Abstract
Although much is known about the signals and mechanisms that lead to pathogenic interactions between plants and fungi, comparatively little is known about fungus–plant mutualistic symbioses. We describe a novel role for reactive oxygen species (ROS) in regulating the mutualistic interaction between a clavicipitaceous fungal endophyte, Epichloë festucae, and its grass host, Lolium perenne. In wild-type associations, E. festucae grows systemically in intercellular spaces of leaves as infrequently branched hyphae parallel to the leaf axis. A screen to identify symbiotic genes isolated a fungal mutant that altered the interaction from mutualistic to antagonistic. This mutant has a single-copy plasmid insertion in the coding region of a NADPH oxidase gene, noxA. Plants infected with the noxA mutant lose apical dominance, become severely stunted, show precocious senescence, and eventually die. The fungal biomass in these associations is increased dramatically, with hyphae showing increased vacuolation. Deletion of a second NADPH oxidase gene, noxB, had no effect on the E. festucae–perennial ryegrass symbiosis. ROS accumulation was detected cytochemically in the endophyte extracellular matrix and at the interface between the extracellular matrix and host cell walls of meristematic tissue in wild-type but not in noxA mutant associations. These results demonstrate that fungal ROS production is critical in maintaining a mutualistic fungus–plant interaction.
INTRODUCTION
The ability to form mutualistic symbiotic associations with microorganisms is one of the most successful strategies that plants have evolved to adapt to the diverse range of biotic and abiotic stresses they encounter in terrestrial environments. The best documented of these symbioses are the nitrogen-fixing root–nodule associations between rhizobia and leguminous plants and the enhanced nutrient uptake (principally phosphate) associations between arbuscular mycorrhizal fungi (Glomeromycota) and a diverse range of monocotyledonous and dicotyledonous plants (Lodwig and Poole, 2003; Parniske, 2004). Less well known, but possibly equally important because of the associated bioprotective benefits, are the symbioses between filamentous fungi and plant aerial tissues (Scott, 2001; Clay and Schardl, 2002; Arnold et al., 2003; Clay, 2004; Schardl et al., 2004). Identification and characterization of the genes from each partner required to initiate and maintain these mutualistic associations are crucial to understanding how these microorganisms interact with their hosts. Recent studies have greatly advanced our understanding of how molecular signals are exchanged and perceived during the establishment of rhizobial and mycorrhizal plant root symbioses and how metabolite exchange occurs between partners for mutual benefit (Lodwig and Poole, 2003; Oldroyd and Downie, 2004; Akiyama et al., 2005; Karandashov and Bucher, 2005). By contrast, very little is known about the signaling mechanisms and biological processes that underlie the mutualistic symbiotic interactions between fungal endophytes and aerial tissues of plants.
Epichloë/Neotyphodium endophytes (Clavicipitaceae, Ascomycota) are biotrophic fungi that systemically colonize the intercellular spaces of leaf primordia, leaf sheaths and blades of tillers, and the inflorescence tissues of reproductive tillers to form symbiotic associations (symbiota) with temperate grasses of the subfamily Pooideae (Schardl, 2001; Scott, 2001; Clay and Schardl, 2002). Host benefits from the symbiosis include improved growth and persistence through improved nutrient acquisition and enhancement of plant tolerance to a range of biotic and abiotic stresses, including drought, disease, and animal herbivory. Endophyte benefits include access to nutrients from the plant apoplastic space and a mode of dissemination by vertical transmission through the seed (Scott, 2001; Schardl et al., 2004). Protection of the symbiotum from insect and small animal herbivory through endophyte synthesis of bioprotective secondary metabolites (alkaloids) is the best-studied benefit of this association (Bush et al., 1997; Lane et al., 2000). Genes for the biosynthesis of the four main classes of bioprotective alkaloids have now been cloned, including genes for the biosynthesis of ergot alkaloids (Panaccione et al., 2001; Wang et al., 2004), lolines (Spiering et al., 2002, 2005), peramine (Tanaka et al., 2005), and indole-diterpenes (Young et al., 2005). Expression studies have shown that the genes for all four pathways are preferentially and highly expressed in planta (Spiering et al., 2002; Tanaka et al., 2005; Young et al., 2005), suggesting that host-specific signaling is required for the induction of these pathways.
