Abstract
Δ22-Unsaturated sterols, containing a double bond at the C-22 position in the side chain, occur specifically in fungi and plants. Here, we describe the identification and characterization of cytochrome P450s belonging to the CYP710A family as the plant C-22 desaturase. Recombinant proteins of CYP710A1 and CYP710A2 from Arabidopsis thaliana and CYP710A11 from tomato (Lycopersicon esculentum) were expressed using a baculovirus/insect system. The Arabidopsis CYP710A1 and tomato CYP710A11 proteins exhibited C-22 desaturase activity with β-sitosterol to produce stigmasterol (CYP710A1, Km = 1.0 μM and kinetic constant [kcat] = 0.53 min−1; CYP710A11, Km = 3.7 μM and kcat = 10 min−1). In Arabidopsis transgenic lines with CYP710A1 and CYP710A11 overexpression, stigmasterol levels increased by 6- to 32-fold. Arabidopsis CYP710A2 was able to produce brassicasterol and stigmasterol from 24-epi-campesterol and β-sitosterol, respectively. Sterol profiling analyses for CYP710A2 overexpression and a T-DNA insertion event into CYP710A2 clearly demonstrated in planta that CYP710A2 was responsible for both brassicasterol and stigmasterol production. Semiquantitative PCR analyses and promoter:β-glucuronidase transgenic approaches indicated strict tissue/organ-specific regulation for each CYP710A gene, implicating differential tissue distributions of the Δ22-unsaturated sterols in Arabidopsis. Our results support the possibility that the CYP710 family may encode P450s of sterol C-22 desaturases in different organisms.
INTRODUCTION
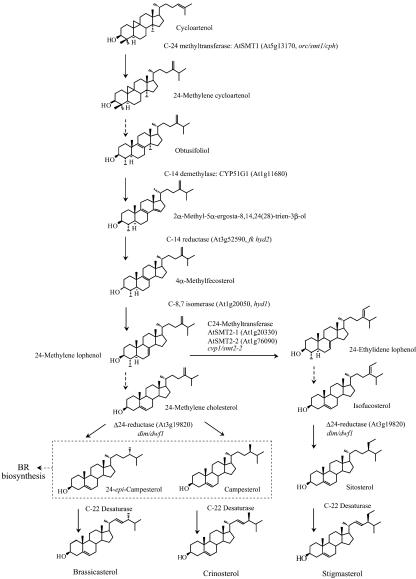
Sterols, isoprenoid-derived lipids produced via the mevalonate pathway, are involved in various cellular functions in eukaryotes, such as maintenance of membrane integrity and biosynthetic precursors of steroid hormones. One of the most remarkable differences in sterol composition among biological kingdoms is the specific occurrence of Δ22-sterols (Figure 1) containing a double bond at C-22 in the sterol side chain in fungi (ergosterol) and plants (stigmasterol). The plant C-22 desaturase has yet to be clarified.
Figure 1.
Sterol Biosynthesis in Arabidopsis.
The pathway from cycloartenol to the Δ22-sterols is shown. 24-Methyl sterols are a mixture of (24R)- and (24S)-epimers of campesterol (Schaller, 2003). The Δ22-desaturases catalyze the C22-desaturation reaction to yield stigmasterol, crinosterol, and brassicasterol from their immediate precursors. Dashed arrows indicate several enzymatic steps not shown here.
In plants, a vast majority of sterols are present in membranes as a mixture of several major molecular species, including β-sitosterol, stigmasterol, campesterol, and cholesterol (Schaller, 2003; Benveniste, 2004). These bulk sterols modulate membrane permeability and fluidity. Furthermore, recent studies (Mongrand et al., 2004; Borner et al., 2005) have suggested that phytosterols, as well as cholesterol in mammals and ergosterol in yeast, are involved in the formation of membrane microdomains that serve as a platform for crucial physiological processes, such as signal transduction for cellular proliferation and differentiation, vesicular trafficking, and cytoskeleton organization. Sterols also serve as precursors for the biosynthesis of steroid hormones involved in many different cellular processes (Schaller, 2003). In plants, brassinosteroids (BRs) are the only class of steroidal phytohormones, functioning in a variety of postembryonic events, including cell division and expansion, responses to light and dark, morphogenesis, apical dominance, and gene expression (Clouse, 2002; Nemhauser and Chory, 2004).
Recent findings with the aid of molecular genetics approaches have suggested possible involvement of phytosterols in BR-independent processes of embryonic and postembryonic development (Clouse, 2000; Lindsey et al., 2003). Thus, Arabidopsis thaliana mutants in the sterol biosynthetic pathway (Figure 1) display severe impairment in embryogenesis and development at specific stages, which cannot be rescued by the application of exogenous BRs. Arabidopsis mutants, sterolmethyltransferase1 (smt1/orc), are defective in cycloartenol C-24 methyltransferase (Diener et al., 2000; Willemsen et al., 2003), and fackel (fk) phenotype is ascribed to the mutation of the C-14 reductase (Jang et al., 2000; Schrick et al., 2000). The hydra1 (hyd1) mutants carry the mutation in the C8-C7 isomerase gene (Souter et al., 2002). Other embryonic mutants of cephalopod (cph) and hyd2 have been found to be allelic to smt1 and fk, respectively (Schrick et al., 2002; Souter et al., 2002). The smt1/orc/cph mutants exhibit developmental abnormalities with altered sterol compositions (Lindsey et al., 2003). It has also been reported that the smt1/orc/cph mutation influenced pattern formation mostly by interfering with the polar localization of PIN proteins (Willemsen et al., 2003), while PIN protein trafficking was not affected in either the hyd1 or hyd2/fk mutant (Souter et al., 2002), implicating differential developmental roles of membrane sterols. It has been reported that unusual sterols accumulated in fk mutant plants, while it is not clear whether or not such sterols have specific roles in BR-independent developmental processes (He et al., 2003; Schrick et al., 2004). On the other hand, smt2 and cotyledon vascular pattern1 (cvp1) are due to mutations in the methyltransferase gene SMT2 encoding the C-24 methytransferase. They produce 24-ethylidene lophenol from 24-methyl lophenol (Schaeffer et al., 2001; Carland et al., 2002). cvp1 plants displayed postembryonic vascular patterning defects (Carland et al., 2002), and SMT2 cosuppression lines contained higher levels of campesterol and lower levels of sitosterol, exhibiting developmental abnormalities such as reduced apical dominance and reduced fertility that cannot be restored by exogenous BRs (Schaeffer et al., 2001). Furthermore, it has recently been reported that a T-DNA insertion mutagenesis within the obtusifoliol 14α-demethylase gene CYP51G1 caused defects in membrane integrity and hypocotyl elongation, leading to postembryonic seedling lethality (Kim et al., 2005). These results indicate that altered sterol profiles due to the mutations both upstream and downstream of the SMT2 step affect membrane properties, thereby influencing signaling cascades involved in normal plant growth, such as cell polarity, auxin efflux, and ethylene signaling. However, no molecular basis for the phytosterol-dependent developmental processes has been established. Thus, complete elucidation of the sterol biosynthetic pathway is a prerequisite to understanding the mechanisms for maintaining correct membrane sterol compositions required for normal plant growth.
Specifically, biosynthesis and physiology of the Δ22-sterols are not clearly understood. In higher plants, membrane sterols are present as a complex mixture of 24-ethyl sterols and 24-methyl sterols with cholesterol as a minor component (Schaller, 2003; Benveniste, 2004). The ratio of 24-ethyl sterols (up to 60% of total sterol) and 24-methyl sterols (<40% of total sterol) is determined by the two sterol methyltransferase activities (Schaller, 2003; Benveniste, 2004). The configuration at C-24 is 100% R in β-sitosterol, while 24-methyl sterols are a mixture of campesterol (campest-5-en-3β-ol) and its C-24 methyl epimer, 24-epi-campesterol (ergost-5-en-3β-ol). Plants in the family Brassicaceae, including Arabidopsis, contain a 24-ethyl-Δ22-sterol, stigmasterol, and 24-methyl-Δ22-sterols, a mixture of brassicasterol [(22E)-ergost-5,22-dien-3β-ol] and crinosterol [(22E)-campest-5,22-dien-3β-ol] (Matsumoto et al., 1983; Benveniste, 2004). Brassicasterol (24R-epimer) and crinosterol (24S-epimer) are thought to be derived from 24-epi-campesterol and campesterol, respectively (Figure 1). However, no sequence has thus far been reported for any higher plant enzyme catalyzing the C-22 desaturase reaction (Benveniste, 2004).
A fungal cytochrome P450 monooxygenase, CYP61, is known as the sterol C-22 desaturase to produce ergosta-5,7,22,24(28)-tetraenol from the immediate precursor, ergosta-5,7,24(28)-trienol, at the penultimate step in the ergosterol biosynthetic pathway (Skaggs et al., 1996; Kelly et al., 1997). A sequence comparison (Figure 2) revealed striking sequence conservation as [FLFA(A/S)QDAS(T/S)S] between plant CYP710A proteins and fungal CYP61 proteins, while the overall sequence similarity is only ∼30%. The conserved sequence is located at one of the putative substrate recognition sites of P450s (Gotoh, 1992) on the I helix (SRS4) that is positioned on the distal side of the heme group. Within this I helix, two Ala residues, corresponding to Ala-295 and Ala-299 in Arabidopsis CYP710A1 (Figure 2, At 710A1), are absolutely conserved between the CYP710 and CYP61 family proteins. By contrast, most P450s have a conserved Thr residue at the second of these two positions that is known to participate in the oxygen activation and proton delivery for hydroxylation reactions catalyzed by P450s (Meunier et al., 2004). These observations suggested the possibility of CYP710A family proteins as the C-22 desaturases in plants (Benveniste, 2004). On the basis of this sequence information, we initiated an investigation to clarify whether plant CYP710A proteins were the functional homologs of fungal CYP61 proteins.
Figure 2.
Alignment of Amino Acid Sequences of CYP710 Proteins.
The deduced amino acid sequences of CYP710A1 (At 710A1) and CYP710A11 from tomato (Se 710A11) proteins were aligned with CYP710A5 (Os 710A5) from O. sativa, CYP710B (Cr 710B) from C. reinhardtii, CYP61 (Sc 61) from S. cerevisiae, and CYP524 (Dicty 524) from D. discoideum. The double-headed arrow above the sequences indicates the conserved amino acids in the putative substrate recognition site (SRS4) of P450s (Gotoh, 1992). The conserved Ala-299 residue (numbered as in CYP710A1) is boxed in green. The arrowhead points to the heme ligand Cys residues. The conserved residues are boxed in red.
