′). The fragment was cloned in the SmaI site of pTRV-RNA2 vector (Liu et al., 2002a). As a control, the pTRV-RNA2 empty vector and the pTRV-RNA2 vector containing a 150-bp Cf-9 fragment (Rowland et al., 2005) were used. Silencing of tomato plants and C. fulvum race 4 GUS infections were performed as before (Rowland et al., 2005). Fungal growth was scored 3 weeks later by GUS staining and light microscopy (Axiophot; Zeiss).
Stable Transformation of Tobacco and Tomato with a Hairpin-Driven RNAi Construct
For hairpin-induced RNAi in tomato and tobacco, the pHellsgate8 vector was used (Helliwell et al., 2002). A fragment of 400 bp was amplified using the primers Le74RNAi5 (5′-ACCGTCGACAGTGACTTTATCAACAGGGATT-3′) and Le74RNAi3 (5′-ACCCTCGAGGCATTTTTCGCGTTTTTCCC-3′) for tomato and NtCMPGF8 (5′-ACCGTCGACAGTGACCTTGTCAACAGGGATC-3′) and NtCMPGR8 (5′-ACCCTCGAGCGATTTCTCATATTATTTCC-3′) for tobacco CMPG1. The amplification products were digested with SalI and cloned into the gateway-compatible vector pENTR4 (Invitrogen) previously digested with SalI and EcoRV to give pEntry4LeCMPG1 and pEntry4NtCMPG1, respectively. An in vitro BP clonase recombination reaction was performed with pEntry4LeCMPG1, pEntry4NtCMPG1, and pHellsgate8 according to the manufacturer's instructions (Invitrogen). The recombination reaction product was transformed into DH5α E. coli. The positive clones were digested to check the intron orientation and sequenced. The selected clones were transformed into agrobacteria for transformation of tomato plants as described (Hammond-Kosack et al., 1998).
Agrobacteria-Mediated Transient Expression in Tobacco
For analysis of Pto- and Inf1-dependent HR production, agrobacteria carrying the binary construct containing Pto/AvrPto and Inf1 (Peart et al., 2002) were infiltrated at different optical densities in 7-week-old tobacco leaves. HR development was observed 3 and 4 d after infiltration.
Accession Numbers
The accession numbers of the sequences included in the alignment are as follows: parsley CMPG1 (AAK69402), tobacco CMPG1 (AAP03884), Arabidopsis PUB20 (AAG51307), and Arabidopsis PUB21 (NP_198565). Sequence data for tomato Cmpg1 have been deposited in the GenBank data library under accession number DQ118759.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Protein levels and Phenotypes of Tobacco Lines Carrying 35S:Nt CMPG1 Construct.
Supplemental Figure 1. The Presence of High Levels of Tobacco CMPG1W64A Correlates with Reduced HR Production.
Supplemental Figure 2. Tobacco CMPG1 Is Involved in Inf1-Mediated HR Production.
Supplemental Figure 3. Correlation between Tomato Cmpg1 mRNA Levels and Defense Responses in Tomato Hairpin Lines.
Supplementary Material
Acknowledgments
We thank Mathew Smoker for tobacco and tomato transformation. We also thank David Baulcombe's laboratory for the TRV vectors used for VIGS in N. benthamiana, Dinesh-Kumar for the TRV vector used for VIGS in tomato, and David Studholme for help with the bioinformatics analysis. We thank Normand Brisson and Kamal Bouarab for useful comments on the manuscript. This research was supported by the Cross-Talk in Signaling in Plants project (EC Grant HRPN/CT-2000-00093) and by the Gatsby Charitable Foundation. R.G.-L. was supported by a European Community Young Researcher Training fellowship. D.I.T. was supported by EU FP6 STREP Grant 511983-TransDeath.
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.040998.