The growth of Epichloë endophytes within host grasses appears to be tightly regulated and synchronized with the growth of host leaves (Tan et al., 2001; Christensen et al., 2002). Hyphae of these endophytes in leaf sheaths are rarely branched, and mostly aligned parallel to the leaf axis. Growth of the hyphae is rapid in expanding leaves but ceases as elongation of the leaf stops. A striking feature of these symbiotic associations is the apparent lack of any host defense reaction to the presence of the endophyte. However, introduction of Epichloë endophytes into related nonnatural hosts frequently results in a host defense reaction that can vary in severity, from a high level of seedling mortality and stunting of the surviving plants (Christensen et al., 1997), to more localized responses such as the death of host cells in the stem apex (Christensen, 1995), to premature death of the hyphae (Koga et al., 1993). This loss of regulated endophyte growth and compatible interaction with the host in some novel associations reveals an underlying signaling mechanism between host and endophyte that maintains the mutualistic interaction. Two outcomes from this crosstalk must be achieved. First, there must be suppression or evasion of potential plant defense responses; second, there must be strict control of fungal growth throughout the host plant.
To identify these signaling mechanisms, we adopted a forward genetics approach. Specifically, we initiated plasmid and T-DNA insertional mutagenesis screens of Epichloë festucae to isolate mutants that are defective in their ability to either initiate or establish a mutualistic interaction with perennial ryegrass (Lolium perenne). In a first screen using restriction enzyme–mediated integration mutagenesis (Schiestl and Petes, 1991; Sánchez et al., 1998), we isolated an E. festucae noxA mutant that is unable to synchronize its growth in planta with that of the plant host. We demonstrate that this phenotype is caused by a defect in an NADPH oxidase that results in the formation of reactive oxygen species (ROS).
RESULTS
Isolation of an E. festucae Mutant That Causes Severe Stunting on Its Grass Host
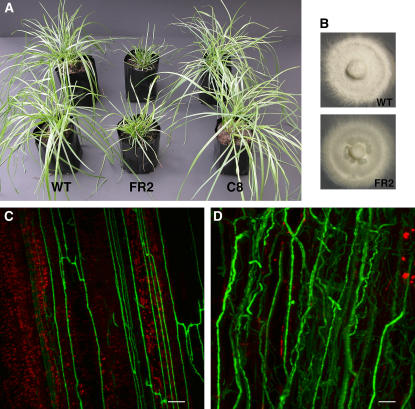
From a plant screen of 220 independent pAN7-1 plasmid insertion mutants of E. festucae strain Fl1, one mutant, designated FR2, was identified that disrupted the mutualistic symbiotic interaction with the perennial ryegrass host. In contrast with the normal growth of host plants infected with wild-type Fl1, perennial ryegrass infected with FR2 becomes severely stunted, has an increase in tiller number, shows precocious senescence, and eventually dies (Figure 1A; see Supplemental Figure 1 online). In axenic culture, the radial growth of FR2 was the same as that of Fl1, but there were fewer aerial hyphae (Figure 1B).
Figure 1.
Symbiotic and Axenic Culture Phenotypes of E. festucae FR2.
(A) Phenotypes of perennial ryegrass infected with E. festucae wild-type Fl1, symbiotic mutant FR2, and complemented strain C8. The photograph was taken 9 weeks after inoculation.
(B) Colony morphology of E. festucae wild-type Fl1 and symbiotic mutant FR2 on complete medium agar after 10 d.
(C) Confocal depth series image of hyphal growth of wild-type WG11 expressing GFP in the leaf sheath of perennial ryegrass. The photograph was taken 10 weeks after inoculation. Bar = 20 μm.
(D) Confocal depth series image of hyphal growth of symbiotic mutant FR2G6 expressing GFP in the leaf sheath of perennial ryegrass. The photograph was taken 10 weeks after inoculation. Bar = 20 μm.