RESULTS
CYP710A Sequences
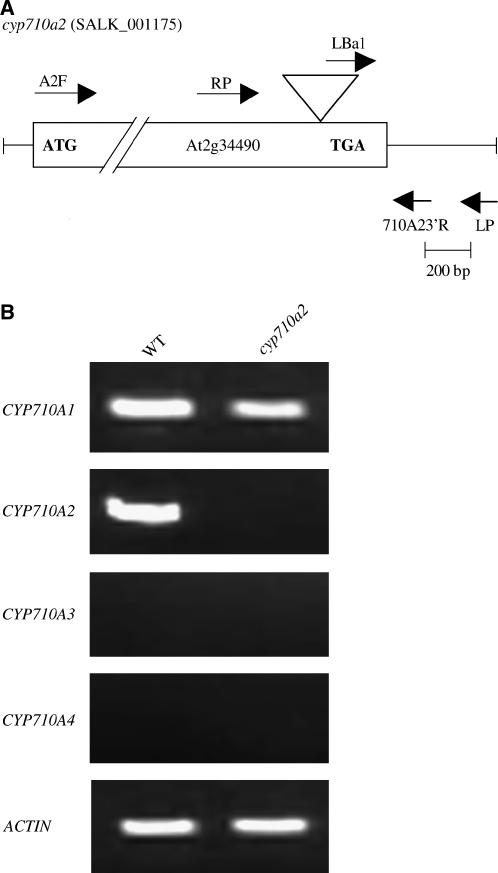
Arabidopsis contains four genes encoding putative P450 proteins belonging to the CYP710A subfamily (The Arabidopsis Information Resource, http://www.arabidopsis.org/): CYP710A1, CYP710A2, CYP710A3, and CYP710A4. No introns were found in these CYP710A genes in Arabidopsis, and we amplified by PCR the putative coding sequences of CYP710A1 and CYP710A2 genes encoding polypeptides of 495 and 499 amino acids, respectively, with the calculated molecular masses of 55,723 and 56,344 D, respectively. The predicted amino acid sequence of CYP710A1 is 81.9, 77.7, and 76.1% identical to those of CYP710A2, CYP710A3, and CYP710A4, respectively. The CYP710A3 protein sequence is 73.9 and 93.9% identical to those of CYP710A2 and CYP710A4 proteins, respectively. Using the amino acid sequence of CYP710A1 as the bait for a tBLASTn search at The Institute for Genomic Research (TIGR) Gene Indices (http://tigrblast.tigr.org/tgi/), we identified a tomato (Lycopersicon esculentum) EST clone containing a putative entire coding region for tomato CYP710A11. The deduced primary structure consists of 501 amino acids with a calculated molecular mass of 57,511 D, and the amino acid sequence is 59.0% identical to that of CYP710A1. Both the entire coding sequences of CYP710A1 and CYP710A2 and the full-length EST clone of CYP710A11 were analyzed for further experiments.
Insect Cell Expression of CYP710A1, CYP710A2, and CYP710A11
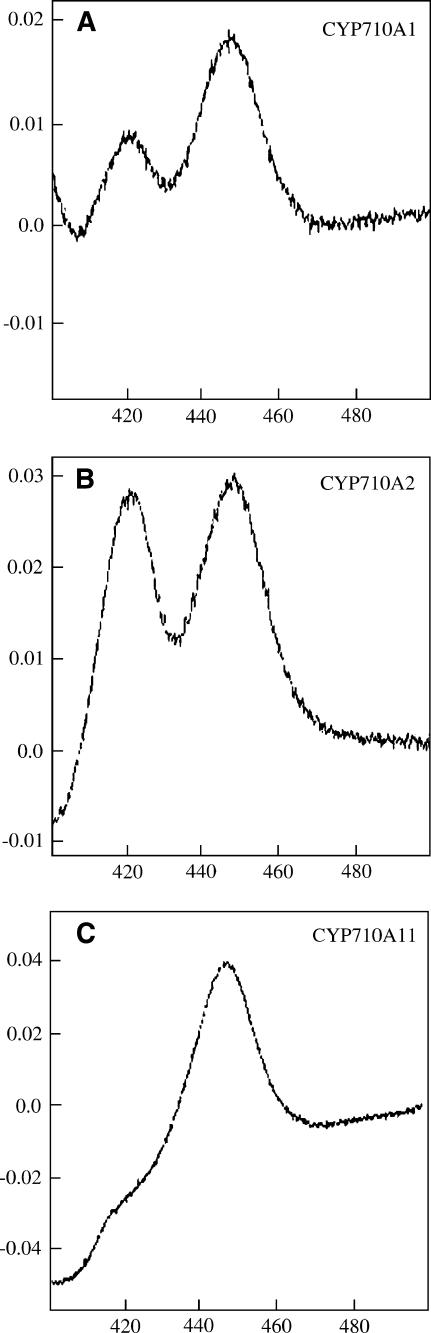
To clarify the C-22 desaturation activity, we characterized the enzymatic properties of CYP710A1, CYP710A2, and CYP710A11 proteins. The entire coding sequences of the Arabidopsis CYP710A genes (CYP710A1 and CYP710A2) and the tomato CYP710A11 cDNA were expressed in insect cells using a baculovirus expression vector system. SDS-PAGE analysis (see Supplemental Figure 1 online) demonstrated that protein bands of ∼55 kD appeared in the microsomal fractions in the insect cells infected with the recombinant viruses carrying the expression cassettes of CYP710A1, CYP710A2, and CYP710A11, respectively. The apparent molecular masses of these expressed proteins were in good agreement with those calculated from the deduced primary structures of CYP710A1, CYP710A2, and CYP710A11 proteins. These recombinant proteins were recovered in the microsomal membrane fractions (100,000g pellet) and used for spectrophotometric determination of P450 (Figure 3). The microsomal fractions from insect cells expressing the recombinant CYP710A proteins showed the expected reduced CO difference spectrum with the absorption maximum at 449 nm. The specific contents of the recombinant CYP710A1, CYP710A2, and CYP710A11 P450 proteins were 100, 71, and 230 pmoles P450/mg microsomal protein, respectively. For the expression of CYP710A1 and CYP710A2 proteins in insect cells, we used the PCR-amplified genomic DNA fragments containing the putative coding sequences of CYP710A1 and CYP710A2 genes. The successful accumulation of P450 proteins with the CO difference spectra (Figure 3) indicated that these Arabidopsis CYP710A genes contain no intron as predicted. No significant P450 accumulation could be detected in the mock-infected insect cells under the same experimental conditions (data not shown).
Figure 3.
Heterologous Expression of Recombinant CYP710A Proteins in Insect Cells.
Reduced CO difference spectra of recombinant CYP710A1, CYP710A2, and CYP710A11. The recombinant P450 samples of 2 mg microsomal protein/mL of CYP710A1 (A) and CYP710A11 (C) and 4.6 mg microsomal protein/mL of CYP710A2 (B) were used for the spectrophotometric analyses.
Enzyme Assay
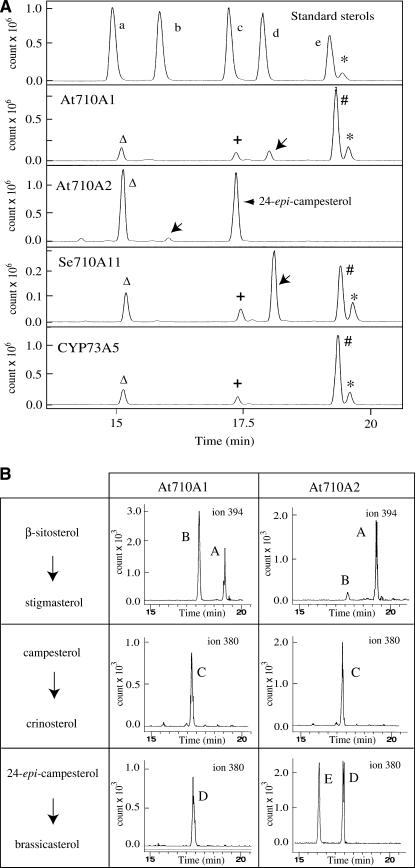
The microsomal fractions from the insect cells expressing the recombinant CYP710A proteins were used for P450 functional characterization using the potential substrates (β-sitosterol, campesterol, and 24-epi-campesterol) for the C-22 desaturation reactions (Figure 1). The gas chromatography analyses of the reaction products from the Arabidopsis CYP710A1 and tomato CYP710A11 microsomes with β-sitosterol as the substrate showed specific peaks at the same retention time (18.5 min, indicated by the arrows in Figure 4A) as that of stigmasterol. A very weak level of stigmasterol production was also detected from the Arabidopsis CYP710A2 assay (Figure 4B). The gas chromatography–mass spectrometry (GC-MS) fragmentation patterns obtained from the peaks in the selective ion mode (SIM) (Figure 4B) demonstrated that the putative reaction products were actually stigmasterol, indicating that the CYP710A1, CYP710A2, and CYP710A11 proteins catalyzed the C-22 desaturase reaction to produce stigmasterol from β-sitosterol in vitro.
Figure 4.
GC-MS Analysis of the Enzyme Reaction.
(A) Reaction products from the recombinant CYP710A assays were analyzed in GC-MS total ion chromatograms. The top panel represents the total ion chromatogram of standard sterols at specific retention times (RTs): a, cholesterol-trimethylsilyl (TMS) (RT = 15.5 min; mass-to-charge value [m/z] = 458); b, brassicasterol-TMS (RT = 16.4 min; m/z = 380); c, campesterol-TMS (RT = 17.4 min; m/z = 383); d, stigmasterol-TMS (RT = 18.5; m/z = 394); e, β-sitosterol (RT = 19.1; m/z = 396). The assays of the At 710A1 (with 10 pmoles of CYP710A1 P450/mL) and the 710A11 (with 20 pmoles of CYP710A11 P450/mL) proteins were done with β-sitosterol as the substrate (60-min reactions), and the At 710A2 assay (90-min reaction with 100 pmoles of CYP710A2 P450/mL) was performed using the synthesized 24-epi-campesterol (containing 30% campesterol as impurity) as the substrate (indicated by the arrowhead). The positions of the reaction products are indicated by the arrows. Sterol structures were identified by reference to relative RT and mass spectra. The pattern of fragment ions with m/z values of 484, 394, 255, and 129 were attributed to stigmasterol, and the fragment ions (m/z = 470, 380, 365, and 129) were used to identify brassicasterol/crinosterol. The 24-epimers were not separately analyzed under our experimental conditions. Campesterol and an unknown compound, both contaminants in β-sitosterol (substrate), are shown by a plus sign and an asterisk, respectively. Cholesterol (from the insect cell microsomes) and β-sitosterol (substrate) are indicated by open triangles (RT = 15.1 min) and pound signs, respectively. Recombinant CYP73A5 microsomes were used for the assay with β-sitosterol (bottom panel).