References
- Aebersold, D.M., Shaul, Y.D., Yung, Y., Yarom, N., Yao, Z., Hanoch, T., and Seger, R. (2004). Extracellular signal-related kinase 1c (ERK1c), a novel 42-kilodalton ERK, demonstrates unique modes of regulation, localization, and function. Mol. Cell. Biol. 24 10000–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira, S., and Takeda, K. (2004). Toll-like receptor signaling. Nat. Rev. Immunol. 4 499–511. [DOI] [PubMed] [Google Scholar]
- Amador, V., Monte, E., García-Martínez, J.L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 343–354. [DOI] [PubMed] [Google Scholar]
- Andersen, P., Kragelund, B.B., Olsen, A.N., Larsen, F.H., Chua, N., Poulsen, F.M., and Skriver, K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279 40053–40061. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (2000). The U box is a modified RING finger - A common domain in ubiquitination. Curr. Biol. 10 R132–R134. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefer, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6 354–358. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R., Grabov, A., Brearley, J., Hammond-Kosack, K.E., and Jones, J.D.G. (1999). K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. Plant J. 19 453–462. [DOI] [PubMed] [Google Scholar]
- Bray, S., Musisi, H., and Bienz, M. (2005). Bre1 is required for Notch signaling and histone modification. Dev. Cell 8 279–286. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Martin-Hernandez, A.M., Jin, H., Chen, J., Baulcombe, D.C., Baker, B., and Jones, J.D. (2004). Virus-induced gene silencing in Solanum species. Plant J. 39 264–272. [DOI] [PubMed] [Google Scholar]
- Chuang, T.H., and Ulevitch, R. (2004). Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat. Immunol. 5 495–502. [DOI] [PubMed] [Google Scholar]
- Cornelissen, B.J.C., Horowitz, J., Van Kan, J.A.L., Goldberg, R.B., and Bol, J.F. (1986). A tobacco mosaic virus-induced tobacco protein is homologous to the sweet-tasting protein thaumatin. Nature 321 531–532. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. (1998). Jpred: A consensus secondary structure prediction server. Bioinformatics 14 892–893. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15 435–467. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Musket, P.R., and Shirasu, K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6 307–311. [DOI] [PubMed] [Google Scholar]
- Downes, B., and Vierstra, R.D. (2005). Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem. Soc. Trans. 33 393–399. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, D., Ciechanover, A., and Varshavsky, A. (2004). Ubiquitin as a central cellular regulator. Cell S116 S29–S32. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin, L.K., Krishnamurthy, N., Tor, M., Sjolander, K.V., and Jones, J.D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., and Ciechanover, A. (2001). The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82 372–428. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 575–607. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., Tang, S., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product Avr9. Plant Cell 10 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P.D., Seo, H.S., Yang, J.Y., and Chua, N.H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6 453–462. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K.I. (2001). U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276 33111–33120. [DOI] [PubMed] [Google Scholar]
- Heise, A., Lippok, B., Kirsch, C., and Hahlbrock, K. (2002). Two immediate-early pathogen-responsive members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proc. Natl. Acad. Sci. USA 13 9049–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejgaard, J., Jacobsen, S., and Svendsen, I. (1991). Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 29 127–131. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Helliwell, C., and Waterhouse, P. (2003). Construct and methods for high-throughput gene silencing in plants. Methods 30 289–295. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Wesley, S.V., Wielopolska, A.J., and Waterhouse, P.M. (2002). High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29 1217–1225. [DOI] [PubMed] [Google Scholar]
- Hershko, A., Heller, H., Elias, S., and Ciechanover, A. (1983). Components of ubiquitin protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258 8206–8214. [PubMed] [Google Scholar]
- Joazeiro, C.A.P., Wing, S.S., Huang, H.K., Leverson, J.D., Hunter, T., and Liu, Y.C. (1999). The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286 309–311. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kamoun, S., van West, P., Vlesshouwers, V., de Groot, K.E., and Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10 1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Nam, J., Boyes, D.C., Holt, B.H., Hubert, D.A., Wiig, A., and Dangl, J.L. (2005). A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J. 44 258–270. [DOI] [PubMed] [Google Scholar]
- Kim, D.W., Lenzen, G., Page, A.L., Legrain, P., Sansonetti, P.J., and Parsot, C. (2005). The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 102 14046–14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Cho, H.S., Kim, D.M., Lee, J.L., and Pai, H.S. (2003). CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim. Biophys. Acta 1651 50–59. [DOI] [PubMed] [Google Scholar]
- Kirsch, C., Logemann, E., Lippok, B., Schmelzer, E., and Hahlbrock, K. (2001). A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J. 26 217–227. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4 21–33. [DOI] [PubMed] [Google Scholar]