To examine the differences in host colonization by wild-type strain Fl1 and mutant FR2 in living plant tissue, constitutively expressing green fluorescent protein (GFP) was introduced into Fl1 and FR2, by transforming pPN82 and pPN83, respectively. The GFP-expressing transformants WG11 and FR2G6 from Fl1 and FR2, respectively, were selected to infect perennial ryegrass seedlings, and the hyphal growth of these transformants in plants was observed by confocal microscopy. Plants infected with FR2G6 showed the typical stunted phenotype previously observed for FR2, whereas WG11 maintained a mutualistic interaction with the host plants, like Fl1. The hyphae of WG11 showed limited branching and were mostly orientated parallel to the longitudinal axis in the intercellular spaces of the leaf (Figure 1C). On the other hand, extensive hyphal colonization was observed in leaves infected with FR2G6. The hyphae of FR2G6 were numerous and occasionally convoluted, and the biomass of the fungus increased significantly compared with the wild type (Figure 1D). The hyphal number of FR2G6 increased in older leaves, whereas similar numbers of WG11 hyphae were present in both young and older leaves. This lack of regulated growth was also evident for hyphae of FR2 growing on the surface of the leaves, compared with the more restricted epiphyllous growth of hyphae in wild-type infected plants (see Supplemental Figure 2 online).
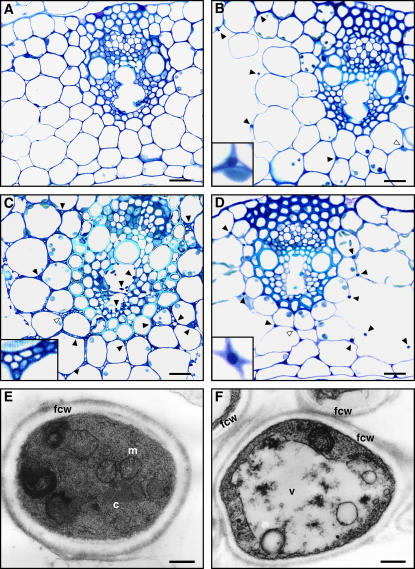
Light microscopic analysis of toluidine blue–stained cross sections of pseudostem tissue showed that large numbers of hyphae are present in older leaves of plants infected with the FR2 mutant, with extensive colonization of the vascular bundles, including both xylem and phloem (Figure 2C). In leaves infected with wild-type Fl1, fungal hyphae are much less frequent and rarely found in vascular bundles of both young and old plant tissue (Figure 2B). No hyphae were observed in uninfected plants (Figure 2A). In addition, wild-type hyphae were well stained compared with mutant hyphae (Figures 2B and 2C, insets), indicating that FR2 hyphae are relatively devoid of cytoplasm. Transmission electron microscopy analysis of hyphae in the outer leaf sheath revealed that mutant cells were frequently irregular in shape, contained large vacuoles, and lacked crystalline aggregations that are typically found in wild-type cells (Figures 2E and 2F).
Figure 2.
In Planta Phenotype of E. festucae Symbiotic Mutant.
(A) to (D) Light micrographs of transverse sections of the outer leaf of perennial ryegrass stained with toluidine blue. Hyphae of endophyte are indicated by arrowheads. Insets in (B) to (D) show higher magnification images of the endophyte hyphae indicated by open arrowheads in the main panels. Bars = 20 μm.
(A) Endophyte-free leaf of perennial ryegrass.
(B) Leaf infected with wild-type Fl1.
(C) Leaf infected with symbiotic mutant FR2.
(D) Leaf infected with complemented strain C8.
(E) and (F) Transmission electron micrographs of cross sections of endophyte hyphae in the intercellular space of the host plant. c, crystalline aggregation; fcw, fungal cell wall; m, mitochondrion; v, vacuole. Bars = 0.25 μm.
(E) Wild type.
(F) FR2 mutant.
Plant associations established between FR2 and tall fescue (Festuca arundinacea) and FR2 and meadow fescue (Festuca pratensis) also showed a stunted phenotype compared with that of plants infected with wild-type E. festucae, confirming that the altered symbiotic phenotype observed for this E. festucae mutant is not limited to one host (see Supplemental Figure 3 online).