(B) GC-MS in SIM. A, β-sitosterol; B, stigmasterol; C, campesterol; D, 24-epi-campesterol (containing 30% campesterol as impurity); E, brassicasterol. The m/z value of 394 was used for SIM analysis of stigmasterol production, and m/z of 380 was used for the detection of brassicasterol/crinosterol.
It has been known that Brassicaceae family plants contain 24-methyl-Δ22-sterols as a mixture of brassicasterol (24R-epimer) and crinosterol (24S-epimer), which are derived from 24-epi-campesterol and campesterol, respectively (Matsumoto et al., 1983; Benveniste, 2004). When campesterol was used as the substrate for the CYP710A1 and CYP710A2 assays, no crinosterol production was detected (Figure 4B). Then, we synthesized 24-epi-campesterol to clarify whether the stereoconfigurations of the 24-methyl group might be distinguished by the Arabidopsis CYP710A proteins. When the CYP710A2 microsomes were assayed with the synthesized 24-epi-campesterol (containing 30% campesterol as impurity), a new product peak appeared at the same retention time (Figure 4A, 16.4 min, indicated by the arrowhead) as that of brassicasterol (Figures 4A and 4B). The fragmentation pattern perfectly matched that of brassicasterol. Interestingly, 24-epi-campesterol could not be the substrate of CYP710A1, and no corresponding Δ22-sterol was produced (Figure 4B). These results indicated that Arabidopsis CYP710A2 was the desaturase responsible for the production of brassicasterol from 24-epi-campesterol (Figure 1). A weak activity of crinosterol production was detected from the CYP710A11 assay with campesterol as the substrate (see Supplemental Figure 2 online), suggesting that tomato plants might produce crinosterol. No reaction products were detected when 24-epi-campesterol was added for the CYP710A11 assay (see Supplemental Figure 2 online).
These recombinant CYP710A reactions were completely dependent on the addition of NADPH, and NADH was not a substitute for NADPH (data not shown). The microsomal assays for the desaturase reactions proceeded even without adding recombinant NADPH-P450 reductase (Mizutani and Ohta, 1998), indicating that the endogenous NADPH-P450 reductase in insect cells could support the desaturase reactions of these recombinant CYP710A proteins to some extent. These results were consistent with our previous results from microsomal assays of Arabidopsis CYP707A proteins that are abscisic acid 8′-hydroxylases (Saito et al., 2004). When the CYP710A1, CYP710A2, and CYP710A11 proteins were assayed with cholesterol and fucosterol, no reaction products were detected under our experimental conditions (data not shown), and no oxidation products (Figure 4A) were detected from the insect cells expressing CYP73A5, which is a cinnamate 4-hydroxylase (Mizutani et al., 1997). In addition, CYP710A1 failed to functionally complement the sterol C-22 desaturation activity with ergosta-5,7,24(28)-ergostatrienol as the substrate in a yeast strain of CYP61 (ERG5) gene disruption (T. Morikawa, unpublished results). These results indicated that the substrate specificities of Arabidopsis CYP710A1 and tomato CYP710A11 proteins were rather strict toward β-sitosterol and that Arabidopsis CYP710A2 was capable of producing brassicasterol and stigmasterol from 24-epi-campesterol and β-sitosterol, respectively.
Enzymatic Properties of CYP710A1, CYP710A2, and CYP710A11
The C-22 desaturase reactions were further investigated using the CYP710A1, CYP710A2, and CYP710A11 microsomes (Table 1). The Km values for β-sitosterol of CYP710A1 and CYP710A11 were estimated to be 1.0 and 3.7 μM, respectively, and the kinetic constant (kcat) values were calculated as 0.53 and 10 min−1, respectively. The Km values indicated that both CYP710A1 and CYP710A11 have high affinity toward β-sitosterol as the substrate. The CYP710A2 activity (Figure 4) was not high enough for reproducible kinetic studies to determine the Km values for 24-epi-campesterol and β-sitosterol. The prominent peak at 420 nm observed with the reduced CO spectrum (Figure 3B) implicated the possibility that processes of correct folding and appropriate heme integration might not have efficiently proceeded with the CYP710A2 in the insect cells. This might be the case for CYP710A1 as well (Figure 3A). Furthermore, the synthesized 24-epi-campesterol contained 30% campesterol as impurity, and the correct Km value for 24-epi-campesterol could not be determined. Relative activities were thus obtained to compare the substrate specificities of CYP710A2. The activities of brassicasterol production and stigmasterol production were 0.028 and 0.0027 nmoles/nmol P450/min, respectively, at 10 μM of the substrate concentrations. No reaction product was detected from the CYP710A2 assay when campesterol alone was used as the substrate (Figure 4B). The brassicasterol production activity level determined with CYP710A2 was <5% of the stigmasterol production activity of CYP710A1 under the same experimental conditions. These enzyme assay results indicated that the substrate specificities of CYP710A proteins were fairly strict in terms of the sterol side chain structures, whereas the catalytic efficiencies were not notably higher than other recombinantly expressed P450s (Mizutani et al., 1997; Saito et al., 2004).
Table 1.
Enzymatic Properties of Recombinant P450s of Arabidopsis CYP710A1 and Tomato CYP710A11
| Activity
|
|||
|---|---|---|---|
| Enzyme | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) |
| CYP710A1 | 1.0 ± 0.090 | 0.53 ± 0.021 | 0.53 ± 0.026 |
| CYP710A11 | 3.7 ± 0.11 | 10 ± 0.22 | 2.8 ± 0.023 |
C-22 Desaturase Activities in Plants
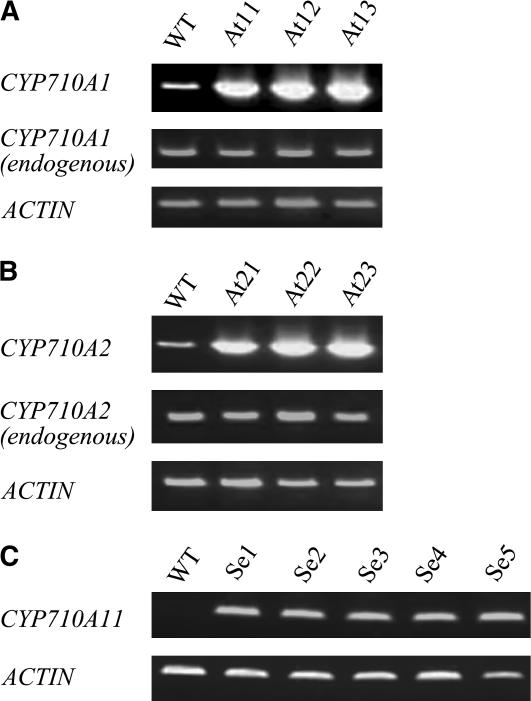
Functions of CYP710 proteins in plants were investigated by transgenic approaches. The full-length coding sequences of the Arabidopsis CYP710A genes (CYP710A1 and CYP710A2) and the tomato CYP710A11 cDNA were put under the control of the cauliflower mosaic virus 35S promoter on a pBIN-derived binary vector, and the constructs (pBINCYP710A1, pBINCYP710A2, and pBINCYP710A11) were then subjected to Agrobacterium tumefaciens–mediated transformation of Arabidopsis plants using the floral dip method (Clough and Bent, 1998). Based on the screening for kanamycin resistance, we identified 15, 7, and 22 independent T1 lines from the transformation experiments of CYP710A1, CYP710A2, and CYP710A11, respectively. T-DNA integration events were confirmed through PCR using genomic DNA (data not shown). We obtained three, three, and five independent transgenic T2 lines as the overexpressors of 35S:CYP710A1 (At11, At12, and At13), 35S:CYP710A2 (At21, At22, and At23), and 35S:CYP710A11 (Se1, Se2, Se3, Se4, and Se5), respectively.
The correlation between P450 transgene expression levels (Figure 5) and sterol compositions (Table 2) were analyzed in these transgenic plants. It should be noted that the 24-epimers were not separately analyzed in this study. In the 35S:CYP710A1-, 35S:CYP710A2-, and 35S:CYP710A11-transgenic lines, the large accumulations of the transgene transcripts were evident (Figure 5). No significant differences in the endogenous CYP710A1 and CYP710A2 expression levels were observed between wild-type plants and transgenic lines. All the 35S:CYP710A1 lines contained stigmasterol at higher levels ranging from 21 to 110 μg/g fresh weight, corresponding to an ∼6- to 32-fold increase compared with that in the wild-type plants (Table 2). Six- to 28-fold increases in the stigmasterol levels (22 to 98 μg/g fresh weight) were also demonstrated in the 35S:CYP710A11 lines (Table 2). No significant changes were observed in the brassicasterol/crinosterol (24-methyl-Δ22-sterols) and 24-epi-campesterol/campesterol (24-methyl-sterols) levels in both the 35S:CYP710A1 and 35S:CYP710A11 lines (Table 2). The weak activity of tomato CYP710A11 toward campesterol detected in the enzyme assay (see Supplemental Figure 2 online) was not reflected in the crinosterol production in Arabidopsis plants (Table 2). In these transformants, β-sitosterol levels decreased, while the ratio of 24-methyl sterols to 24-ethyl sterols remained largely unchanged. In the 35S:CYP710A2 lines, brassicasterol/crinosterol levels increased by ∼18 times from that in wild-type plants (Table 2). The elevated levels of brassicasterol/crinosterol were accompanied by the decreases in 24-epi-campesterol/campesterol levels (Table 2), supporting in planta that Arabidopsis CYP710A2 was involved in the desaturase reaction to produce brassicasterol/crinosterol from 24-epi-campesterol/campesterol. Stigmasterol levels also dramatically increased, and this increase was associated with the decrease in β-sitosterol levels (Table 2), indicating that CYP710A2 was also implicated in the stigmasterol production from β-sitosterol (Figure 4). The accumulation levels of the Δ22-sterols in the 35S:CYP710A2 lines were comparable to those in the 35S:CYP710A1 and 35S:CYP710A11 lines (Table 2), implicating the possibility that the activity of CYP710A2 (Figure 3) might not be fully expressed in the enzyme assay.