- Kraft, E., Stone, S.L., Ma, L., Su, N., Gao, Y., Lau, O., Deng, X., and Callis, J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Peer, B. (2004). SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32 D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002. b). Role of the SCF ubiquitin ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.L., Schiff, M., and Dines-Kumar, S.P. (2002. a). Virus-induced gene silencing in tomato. Plant J. 31 777–786. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J., Bryson, K., and Jones, D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16 404–405. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kaushik, S., and Nandety, R.S. (2005). Evolving disease resistance genes. Curr. Opin. Plant Biol. 8 129–134. [DOI] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil, Y., Shiu, S., Stone, S.L., Salt, J.N., and Goring, D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-Box E3 ubiquitin ligase family. Plant Physiol. 134 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani, M., and Tansey, W.P. (2003). How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4 192–201. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, Z., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki, T., Mitsuhara, I., Seo, S., Ohtsubo, N., and Ohashi, T. (1998). Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39 500–507. [Google Scholar]
- Ohi, M.D., Vander Kooi, C.W., Rosenberg, J.A., Chazin, W.J., and Gould, K.L. (2003). Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panter, S.N., Hammond-Kosack, K.E., Harrison, K., Jones, J.D., and Jones, D.A. (2002). Developmental control of promoter activity is not responsible for mature onset of Cf-9B-mediated resistance to leaf mold in tomato. Mol. Plant Microbe Interact. 15 1099–1107. [DOI] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B., and Jones, J.D. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 12 821–832. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.F., and Martin, G.M. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41 215–243. [DOI] [PubMed] [Google Scholar]
- Piedras, P., Hammond-Kosack, K.E., Harrison, K., and Jones, J.D.G. (1998). Rapid Cf-9- and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol. Plant Microbe Interact. 11 1155–1166. [Google Scholar]
- Pringa, E., Martinez-Noel, G., Müller, U., and Harbers, K. (2001). Interaction of the RING finger-related U-box motif of a nuclear dot protein with ubiquitin-conjugating enzymes. J. Biol. Chem. 276 19617–19623. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus is a vector for analysis of gene function by silencing. Plant J. 25 237–245. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. (1999). A generic purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rivas, S., Romeis, T., and Jones, J.D.G. (2002). The Cf-9 disease resistance protein is present in an ∼420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas, S., Rougon-Cardoso, A., Smoker, M., Schauser, L., Yoshioka, H., and Jones, J.D.G. (2004). CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 23 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas, S., and Thomas, C.M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43 395–436. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Ludwig, A.A., Martin, R., and Jones, J.D.G. (2001). Calcium-dependent protein kinases play an essential role in a plant defense response. EMBO J. 20 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., and Jones, J.D.G. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S., Klessig, D.F., Hirt, H., and Jones, J.D.G. (1999). Rapid Avr9- and Cf9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, O., Ludwig, A.A., Merrick, C., Baillieul, F., Tracy, F., Durrant, W.E., Fitz-Laylin, L., Nekrasov, V., Yoshioka, H., and Jonathan, J.D.G. (2005). Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, J.D., and Hicke, L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278 35857–35860. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Poting, C.P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant, F., Bru, C., Carrere, S., Courcelle, E., Gouzy, J., Peyruc, D., and Kahn, D. (2002). ProDom: Automated clustering of homologous domains. Brief. Bioinform. 3 246–251. [DOI] [PubMed] [Google Scholar]
- Sharma, P.C., Ito, A., Shimizu, T., Terauchi, R., Kamoun, S., and Saitoh, H. (2003). Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol. Genet. Genomics 269 583–591. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Dixon, M.S., Parniske, M., Golstein, C., and Jones, J.D.G. (1998). Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Tang, S., Hammond-Kosack, K., and Jones, J.D.G. (2000). Comparison of the hypersensitive response induced by the tomato Cf-4 and Cf-9 genes in Nicotiana spp. Mol. Plant Microbe Interact. 13 465–469. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman, H., Levkowitz, G., Alroy, I., and Yarden, Y. (1999). The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 274 22151–22154. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J., Wang, J., Li, Q., Hwang, J.R., Patterson, C., and Zhang, H. (2003). AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 132 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.-W., González-Lamothe, R., Ewan, R.A., Rowland, O., Yoshioka, H., Shenton, M., Ye, H., O'Donnell, E., Jones, J.D.G., and Sadanandom, A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., Xie, Q., Nahm, B.H., Leung, H., and Wang, G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., Wang, P., Jeffrey, P.D., and Payletich, N.P. (2000). Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102 533–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.