Mutant FR2 Contains a Plasmid Insertion in an NADPH Oxidase Gene
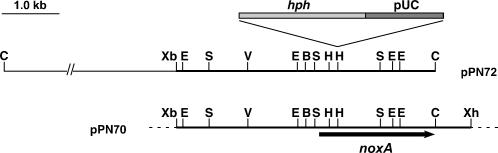
DNA gel blot analysis of genomic digests of FR2 probed with pAN7-1 indicated that the plasmid was present as a single copy contained within a 15-kb ClaI fragment (Figure 3). This fragment was recovered by plasmid rescue as pPN72, and the sequence of the E. festucae junctions was determined. Analysis of this sequence showed that pAN7-1 had disrupted an open reading frame encoding a protein with significant sequence similarity to gp91phox (otherwise known as NOX2), the transmembrane catalytic subunit of the mammalian NADPH oxidase that catalyzes the conversion of molecular oxygen to superoxide (Harper et al., 1985; Wallach and Segal, 1997; Lambeth, 2004). To isolate a genomic clone of noxA, a cosmid library of Fl1 was screened and eight positive clones, all of which mapped to a single genomic locus, were isolated. The sequence of a subclone, pPN73, of one of these cosmids, pPN70, was determined and shown to be identical to that of pPN72, confirming that there was no deletion of flanking genomic DNA at the pAN7-1 insertion site in FR2. These results indicate that the altered symbiotic phenotype of FR2 is caused by a mutation in the E. festucae noxA gene.
Figure 3.
Physical Maps of Plasmid-Tagged Mutant and Wild-Type Loci.
The pAN7-1–tagged locus in the FR2 mutant (pPN72) and the corresponding wild-type genomic region (pPN70) showing the plasmid insertion site in the mutant and restriction enzyme sites for BglII (B), ClaI (C), EcoRI (E), EcoRV (V), HindIII (H), SalI (S), XbaI (Xb), and XhoI (Xh). The arrow identifies the E. festucae noxA gene. Sequence analysis of the junctions between pAN7-1 and E. festucae noxA in FR2 showed that the HindIII site at the right junction of the plasmid and genomic DNA was lost.
E. festucae noxA Encodes a Homolog of gp91phox
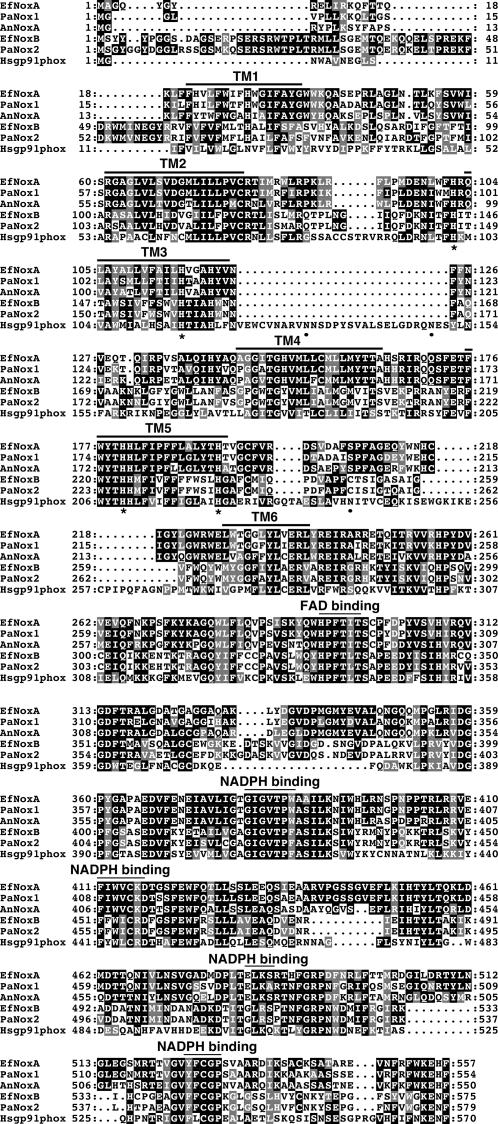
Sequence analysis of noxA predicts the presence of two introns that were confirmed by sequencing two independent cDNA clones generated by RT-PCR using RNA isolated from mycelium of wild-type Fl1 grown in axenic culture. NADPH oxidase A (NoxA) shares 82 and 72% amino acid identity with Podospora anserina Nox1 and Aspergillus nidulans NoxA, respectively (Figure 4) (Lara-Ortíz et al., 2003; Malagnac et al., 2004). The deduced polypeptide sequence of NoxA possesses features considered critical for NADPH oxidase function, namely six hydrophobic segments within the N-terminal region that are proposed to be membrane-spanning domains involved in transmembrane electron transport, as well as motifs corresponding to binding sites for heme, flavin adenine dinucleotide (FAD), and NADPH (Lambeth, 2004). This analysis predicts that NoxA is membrane-localized. The third and fifth hydrophobic segments each contains two conserved His residues (Figure 4), which are thought to serve as coordination sites for two heme moieties, based on analyses of human gp91phox and yeast ferric reductase (Finegold et al., 1996). Analysis of NoxA for potential N-glycosylation sites (N-X-S/T) (Kornfeld and Kornfeld, 1985) revealed a complete absence of this motif (Figure 4), in contrast with the presence of five such motifs in human gp91phox, three of which have been shown experimentally to be N-glycosylated (Harper et al., 1985; Wallach and Segal, 1997). Two of three motifs required for the interaction of human gp91phox with p47phox (87STRVRRQL, 451FEWFADLL, and 555ESGPRGVHFIF) (DeLeo et al., 1995) are absent in the fungal NADPH oxidases, consistent with the fact that no p47phox homolog can be identified within fungal genome databases.