Figure 5.
Overexpression of CYP710A Genes in Arabidopsis Transgenic Lines.
(A) RT-PCR analyses of the expression levels of the CYP710A1 transgene in the 35S:CYP710A1 lines (At11 to At13).
(B) RT-PCR analyses of the expression levels of the CYP710A2 transgene in the 35S:CYP710A2 lines (At21 to At23).
(C) RT-PCR analyses of the expression levels of the CYP710A11 transgene in the 35S:CYP710A11 lines (Se1 to Se5).
Table 2.
Sterol Compositions in 35S:CYP710A1-, 35S:CYP710A2-, and 35S:CYP710A11-Transgenic Arabidopsis Lines
| Sample | 24-Methyl-Δ22-Sterolsa | 24-Methyl-Sterolsa | Stigmasterol | β-Sitosterol |
|---|---|---|---|---|
| Wild type | 0.85 ± 0.062 | 23 ± 2.4 | 3.4 ± 0.52 | 250 ± 20 |
| At11 | 1.9 ± 0.62 | 19 ± 1.5 | 110 ± 9.9 | 81 ± 3.7 |
| At12 | 0.22 ± 0.052 | 19 ± 2.1 | 21 ± 4.5 | 160 ± 6.9 |
| At13 | 0.50 ± 0.010 | 21 ± 3.2 | 86 ± 16 | 140 ± 9.1 |
| At21 | 15 ± 1.4 | 4.5 ± 0.33 | 81 ± 7.2 | 170 ± 3.8 |
| At22 | 16 ± 0.22 | 2.5 ± 0.20 | 83 ± 9.1 | 110 ± 13 |
| At23 | 14 ± 1.8 | 4.1 ± 0.44 | 50 ± 7.7 | 120 ± 5.8 |
| Se1 | 0.66 ± 0.16 | 17 ± 0.43 | 27 ± 5.8 | 190 ± 27 |
| Se2 | 0.63 ± 0.10 | 20 ± 2.6 | 27 ± 3.6 | 190 ± 27 |
| Se3 | 1.20 ± 0.20 | 26 ± 0.28 | 98 ± 29 | 190 ± 61 |
| Se4 | 0.96 ± 0.067 | 20 ± 3.5 | 46 ± 2.5 | 170 ± 33 |
| Se5 | 0.60 ± 0.053 | 16 ± 1.2 | 22 ± 4.9 | 140 ± 20 |
| cyp710a2 | n.d.b | 20 ± 1.1 | 3.7 ± 0.72 | 170 ± 14 |
The same plant samples were divided into two parts for the sterol analyses and the RT-PCR to monitor the transgene expression levels (Figure 5). Data are the means ± sd of triplicate determinations. Values are given in μg/g fresh weight.
The C24-epimers were not separately analyzed. 24-Methyl-Δ22-sterols are a mixture of brassicasterol and crinosterol, and 24-methyl-sterols consist of 24-epi-campesterol and campesterol.
n.d., not detected.
In addition to the overexpression studies, the brassicasterol/crinosterol production activities of CYP710A2 were further investigated using plants carrying a T-DNA insertion within CYP710A2 (Figure 6A, cyp710a2 line). Clear phenotypic alterations were not observed in the cyp710a2 plants grown in soil under continuous light at 22°C.The T-DNA insertion event was in the coding sequence of CYP710A2 (Figure 6A), and no apparent CYP710A2 transcript accumulation was detected (Figure 6B). In the cyp710a2 plants, the brassicasterol/crinosterol production activity was almost completely abolished (Table 2). On the other hand, the stigmasterol content in the cyp710a2 plants was maintained at the same level as that of wild-type plants, which was in agreement with the finding that CYP710A1, as well as CYP710A2, was responsible for the stigmasterol production. Thus, the activities of CYP710A1 and CYP710A2 proteins were redundant in terms of the production of stigmasterol from β-sitosterol. The enzyme assay results showed that CYP710A2 was highly specific for 24-epi-campesterol for producing brassicasterol (Figure 4), and campesterol was not accepted as the substrate. In addition, the cyp710a2 plants contained no 24-methyl-Δ22-sterols (Table 2). These results suggested that Arabidopsis plants might produce brassicasterol from 24-epi-campesterol as the major 24-methyl-Δ22-sterol through the enzyme activity of CYP710A2. No significant difference was observed in the 24-methyl-sterol levels (Table 2) between wild-type plants and the cyp710a2 plants. The first committed step in the BR biosynthesis is the enzymatic conversion of campesterol/24-epi-campesterol into campestanol via the formation of 24-methyl-cholest-4-en-3β-ol and 24-methyl-cholest-4-en-3-on (Noguchi et al., 1999), and brassicasterol/crinosterol are not involved in the BR biosynthetic pathway (Figure 1). In the cyp710a2 plants, campesterol/24-epi-campesterol accumulated at the same level as that in the wild type (Table 2), while brassicasterol/crinosterol was depleted. Thus, BR biosynthesis should not be affected in this T-DNA mutant line.
Figure 6.
T-DNA Insertion Mutagenesis.
(A) Schematic diagram of the T-DNA insertion event within CYP710A2 in the cyp710a2 line (SALK_001175). The primers (Table 3) used for the analysis of the T-DNA insertion are shown.
(B) Expression levels of CYP710A genes (CYP710A1, CYP710A2, CYP710A3, and CYP710A4) in the cyp710a2 line.
Expression Profiles of CYP710A Family Genes in Arabidopsis
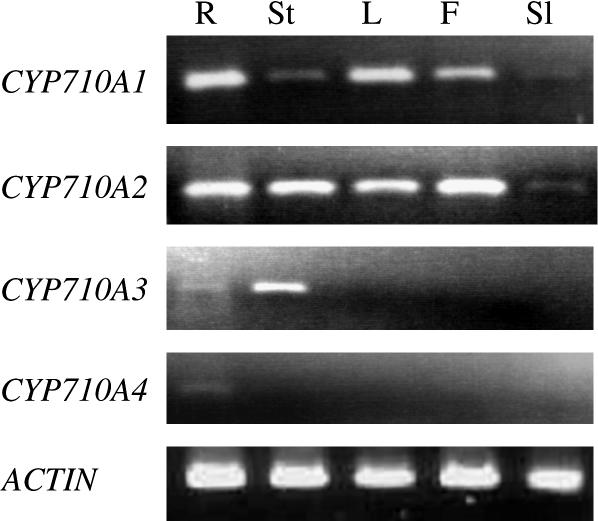
We determined tissue-specific expression profiles of the CYP710A genes in Arabidopsis by RT-PCR (Figure 7). Transcript accumulation from CYP710A1 expression was detected at higher levels in various organs, including roots, leaves, and flowers, while the levels in stems and mature siliques were found to be very low. The expression of CYP710A2 was ubiquitous except for a weak signal in mature siliques. CYP710A3 expression was specific for the stem, and very weak expression of CYP710A4 was detected in roots. These results indicated specific regulation mechanisms for the expression of CYP710A genes in different tissues and organs.
Figure 7.
Expression Profiles of CYP710A Genes in Arabidopsis.
Endogenous expression levels of CYP710A1, CYP710A2, CYP710A3, and CYP710A4 genes were analyzed in wild-type Arabidopsis plants. R, root; St, stem; L, leaf; F, inflorescence stem; Sl, mature silique.
We generated transgenic Arabidopsis lines of promoter:β-glucuronidase (GUS) fusions to monitor the expression profiles of CYP710A1 (A1P:GUS), CYP710A2 (A2P:GUS), CYP710A3 (A3P:GUS), and CYP710A4 (A4P:GUS). In the A1P:GUS plants (Figure 8), the reporter expression was restricted to the vascular tissues in both shoots and roots. The GUS staining was seen in sepals, and both primary and lateral root tips were also GUS stained. It should be noted that only older inflorescences were consistently stained, while no staining was observed in younger tissues. The staining in the A2P:GUS plants was observed in both root vascular tissues and leaf tissues with higher levels in younger leaves. The staining was detected in inflorescence stems, together with carpels and seeds as well. The GUS staining also suggested significant levels of CYP710A2 expression in siliques, whereas only a lower level of mRNA accumulation was detected by semiquantitative PCR analysis (Figure 7). On the other hand, it was suggested that the CYP710A3 expression was restricted to the primary root caps and immature petals, and no GUS staining was detected in other tissues. In the A4P:GUS plants, the GUS staining was only seen in the roots, including root hair, but no staining was observed in the root tips. These results suggest that CYP710A genes in Arabidopsis may be under strict regulations for tissue and developmental stage-specific expression.
Figure 8.
Promoter:GUS Staining.
Promoter:GUS fusion analyses for differential expression patterns of CYP710A1 (A), CYP710A2 (B), CYP710A3 (C), and CYP710A4 (D). Tissue-specific expression patterns are shown for CYP710A1 ([E] to [I]), CYP710A2 ([J] to [N]), CYP710A3 ([O] to [Q]), and CYP710A4 ([R] and [S]).
DISCUSSION
CYP710 Family P450 Genes
We have demonstrated both in vitro and in planta that the P450 enzymes, CYP710A1 from Arabidopsis and CYP710A11 from tomato, were responsible for the C-22 desaturation reaction with β-sitosterol as the substrate, yielding stigmasterol as the product (Figure 1), and that CYP710A2 from Arabidopsis was responsible for the production of brassicasterol and stigmasterol from 24-epi-campesterol and β-sitosterol (Figure 1), respectively. The T-DNA mutagenesis study demonstrated that CYP710A2 was essential for the brassicasterol/crinosterol production in Arabidopsis, indicating that unidentified desaturases were not able to compensate for the C-22 desaturase reaction catalyzed by CYP710A2. While we presented evidence for the C-22 desaturases only from Arabidopsis and tomato, it is possible that the desaturation reactions in other plant species may be also catalyzed by the CYP710A family members.
CYP710A genes are present in diverse plant species from gymnosperms to monocots and dicots (Nelson et al., 2004), and CYP710 family sequences are also found as CYP710B in lower photosynthetic unicellular eukaryotes, including red algae (Cyanidioschyzon merolae) and green algae (Chlamydomonas reinhardtii). The deduced amino acid sequence of Arabidopsis CYP710A1 is 40.4 and 37.9% identical to that of CYP710B from the green alga, C. reinhardtii. These CYP710 proteins from plants and lower eukaryotes contained the functional domains, such as the N-terminal hydrophobic membrane-anchoring region of 30 to 50 amino acids and a conserved Cys residue in the C-terminal portion involved in binding the heme iron in the fifth coordination site (Schuler and Werck-Reichhart, 2003). The sequence conservation of F(L/M)FA(A/S)QDA(S/T)(S/T)S at the substrate recognition site (SRS4) on the putative I helix region (Gotoh, 1992) is characteristic of the CYP710 proteins and is thought to be crucial for C-22 desaturation activity.
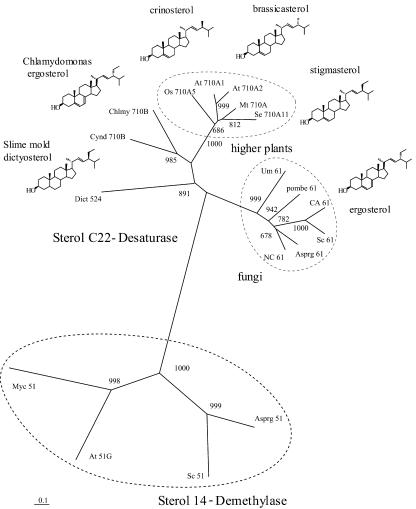
Figure 2 shows the alignment of the deduced primary structures of Arabidopsis CYP710A1 and tomato CYP710A11 proteins with those of CYP710A5 from Oryza sativa, CYP710B from C. reinhardtii, CYP524 from Dictyostelium discoideum, and CYP61 from Saccharomyces cerevisiae. It has been reported that C. reinhardtii produces ergosterol and 7-dehydroporiferasterol as the major end sterols (Salimova et al., 1999), suggesting that CYP710B may be the C-22 desaturase in this algae. The CYP710 family is one of four P450 families (CYP51, CYP97, CYP710, and CYP711) conserved between higher plants and algae, implying that the sterol C-22 desaturation reaction may be essential in all plant taxa. A partial CYP710 sequence was also identified from Physcomitrella patens (contig629 at PHYSCObase, http://moss.nibb.ac.jp/), suggesting that the C-22 desaturase may also function in bryophytes, the ancestors of vascular plants.
A phylogenetic tree (Figure 9; see Supplemental Figure 3 online) was obtained with CYP710A proteins from higher plants, CYP710B proteins from C. reinhardtii and C. merolae, and fungal CYP61 sequences including S. cerevisiae and S. pombe. The higher plant CYP710 proteins formed a cluster distinct from the fungal CYP61 cluster. These P450 sequences of CYP710 proteins and CYP61 proteins were distant from those of another P450 monooxygenase, CYP51, involved in the sterol biosynthetic pathway catalyzing the sterol 14-demethylase reaction (Benveniste, 2004). It should be noted that CYP524 from a cellular slime mold (D. discoideum), containing the sequence conservation in the substrate recognition site (Figure 2), was located on the same branch of CYP710 proteins in the phylogenetic tree (Figure 9). D. discoideum produces 4α-methylergostanol, 24β-ethycholesta-8,22-enol, and dictyosterol as Δ22-sterols (Nes et al., 1990), suggesting the possibility that these different subfamily P450s may be involved in the same catalytic function. Sterol analysis indicated that the slime mold evolved from algal rather than from fungal ancestors (Nes et al., 1990), which is consistent with the close location of CYP524 and CYP710B in the phylogenetic tree (Figure 8).
Figure 9.
Phylogenetic Tree for CYP710 Proteins and Major Δ22-Sterols.
Phylogenetic tree of CYP710-related sequences were obtained with CYP710A1 (At 710A1) and CYP710A2 (At 710A2) from Arabidopsis, CYP710A11 (Se 710A11) from tomato, CYP710A (Mt 710A) from M. truncatula, CYP710A5 (Os 710A5) from O. sativa, CYP710B (Chlmy 710B) from C. reinhardtii, CYP710B (Cynd 710B) from C. merolae, CYP524 (Dict 524) from D. discoideum, CYP61 (pombe 61) from S. pombe, CYP61 (NC 61) from N. crassa, CYP61 (CA 61) from C. albicans, CYP61 (Sc 61) from S. cerevisiae, CYP61 (Asprg 61) from A. fumigatus, CYP61 (Um 61) from U. maydis, CYP51F1 (Asprg 51) from A. nidulans, CYP51F1 (Sc 51) from S. cerevisiae, CYP51G1 (At 51G) from Arabidopsis, and CYP51B1 (Myc 51) from M. tuberculosis. Major Δ22-sterols from higher plants (stigmasterol, brassicasterol, and crinosterol), C. reinhardtii (ergosterol), and D. discoideum (dictyosterol) are shown next to the corresponding genes. Values at branch nodes indicate numbers of bootstrap trials out of 1000 that produced each node.
Δ22-Sterols
The Arabidopsis CYP710A1 and tomato CYP710A11 proteins catalyzed the C-22 desaturation reaction with β-sitosterol as the primary substrate. In the 35S:CYP710A1- and 35S:CYP710A11- transgenic lines, the stigmasterol levels increased by 6- to 32-fold and 6- to 28-fold compared with those of wild-type plants, respectively. These results substantiated the fact that the C-22 desaturation activity of the CYP710A1 and CYP710A11 proteins were also functioning in plants to produce stigmasterol from β-sitosterol.
CYP710A2 was able to act on 24-epi-campesterol and β-sitosterol to yield brassicasterol and stigmasterol, respectively. These enzyme activities were confirmed using the 35S:CYP710A2-transgenic plants and the cyp710a2 line containing the T-DNA insertion within CYP710A2 (Table 2). In the enzyme assay experiments, CYP710A2 was specific for 24-epi-campesterol, indicating that brassicasterol should be the major form of the 24-methyl-Δ22-sterol in Arabidopsis. This is consistent with the finding that the seeds of Brassica plants (rape [Brassica napus] and mustard [Brassica juncea]) contained the C-22 unsaturated forms of the 24-methyl sterols as a mixture of brassicasterol and crinosterol (10 to 30%) as the minor component (Matsumoto et al., 1983). In this study, the stereoconfigurations of the 24-methyl group were not distinguished, and we cannot rule out the possibility that campesterol may be a very weak substrate for the CYP710A2 reaction to accumulate crinosterol in planta. The ratio of brassicasterol to crinosterol should be clarified in Arabidopsis plants. The enzyme reactions catalyzed by CYP710A3 and CYP710A4, of which recombinant enzymes could not be functionally expressed in our system (data not shown), also should be elucidated for complete understanding of the biosynthesis of the Δ22-sterols in Arabidopsis.
It has been reported that proper ratios of β-sitosterol to campesterol are imperative for normal plant development and sexual reproduction (Schaller, 2003; Benveniste, 2004). While the CYP710A1, CYP710A2, and CYP710A11 overexpressors accumulated stigmasterol at ∼6- to 32-fold higher levels, with a concurrent decrease in the β-sitosterol level, of that in wild-type plants, there were no significant changes in the ratio of 24-ethyl sterols (β-sitosterol and stigmasterol) to 24-methyl sterols (campesterol and brassicasterol). The 35S:CYP710A1, 35S:CYP710A2, and 35S:CYP710A11 lines displayed no drastic phenotypic alterations, such as dwarfism or developmental abnormalities, indicating that the accumulation of the Δ22-sterols at the observed levels was not influencing BR biosynthesis and other developmental processes revealed by studies of mutants such as smt1/orc/cph, hyd1, hyd2/fk, smt1, and smt2/cvp1 (Clouse, 2000; Lindsey et al., 2003).
The cyp710a2 plants in which CYP710A2 function was abolished by the T-DNA insertion (Figure 6, Table 2) did not exhibit clear phenotypic alterations. The enzyme assay results indicated that Arabidopsis CYP710A2 was involved in the production of both Δ22-methyl-sterols (brassicasterol/crinosterol) and Δ22-ethyl-sterol (stigmasterol). The cyp710a2 plants did not accumulate brassicasterol/crinosterol, while stigmasterol was produced at a level comparable to that in wild-type plants (Table 2). Thus, Arabidopsis CYP710A2 is thought to be the crucial enzyme for the production of brassicasterol/crinosterol in Arabidopsis, and enzymes other than CYP710A2 also should be involved in stigmasterol production. This is consistent with the findings that both CYP710A1 and CYP710A2 enzymes catalyzed the C-22 desaturation reaction to produce stigmasterol from β-sitosterol in Arabidopsis. In the cyp710a2 plants, campesterol/24-epi-campesterol, the direct precursors of BR biosynthesis (Figure 1), accumulated at the same level as that in wild-type plants, indicating that BR biosynthesis should not be affected. There are different possibilities for the lack of a phenotype in the cyp710a2 line. Brassicasterol and crinosterol specifically occur in the Brassicaceae (Benveniste, 2004), but their physiological functions are not known. It is possible that the loss of brassicasterol/crinosterol production activity may not be detrimental to plants. It is also possible that the physiological roles of the Δ22-sterols may be redundant. If this is the case, the defect in brassicasterol/crinosterol production activity should not be a critical factor for the growth of Arabidopsis containing stigmasterol at a normal level.
The tissue specificity of the Δ22-sterol production activities remains to be clarified. The expression of Arabidopsis CYP710A genes was suggested to be under strict regulation in different tissues and organs (Figure 8), suggesting the possibility that phenotypic alterations may be observed in definite tissues at specific developmental stages. It has been reported that another P450 gene involved in sterol biosynthesis, CYP51G1, was strongly expressed in leaf vascular tissues (Kim et al., 2005), which was closely similar to the expression pattern of CYP710A1 involved in stigmasterol production (Figure 8). CYP51G1 (At1g11680) is the single functional gene (Kim et al., 2005) encoding obtusifoliol 14α-demetylase protein in Arabidopsis (Figure 1). The promoter:GUS experiments suggested that the brassicasterol/crinosterol production activities of CYP710A2 might be higher in younger leaves (Figure 8), implicating greater demands for brassicasterol/crinosterol in younger leaves. Sterol analyses of specific samples, such as mesophyll protoplasts, vascular tissues, and root tips, may provide information to link phenotypic alterations, if any, with differential distributions of the Δ22-sterols in Arabidopsis.