Figure 4.
Alignment of the Predicted Amino Acid Sequences for E. festucae noxA and noxB with Related Proteins.
The amino acid sequences of E. festucae (Ef) NoxA and NoxB are aligned with those of human (Hs) gp91phox (Royer-Pokora et al., 1986), A. nidulans (An) NoxA (Lara-Ortíz et al., 2003), and P. anserina (Pa) Nox1 and Nox2 (Lalucque and Silar, 2003). Amino acids conserved between sequences are boxed in black (identical) or gray (conservative replacements). Six potential transmembrane-spanning domains (TM1 to TM6) and putative FAD and NADPH binding domains are indicated by lines above the aligned sequences. Conserved His residues, demonstrated to bind heme in yeast ferric reductase and human gp91phox (Finegold et al., 1996), are indicated by asterisks. Dots under the sequences indicate amino acid residues involved in the N-glycosylation of human gp91phox (Wallach and Segal, 1997).
E. festucae noxA Is Essential for Maintaining a Mutualistic Interaction with Perennial Ryegrass
To confirm that plasmid insertion in E. festucae noxA was responsible for the altered symbiotic phenotype observed for FR2, plasmid pPN74, containing the full-length noxA gene, was transformed into protoplasts of FR2. Among eight geneticin-resistant transformants analyzed by PCR and DNA gel blot analysis, three were shown to contain an intact copy of noxA. Two of these transformants (C5 and C8) were infected into perennial ryegrass seedlings, and the symbiotic phenotypes were compared with associations of infected wild-type and FR2 genes. In plants infected with the mutant FR2, stunting of the leaves and an increase in the number of tillers were obvious at 5 weeks after inoculation, whereas these changes were not observed in plants containing wild-type and transformant C5 and C8 during a 9-week period (Figure 1A; see Supplemental Figure 1 online). Light microscopic analysis of aniline blue–stained tissue showed that hyphal growth of C5 and C8 in planta was comparable to that of the wild type. Toluidine blue–stained cross sections of pseudostem tissue showed that the number of hyphae in plant leaves infected with C5 and C8 was very similar to that of the wild type. Hyphae had densely staining cytoplasm (Figure 2D, inset), and there was no colonization of the vascular bundle tissue (Figure 2D). Transmission electron microscopy analysis showed that hyphal cells of C5 and C8 have the same cytological features as the wild type (data not shown). These results indicated that pPN74, containing a full-length copy of noxA, restored the wild-type symbiotic phenotype with respect to host plant growth, hyphal distribution, and cellular morphology in host plants. These results confirm that the changes in symbiotic phenotype observed for FR2 are the result of disruption of E. festucae noxA.
Furthermore, we prepared a replacement construct, pPN75, and recombined a PCR-generated linear fragment of this plasmid into the genome of E. festucae strain Fl1 (Figure 5A). Potential noxA replacements were identified by PCR screening of hygromycin-resistant transformants for a noxA deletion. This screen identified two putative replacements, A17 and A44, out of 54 screened (3.7%). DNA gel blot analysis of genomic digests of the transformants probed with a noxA fragment (Figure 5B) confirmed that these transformants contained a replacement at the noxA locus. To determine the symbiotic phenotype of the mutants, perennial ryegrass seedlings were infected with A17, A44, FR2, or wild-type Fl1. Within 8 weeks after infection, all of the plants infected with A17 and A44 showed the same stunted phenotype observed with FR2, including an increased number of tillers and premature senescence of the host plants (Figure 5C). Light and transmission electron microscopy confirmed that these mutants had altered hyphal morphology and growth in the plants, similar to that observed for mutant FR2 (data not shown). Real-time PCR analysis demonstrated that the fungal biomass of mutant (A44) associations were significantly greater than that of wild-type associations (see Supplemental Figure 4 online) Together, these results clearly showed that mutation of noxA is responsible for the disrupted symbiotic interaction between E. festucae mutant FR2 and perennial ryegrass.
Figure 5.
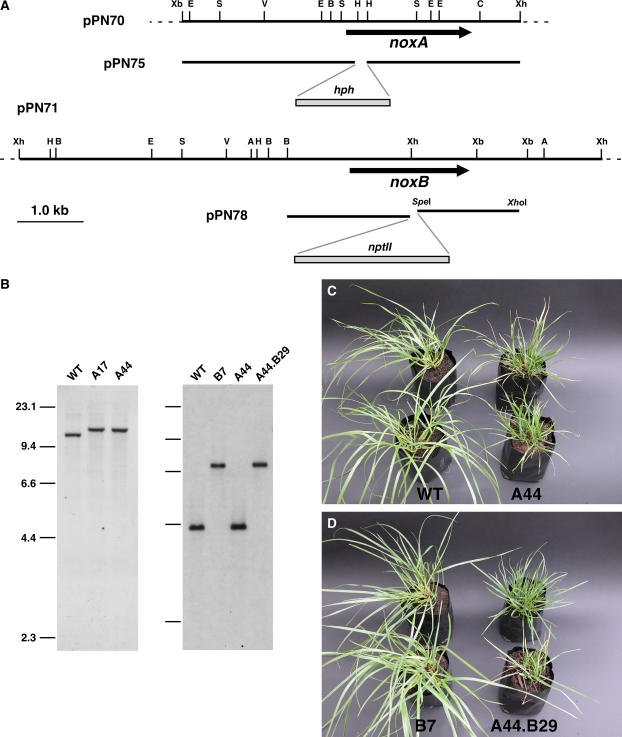
Deletion of E. festucae noxA and noxB.
(A) Physical map of the E. festucae noxA and noxB genomic regions, pPN70 and pPN71, and linear inserts of replacement constructs, pPN75 and pPN78, showing restriction enzyme sites for ApaI (A), BglII (B), ClaI (C), EcoRI (E), EcoRV (V), HindIII (H), SalI (S), XbaI (Xb), and XhoI (Xh).
(B) Autoradiographs of a DNA gel blot of SacII genomic digests (2 μg) of E. festucae wild-type strain Fl1 (WT) and noxA deletion strains A17 and A44 probed with digoxigenin-labeled noxA insert from pPN74 amplified with primers pII99-3 and pII99-4, and of a DNA gel blot of ApaI genomic digests of E. festucae wild-type strain Fl1 (WT), noxB deletion strain B7, noxA deletion strain A44, and noxA noxB double deletion strain A44.B29 probed with digoxigenin-labeled noxB insert from pPN71 amplified with primers nox2a and nox2d. Sizes in kilobases of HindIII-digested λDNA markers are indicated at left.
(C) and (D) Phenotypes of perennial ryegrass plants infected with E. festucae wild-type Fl1 and noxA deletion strain A44 (C) and noxB deletion strain B7 and noxA noxB double deletion strain A44.B29 (D). The photographs were taken 9 weeks after inoculation.
Isolation of E. festucae noxB
With the exception of the Aspergillus species, at least two nox homologs, noxA (Nox1) and noxB (Nox2), can be identified in available ascomycete fungal genome databases (Lara-Ortíz et al., 2003; Malagnac et al., 2004; Aguirre et al., 2005). To isolate the E. festucae noxB homolog, primers were designed to conserved sequences of fungal noxB genes, and a fragment of ∼1.2 kb was amplified by PCR from genomic DNA of Fl1. The deduced amino acid sequence of this fragment showed 83.5% identity to the corresponding region of the P. anserina Nox2 sequence (Malagnac et al., 2004), confirming that the fragment amplified was the E. festucae noxB homolog. Using this PCR product as a probe, three clones containing overlapping cosmids were isolated from an Fl1 cosmid library. The full-length sequence of noxB was generated from subclones pPN76 and pPN77 of cosmid pPN71. Sequence analysis of noxB predicts the presence of two introns that were confirmed by cDNA analysis. NoxB shares 84.6 and 83% identity with P. anserina Nox2 and Neurospora crassa Nox-2 polypeptide sequences, respectively (Malagnac et al., 2004) (Figure 4). E. festucae NoxA and NoxB share 32.6% identity. NoxB contains the same conserved motifs found in NoxA, plus 50 extra amino acids at the N terminus of the protein that are conserved with P. anserina Nox2 (Figure 4).