The physiological roles of Δ22-sterols are not well known. Studies from sterol mutants indicated that correct membrane properties are assured through the maintenance of proper sterol compositions, affecting cell functions such as cell polarity, auxin efflux, and ethylene signaling (Clouse, 2000; Lindsey et al., 2003). For example, the membrane localization of PIN1 and PIN3 proteins is disturbed in the sterol mutant smt1orc (Willemsen et al., 2003), indicating that sterol profiles are important determinants of protein targeting in plants. Furthermore, fk and hyd1 as well as smt1/cph mutants are characterized by altered sterol profiles and accumulation of abnormal sterol intermediates, exhibiting incomplete cell walls and aberrant cell wall thickenings in embryonic and postembryonic tissues together with ectopic callose and lignin deposits (Schrick et al., 2004). In animals and yeasts, lipid microdomains or rafts are defined as specialized detergent-resistant membrane regions enriched in sphingolipids and cholesterol, harboring proteins involved in crucial physiological processes, such as signal transduction for cellular proliferation and differentiation, vesicular trafficking, and cytoskeleton organization (Harder and Simons, 1997; Hakomori et al., 1998). Recently, it has been reported that plant cells also contained lipid microdomains/rafts enriched in β-sitosterol, stigmasterol, and 24-methylcholesterol as well as cholesterol and sphingolipids (Mongrand et al., 2004; Borner et al., 2005). Involvement of Δ22-sterols in the formation and functional properties of lipid rafts/microdomains remains to be clarified.
In this study, we show that CYP710A1 and CYP710A2 from Arabidopsis and CYP710A11 from tomato are the sterol C-22 desaturases. The identification of the sterol C-22 desaturase genes in plants enables the manipulation of the expression levels of CYP710A genes, leading to the direct modification of sterol composition in crop plants. Furthermore, generating transgenic plants lacking either specific Δ22-sterol or all of the Δ22-sterol species by means of gene knockout and RNA interference should shed light on the crucial roles of phytosterols underlying BR-independent processes of embryonic and postembryonic development.
METHODS
Materials
Arabidopsis thaliana ecotype Columbia (Col-0) (Lehle Seeds) seedlings were grown at 22°C under continuous light as described previously (Mizutani and Ohta, 1998). For sterol analysis, RNA isolation, and GUS staining, Arabidopsis seedlings were grown under a sterile condition on 0.8% (w/v) agar plates containing germination medium (GM) supplemented with 1× Murashige and Skoog salts and 1% (w/v) sucrose (Valvekens et al., 1998).
Isolation of CYP710 Coding Sequences from Arabidopsis and Tomato
The entire coding sequences of CYP710A1 and CYP710A2 genes in Arabidopsis contain no intron and were thus amplified by PCR using genomic DNA as the template. Genomic DNA was prepared from 2-week-old Arabidopsis seedlings as described (Sambrook et al., 1989). A primer set of A1F and A1R (Table 3) was used for the amplification of the CYP710A1 coding sequence, and A2F and A2R (Table 3) was used for the amplification of the CYP710A2 coding sequence. The PCR products for the coding sequences of CYP710A1 and CYP710A2 were cloned into a pDrive cloning vector (Qiagen) to give pDCYP710A1 and pDCYP710A2, respectively. A tomato (Lycopersicon esculentum) cDNA clone coding for a putative CYP710A protein was identified through a tBLASTn search at the TIGR Gene Indices (http://tigrblast.tigr.org/tgi/) against the tomato EST database using the amino acid sequence of CYP710A1. Among candidate sequences identified, we obtained an EST of cLEX15L1 from Clemson University Genomics Institute. The cLEX15L1 insert was completely sequenced and shown to contain the putative entire coding region of a tomato CYP710A. The tomato P450 sequence was designated as CYP710A11 (David Nelson, http://drnelson.utmem.edu/CytochromeP450.html).
Table 3.
Primer Sequences
| Primer | Primer Sequence (5′–3′) |
|---|---|
| A1F | CCGTCTAGAGAATTCATGGTTTTCTCTGTTTCTAT |
| A1R | CCGGTCGACCTCGAGTTAGGAAAAGTTGGGATACTT |
| A2F | CGCGGATCCATGGTTTTCTCAGTTTCCAT |
| A2R | CCGCTCGAGTCAGAGGTTCGGATACGTT |
| 710A1QF | AGGCGCGTCGCAAAGTATCCCAACTTTTCC |
| 710A13′R | CTCGTGGGCCACTCACTTCGAGAGA |
| 710A2QF | ATCGTAACGTATCCGAACCTCTGA |
| 710A23′R | CACGATCACGAACAACAATAATAGT |
| 710A34QF | CCGCCTCGTTACCTCTCCTTGA |
| 710A3QR | CAACACATATATTAGGTGAA |
| 710A4QR | AAGAACACGAATTAAATATG |
| 710ToF | ATGGCATCCATTTGGGGTTTGTTATCT |
| 710ToR | TCATCGTGTGCACCTGTGTGCAAGGAAA |
| Act-F | ATGGCTGATGGTGAAGACATTC |
| Act-R | TCAGAAGCACTTCCTGTGAAC |
| A1pF | TGCTCTAGATGGAAGTCTTCGAGACTGAA |
| A1pR | TGCTCTAGAGTTTCTTTGTTTCTAGCTTG |
| A2pF | CCCAAGCTTAAGACTTTTGAACCATGTTA |
| A2pR | CCCAAGCTTCCTTTTTTTTCCTCTTCT |
| A3pF | TGCTCTAGAACCACATGATATCTAATGGA |
| A3pR | TGCTCTAGATGCTCTTTGTTTTCTATTA |
| A4pF | CGCGGATCCTTTGCATTCATATCCGAAAT |
| A4pR | CGCGGATCCTGCTCTTTGTTTTCTATT |
| LP | CGGTTTTGGCTTTCTGTCGTT |
| RP | GGCGGTTTGCTCTTCGATTTT |
| LBa1 | TGGTTCACGTAGTGGGCCATG |
Heterologous Expression in Insect Cells
Recombinant CYP710A proteins were prepared by expressing the full lengths of the Arabidopsis CYP710A1 and CYP710A2 coding sequences and the tomato CYP710A11 cDNA in a baculovirus expression vector system as described previously (Mizutani et al., 1997), using the Bac-to-Bac baculovirus expression system (Invitrogen) and Spodoptera furugiperda cells (Sf9; Invitrogen). Briefly, pDCYP710A1 and pDCYP710A2 were double-digested with EcoRI-XhoI and BamHI-XhoI, respectively, and the tomato CYP710A11 cDNA insert of cLEX15L1 in pBluescript SK− was isolated by digesting with BamHI and XhoI. These CYP710A coding sequences were cloned into a pFastBac1 plasmid digested with appropriate restriction enzymes. The pFastBac1 constructs were then used for the preparation of recombinant bacmid DNAs by transformation of Escherichia coli strain DH10Bac (Invitrogen), and transfection of the insect cells was done according to the manufacturer's instructions (Invitrogen). For expression of the recombinant P450 proteins, Sf9 cells were maintained in Sf900II serum-free medium (Invitrogen) supplemented with 200 μM 5-aminolevulinic acid and 200 μM ferrous citrate to increase the low heme synthetic capacity of the insect cells (Saito et al., 2004).
Enzyme Assays
For preparation of microsomal fractions, the infected cells (60 mL of suspension-cultured cells) were washed with PBS and suspended in buffer A consisting of 20 mM potassium phosphate, pH 7.25, 20% (v/v) glycerol, 1 mM EDTA, and 1 mM DTT. The cells were sonicated, and cell debris was removed by centrifugation at 10,000g for 15 min. The supernatant was further centrifuged at 100,000g for 1 h, and the pellet was homogenized with buffer A as the microsomal fraction. The microsomal fractions were stored at −80°C until use. The complete reaction mixture (0.5 mL) consisted of 50 mM potassium phosphate, pH 7.25, recombinant CYP710A microsomes (50 to 500 μg protein/mL), 100 μM NADPH, and sterol substrates at different concentrations ranging from 1 to 100 μM. To monitor full P450 activities, microsome assays were supplemented with 0.1 unit/mL of a purified recombinant Arabidopsis NADPH-P450 reductase preparation (Mizutani and Ohta, 1998). After 5 to 90 min at 30°C, the reactions were stopped by adding 50 μL of 1 n HCl, and the reaction products were extracted four times with an equal volume of ethyl acetate. The ethyl acetate extracts were evaporated to dryness, and TMS ether derivatives for GC-MS analysis were prepared in a 1:1 mixture (40 μL) of pyridine and N,O-bis(trimethylsilyl)trifluoroacetamide (Wako Pure Chemical Industries) containing 1% (v/v) trimethylchlorosilane (Nacalai Tesque) at 90°C for 1 h. For calculation of enzyme kinetic parameters, linear regression was used in double-reciprocal analyses of activity data (Saito et al., 2004).
Semiquantitative RT-PCR Analysis
RT-PCR analyses were done for detailed comparison of the expression levels of CYP710A1, CYP710A2, CYP710A3, and CYP710A4 genes in Arabidopsis. Total RNA was isolated using an RNeasy plant mini kit (Qiagen), and genomic DNA contamination was eliminated using an RNase free DNase set (Qiagen). First-strand cDNA was synthesized using the Takara RNA PCR kit (AMV) version 3.0 in a 10-μL reaction mixture containing 300 ng of total RNA using an oligo(dT)16 as the reverse primer. The reverse transcription reactions were performed at 30°C for 10 min, 50°C for 30 min, and 95°C for 5 min and then chilled to 5°C for 5 min. The PCR was done using 2 μL of the reverse transcription products as the template in 10 μL of reaction mixture containing 1 mM MgCl2, 0.2 mM deoxynucleotide triphosphate mixture, 0.025 unit/μL Takara Ex Taq HS, and 0.2 μM of primers using the Takara RNA PCR kit (AMV) version 3.0. The PCR was programmed in 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min followed by an extension period of 10 min at 72°C. The gene-specific primer set for CYP710A1 was a pair of 710A1QF and 710A13′R (Table 3). A set of 710A2QF and 710A23′R (Table 3) was used for the analysis of CYP710A2 gene expression. For the CYP710A3 expression analysis, a set of 710A34QF and 710A3QR (Table 3) was prepared, and a set of 710A34QF and 710A4QR (Table 3) was used for CYP710A4 analysis. Arabidopsis ACTIN (Nairn et al., 1988) as the internal control was amplified under the same PCR conditions using the primer pair Act-F and Act-R (Table 3).
Overexpression of CYP710A
The entire coding sequences of CYP710A1 and CYP710A2 were double digested with XbaI-SalI and BamHI-XhoI from pDCYP710A1 and pDCYP710A2, respectively, and cloned into an XbaI-SalI and a BamHI-XhoI double-digested pBIN-based vector (Clontech) to yield pBINCYP710A1 and pBINCYP710A2, respectively. From cLEX15L1, CYP710A11 cDNA was excised with BamHI and XhoI and cloned into a BamHI-XhoI double-digested pBIN vector (pBINCYP710A11). These plasmids were electroporated into Agrobacterium tumefaciens strain EHA105 and transformed into Arabidopsis using the floral dip method (Clough and Bent, 1998). T1 seeds were screened on GM agar plates containing 25 μg/mL kanamycin, and resistant seedlings were transferred to soil and allowed to set seed. 35S:CYP710A1, 35S:CYP710A2, and 35S:CYP710A11 homozygous lines were selected by examining the kanamycin resistance of T3 seedlings.
Transgene expression levels in 35S:CYP710A1, 35S:CYP710A2, and 35S:CYP710A11 plants were analyzed by RT-PCR. Total RNA was extracted from rosette leaves of T2 plants using an RNeasy plant mini kit. The same plant samples were also used for the sterol composition analyses. RT-PCR was done using the Takara RNA PCR kit (AMV) version 3.0. The primer set of A1F and A1R (Table 3) was used to check the expression levels of both the CYP710A1 transgene and endogenous CYP710A1, and the pair of A2F and A2R was used to monitor the transcripts from both the CYP710A2 transgene and endogenous CYP710A2. CYP710A11 expression levels were studied using a set of 710ToF and 710ToR (Table 3). Endogenous expression levels of CYP710A1 and CYP710A2 genes in the transformants were studied using a set of primers of A1F and 710A13′R and a set of A2F and 710A23′R, respectively (Table 3). The primer sequences of 710A13′R and 710A23′R were derived from the 3′-noncoding regions of CYP710A1 and CYP710A2 genes, respectively, and the PCR products thus obtained were ascribed to the transcripts from the expression of CYP710A1 and CYP710A2 genes driven by their own promoters. Arabidopsis ACTIN (Nairn et al., 1988) as the internal control was amplified under the same PCR conditions.
T-DNA Insertion Lines
A T-DNA insertion event within the CYP710A2 coding sequence, cyp710a2 (SALK_001175), was found by the database search with the SIGnAL Arabidopsis Gene Mapping Tool (Alonso et al., 2003; http://signal.salk.edu/cgi-bin/tdnaexpress), and the seeds were obtained from ABRC (Columbus, OH). A gene-specific primer set of LP and RP and a T-DNA border primer LBa1 (Table 3) were used to clarify the T-DNA insertion event. Individual plants homozygous for a T-DNA insertion in CYP710A2 were identified by PCR screening and segregation analysis. The primer pairs of 710A1QF and 710A13′R, A2F and 710A23′R, 710A34QF and 710A3QR, and 710A34QF and 710A4QR (Table 3) were used to check the expression levels of CYP710A1, CYP710A2, CYP710A3, and CYP710A4 genes, respectively. Arabidopsis ACTIN (Nairn et al., 1988) as the internal control was amplified under the same PCR conditions using a primer pair of Act-F and Act-R (Table 3). Other Salk T-DNA insertions related to CYP710A2 were found outside the coding sequence and excluded from the analysis.
Promoter:GUS
For promoter:GUS analysis, promoter regions of Arabidopsis CYP710A genes including 5′-upstream portions were amplified by PCR from genomic DNA as the template. A 2012-bp fragment (A1P) for CYP710A1 was amplified using a primer pair of A1pF and A1pR (Table 3), and a 2008-bp fragment for CYP710A2 (A2P) was obtained using A2pF and A2pR (Table 3). As the CYP710A3 promoter (A3P), a 2002-bp fragment was amplified using A3pF and A3pR (Table 3). A 2060-bp fragment as for CYP710A4 (A4P) was obtained using a primer set of A4pF and A4pR (Table 3). The PCR products were subcloned into a pDrive cloning vector (Qiagen), and the inserts were completely sequenced. The restriction fragments were excised and subcloned into a plant transformation vector pBI101.1 (Clontech) to give pBI101-A1P, pBI101-A2P, pBI101-A3P, and pBI101-A4P. These plasmids were electroporated into A. tumefaciens strain GV3101 and transformed into Arabidopsis using the floral dip method (Clough and Bent, 1998). T1 seeds were screened on medium containing 25 μg/mL kanamycin, and resistant seedlings were transferred to soil and allowed to set to seed. Homozygous lines were selected by examining the kanamycin resistance of T3 seedlings.
Staining of Promoter:GUS Transgenic Plants
After 3 weeks of growth on GM plates, the promoter:GUS transgenic plants (A1P:GUS, A2P:GUS, A3P:GUS, and A4P:GUS) were first treated with 90% (v/v) ice-cold acetone for 1 h and submerged in GUS staining buffer containing 50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 0.1% (w/v) Triton X-100, and 0.5 mg/L 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Nacalai Tesque). The tissues were then infiltrated with staining buffer under vacuum and incubated overnight at 37°C (Jefferson, 1987). After rinsing with water, tissues were cleared with ethanol:acetic acid (9:1, v/v), which was followed with washes using 90, 70, and 50% (v/v) diluted ethanol. The tissues were then left in 50% glycerol for later photography.
Sterol Analysis
Samples (50 mg) were frozen and homogenized in liquid N2, and total sterol fractions were extracted for 30 min at room temperature in 5 mL chloroform/methanol (1:2, v/v). By adding 2 mL of 1% (w/v) KCl and 1 mL of chloroform, sterols were recovered in the organic phase. Two milliliters of methanol/water (10:9, v/v) was then added to the organic phase and evaporated to dryness under a N2 stream. The saponification was done in 2.5 mL of 1 M KOH in methanol at 90°C for 1 h, and 2 mL chloroform and 2.5 mL water were added to recover the organic phase. After adding 1.25 mL of 0.5 M KOH and 6 mL water, the organic phase was evaporated to dryness in vacuo and used for the TMS derivatization. Sterols were analyzed by GC-MS on a Saturn 2200 GC-MS system (Varian) with a CP-SIL5 CB LOW BLEED/MS column (30 m × 0.25 mm) (Varian). The port temperature for split injections (a split ratio of 50%) was 270°C, and helium was used as the carrier gas at a flow rate of 1.5 mL/min. The temperature program was started with a slow rise from 230 to 285°C (2°C/min), from 285 to 320°C (10°C/min), and finally 10 min at 320°C. Standards consisting of cholesterol, brassicasterol, campesterol, stigmasterol, and β-sitosterol were used for quantification and identification. The peak areas were automatically calculated using the 2200 Workstation (Varian), and sterol amounts were determined from the ratio of the peak area of each sterol to that of 5-α-cholestane as an internal standard (Dyer et al., 1995). Sterol structures were identified by reference to relative retention time and mass spectra. The pattern of fragment ions with m/z values of 484, 394, 255, and 129 were attributed to stigmasterol, and the fragment ions (m/z = 470, 380, 365, and 129) were used to identify brassicasterol/crinosterol. The 24-epimers were not separately analyzed under our experimental conditions.
DNA Sequence Analysis
DNA sequences were determined from both strands using CEQTM DTCA-Quick Start kits (Beckman Coulter) and a DNA sequencer, CEQTM2000XL DNA analysis system (Beckman Coulter). DNA and amino acid sequences were analyzed using GENETYX software (Software Development).
Phylogenetic Analysis
Amino acid sequences were aligned at DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/search/clustalw-e.html) using the ClustalW program (Thompson et al., 1994), and phylogenetic trees were determined using the neighbor-joining method (Saitou and Nei, 1987) with bootstrap analysis (1000 replicates) and Kimura's correction for protein distances (Kimura, 1983). The tree was visualized using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The following sequences were analyzed: CYP710A1 and CYP710A2 from Arabidopsis, CYP710A11 from tomato, CYP710A from Medicago truncatula, CYP710A5 from Oryza sativa, CYP710B from Chlamydomonas reinhardtii, CYP710B from Cyanidioschyzon merolae, CYP524 from Dictyostelium discoideum, CYP61 from Schizosaccharomyces pombe, CYP61 from Neurospora crassa, CYP61 from Candida albicans, CYP61 from Saccharomyces cerevisiae, CYP61 from Aspergillus fumigatus, CYP61 from Ustilago maydis, CYP51F1 from Aspergillus nidulans, CYP51F1 from S. cerevisiae, CYP51G1 from Arabidopsis, and CYP51B1 from Mycobacterium tuberculosis.
Chemical Synthesis
Crinosterol was synthesized for identification of reaction products in the enzyme assay experiments. A mixture of (22E,24S)-6β-methoxy-3α,5-cyclo-5α-ergost-22-ene (340 mg, 0.82 mmoles) and a catalytic amount of p-toluenesulfonic acid in an 80% aqueous tetrahydrofuran solution (10 mL) was refluxed for 3 h. The reaction mixture was cooled to room temperature and poured into a saturated aqueous sodium hydrogencarbonate solution (30 mL) followed by extraction with ethyl acetate (1 × 20 mL, 4 × 10 mL). The combined organic layer was washed with brine (15 mL), dried over anhydrous magnesium sulfate, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (hexane:ethyl acetate = 4:1) to yield crinosterol (258 mg, 79% yield) as a colorless solid. NMR δH (300 MHz, CDCl3): 0.69 (3H, s), 0.82 (3H, d, J = 6.9), 0.84 (3H, d, J = 6.9), 0.91 (3H, d, J = 6.6), 1.00 (3H, d, J = 5.7), 1.01 (3H, s), 3.52 (1H, m), 5.16 (2H, m), and 5.35 (1H, m). All physicochemical properties were identical to those reported previously (Lang and Djerassi, 1982; Anastasia et al., 1983).
The 24-epi-campesterol was synthesized as the substrate for the enzyme assay of recombinant CYP710A enzymes. A mixture of (22E,24R)-6β-methoxy-3α,5-cyclo-5α-ergost-22-ene (305 mg, 0.74 mmoles) and platinum(II) oxide (50 mg) in ethyl acetate (25 mL) was stirred overnight under hydrogen atmosphere. The catalyst was removed by decantation, and the solvent was removed in vacuo to give (24S)-6β-methoxy-3α,5-cyclo-5α-ergostan (301 mg). This crude product was purified by flash column chromatography (hexane:ethyl acetate = 4:1) to give 24-epi-campesterol (208 mg, 70% yield for two steps) as a colorless solid. NMR δH (500 MHz, CDCl3): 0.68 (3H, s), 0.776 (3H, d, J = 6.8), 0.783 (3H, d, J = 6.8), 0.86 (3H, d, J = 6.8), 0.92 (3H, d, J = 6.5), 1.01 (3H, s), 3.52 (1H, tt, J = 11.0, 4.7), and 5.35 (1H, m). NMR δC (125 MHz, CDCl3): 11.84, 15.43, 17.58, 18.88, 19.38, 20.51, 21.07, 24.28, 28.18, 30.56, 31.44, 31.65, 31.89 (×2), 33.70, 36.17, 36.49, 37.24, 39.05, 39.75, 42.29 (×2), 50.12, 55.98, 56.74, 71.78, 121.70, and 140.75. NMR analysis showed that our 24-epi-campesterol preparation was contaminated by campesterol (∼30% of the total material), a 24-epimer of the desired product. Although they could not be separated, the structures were established by comparing 1H and 13C NMR spectra with those of authentic campesterol. Khripach et al. (2002) reported inversion of stereochemistry at C-24 during the course of hydrogenation of C-24 substituted Δ22-steroids. The same kind of isomerization might have occurred in our hydrogenation step of (22E,24R)-6β-methoxy-3α,5-cyclo-5α-ergost-22-ene.
NMR spectra were recorded on either the Bruker ARX-500 or the JEOL JNM-AL300. Chemical shifts have been reported in the δ value by ppm unit relative to tetramethylsilane (δ 0 ppm for 1H) and the solvent (δ 77.0 ppm for 13C) as an internal standard in deuteriochloroform (CDCl3) solution. All J-values are given in hertz.
The (22E,24S) and (22E,24R)-6β-methoxy-3α,5-cyclo-5α-ergost-22-ene were prepared from stigmasterol as described (Takatsuto et al., 1997). Flash column chromatography was done using Kieselgel 60 (Merck) as the adsorbent.
Assay Methods
All spectrophotometric determinations were done at room temperature using a Cary 300 spectrophotometer (Varian). P450 was estimated from the CO difference spectrum using an extinction coefficient (Σ = 91 mM−1 cm−1). NADPH-P450 reductase was assayed by measuring its NADPH-cytochrome c reductase activity, and the rate of cytochrome c reduction was calculated from the A550 change using an extinction coefficient (Σ = 21 mM−1 cm−1). Protein was determined using the Coomassie protein assay reagent (Bio-Rad Laboratories).
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: CYP710A1 (AB219423), CYP710A2 (AB233425), CYP710A3 (At2g28850), CYP710A4 (At2g28860), cLEX15L1 (AW622568), CYP710A11 (AB223043), M. truncatula CYP710A (TC101450, http://tigrblast.tigr.org/tgi/), O. sativa CYP710A5 (NM_188395), S. pombe CYP61 (CAB11640), N. crassa CYP61 (EAA32679), C. albicans CYP61 (EAK98020), S. cerevisiae CYP61 (CAA89116), A. fumigatus CYP61 (EAL88107), U. maydis CYP61 (EAK81168), A. nidulans CYP51F1 (EAA59021), S. cerevisiae CYP51F1 (AAB68433), Arabidopsis CYP51G1 (NP_172633), and M. tuberculosis CYP51B1 (NP_215278). C. reinhardtii CYP710B (ChlamyDB, http://www.chlamy.org/chlamydb.html), C. merolae CYP710B (Cyanidioschyzon merolae Genome Project, http://merolae.biol.s.u-tokyo.ac.jp/), and D. discoideum CYP524 (dictyBase, http://dictybase.org/) sequences were retrieved from the database indicated.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heterologous Expression of Recombinant CYP710A Proteins in Insect Cells.
Supplemental Figure 2. GC-MS in the Selected Ion Monitoring Mode for Tomato CYP710A11 Assay.
Supplemental Figure 3. Multiple Sequence Alignment Used for Phylogenetic Analysis Visualized Using CINEMA5 (Parry-Smith et al., 1998, http://umber.sbs.man.ac.uk/dbbrowser/CINEMA2.1/).
Supplementary Material
Acknowledgments
This work was supported by New Energy and Industrial Technology Development (as part of the project called Development of Fundamental Technologies for Controlling the Process of Material Production of Plants). This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 16580281 to D.O.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daisaku Ohta (ohtad@bioinfo.osakafu-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036012.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anastasia, M., Allevi, P., Ciuffreda, P., and Fiecchi, A. (1983). Stereoselective synthesis of crinosterol [(22E,24S)-ergosta-5,22-dien-3β-ol]. J. Chem. Soc. Perkin Trans. 1 2365–2367. [Google Scholar]
- Benveniste, P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55 429–457. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2000). Plant development: A role for sterols in embryogenesis. Curr. Biol. 10 601–604. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2002). Brassinosteroid signal transduction: Clarifying the pathway from ligand perception to gene expression. Mol. Cell 10 973–982. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., Li, H., Zhou, W., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, R.G., Hetherington, C.S., Alberti, K.G.M.M., and Laker, M.F. (1995). Simultaneous measurement of phytosterols (campesterol and β-sitosterol) and 7-ketocholesterol in human lipoproteins by capillary column gas chromatography. J. Chromatogr. B 663 1–7. [DOI] [PubMed] [Google Scholar]
- Gotoh, O. (1992). Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 267 83–90. [PubMed] [Google Scholar]
- Hakomori, S., Yamamura, S., and Handa, A.K. (1998). Signal transduction through glyco(sphingo)lipids. Introduction and recent studies on glyco(sphingo)lipid-enriched microdomains. Ann. N. Y. Acad. Sci. 845 1–10. [DOI] [PubMed] [Google Scholar]
- Harder, T., and Simons, K. (1997). Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol. 9 534–542. [DOI] [PubMed] [Google Scholar]
- He, J.X., Fujioka, S., Li, T.C., Kang, S.G., Seto, H., Takatsuto, S., Yoshida, S., and Jang, J.C. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kelly, S.L., Lamb, D.C., Baldwin, B.C., Corran, A.J., and Kelly, D.E. (1997). Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 272 9986–9988. [DOI] [PubMed] [Google Scholar]
- Khripach, V.A., Zhabinskii, V.N., Konstantinova, O.V., Khripach, N.B., Antonchick, A.P., and Schneider, B. (2002). [3,3]-Claisen rearrangements in 24α-methyl steroid synthesis. Application to campesterol, crinosterol, and Δ25-crinosterol side chain construction. Steroids 67 597–603. [DOI] [PubMed] [Google Scholar]
- Kim, H.B., Schaller, H., Goh, C.-H., Kwon, M., Choe, S., An, C.S., Durst, F., Feldmann, A., and Feyereisen, R. (2005). Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 138 2033–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1983). The Neutral Theory of Molecular Evolution. (Cambridge, UK: Cambridge University Press).
- Lang, R.W., and Djerassi, C. (1982). Stereochemical aspects of acid-catalyzed cyclopropane ring-opening reactions. A stereospecific pathway to crinosterol and brassicasterol. Helv. Chim. Acta 65 407–418. [Google Scholar]
- Lindsey, K., Pullen, M.L., and Topping, J.F. (2003). Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 8 521–525. [DOI] [PubMed] [Google Scholar]
- Matsumoto, T., Shimizu, N., Shigemoto, T., Itoh, T., Iida, T., and Nishioka, A. (1983). Isolation of 22-dehydrocampesterol from the seeds of Brassica juncea. Phytochemistry 22 789–790. [Google Scholar]
- Meunier, B., de Visser, S.P., and Shaik, S. (2004). Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 104 3947–3980. [DOI] [PubMed] [Google Scholar]
- Mizutani, M., and Ohta, D. (1998). Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani, M., Ohta, D., and Sato, R. (1997). Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 113 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J.P., Hartmann, M.A., Bonneu, M., Simon-Plas, F., Lessire, R., and Bessoule, J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279 36277–36286. [DOI] [PubMed] [Google Scholar]
- Nairn, C.J., Winsett, L., and Ferl, R.J. (1988). Nucleotide sequence of an actin gene from Arabidopsis thaliana. Gene 65 247–257. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R., Schuler, M.A., Paquette, S.M., Werck-Reichhart, D., and Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser, J.L., and Chory, J. (2004). BRing it on: New insights into the mechanism of brassinosteroid action. J. Exp. Bot. 55 265–270. [DOI] [PubMed] [Google Scholar]
- Nes, W.D., Norton, R.A., Crumley, F.G., Madigan, S.J., and Katz, E.R. (1990). Sterol phylogenesis and algal evolution. Proc. Natl. Acad. Sci. USA 87 7565–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Takatsuto, S., Sakurai, A., Yoshida, S., Li, J., and Chory, J. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry-Smith, D.J., Payne, A.W.R., Michie, A.D., and Attwood, T.K. (1998). CINEMA—A novel colour INteractive editor for multiple alignments. Gene 221 GC57–GC63. [DOI] [PubMed] [Google Scholar]
- Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., and Mizutani, M. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Salimova, E., Boschetti, A., Eichenberger, W., and Lutova, L. (1999). Sterol mutants of Chlamydomonas reinhardtii: Characterisation of three strains deficient in C24(28) reductase. Plant Physiol. Biochem. 37 241–249. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schaeffer, A., Bronner, R., Benveniste, P., and Schaller, H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25 605–615. [DOI] [PubMed] [Google Scholar]
- Schaller, H. (2003). The role of sterols in plant growth and development. Prog. Lipid Res. 42 163–175. [DOI] [PubMed] [Google Scholar]
- Schrick, K., Fujioka, S., Takatsuto, S., Stierhof, Y.D., Stransky, H., Yoshida, S., and Jurgens, G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38 227–243. [DOI] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jurgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., and Jurgens, G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31 61–73. [DOI] [PubMed] [Google Scholar]
- Schuler, M.A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54 629–667. [DOI] [PubMed] [Google Scholar]
- Skaggs, B.A., Alexander, J.F., Pierson, C.A., Schweitzer, K.S., Chun, K.T., Koegel, C., Barbuch, R., and Bard, M. (1996). Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene 169 105–109. [DOI] [PubMed] [Google Scholar]
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuto, S., Watanabe, T., Fujioka, S., and Sakurai, A. (1997). Synthesis of new naturally occurring 6-deoxo brassinosteroids. J. Chem. Res. Synop. 134–135, Miniprint 901–924.
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Montagu, M.V., and Lusebettens, M.V. (1998). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.