E. festucae noxB Is Not Required to Maintain a Mutualistic Interaction with Perennial Ryegrass
To investigate the biological role of noxB, a replacement construct, pPN78, was prepared, and a PCR-generated linear fragment (6.3 kb) of this plasmid (Figure 5A) was recombined into the genome of Fl1 and the noxA deletion mutant A44. PCR screening identified transformants B7 and A44.B29 from the Fl1 and A44 backgrounds, respectively, that had patterns consistent with targeted replacement events. DNA gel blot analysis of genomic digests of these transformants probed with a noxB fragment confirmed that the transformants contained a replacement at the noxB locus (Figure 5B).
Plants infected with A44.B29 (ΔnoxA ΔnoxB) had the stunted phenotype identical to that of A44 (ΔnoxA) (Figures 5C and 5D). However, plants infected with B7 (ΔnoxB) were identical to those infected with wild-type Fl1 (Figure 5D). Light microscopic analysis of plant material stained with aniline blue confirmed that the hyphal growth and morphology of B7 in planta were comparable to those of Fl1. These results showed that E. festucae NoxB is dispensable for the maintenance of the mutualistic association between E. festucae and perennial ryegrass. Moreover, there was no detectable difference in growth between wild-type Fl1 and B7 in axenic culture.
E. festucae noxA Is Preferentially Expressed in Planta
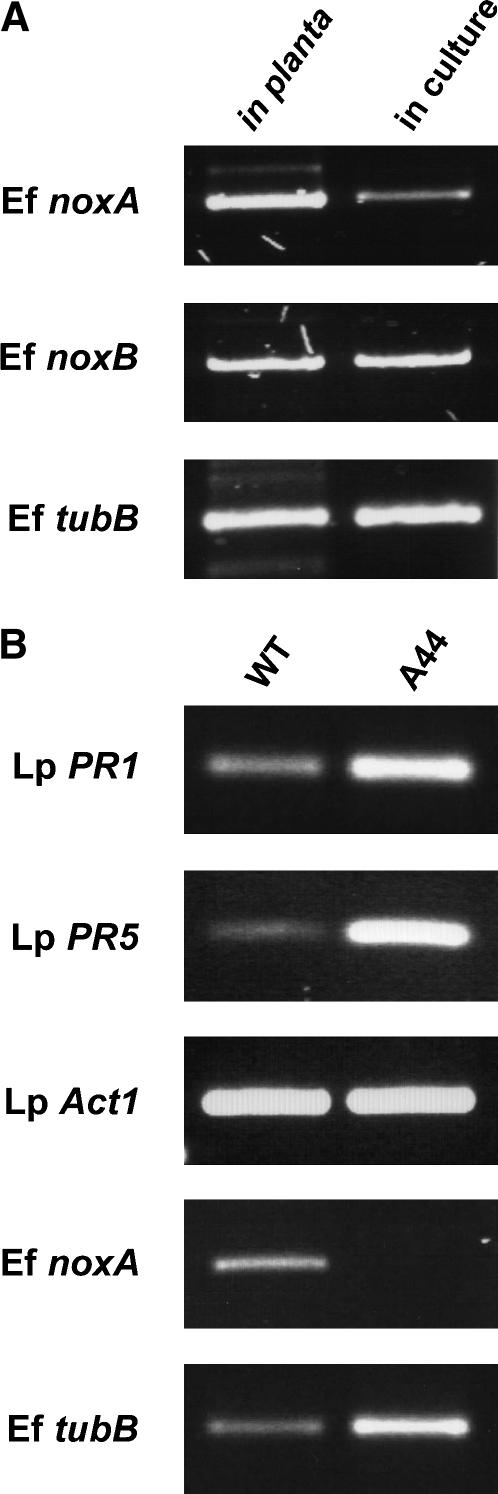
To determine the expression level of nox genes in axenic culture and in host plants, the amount of endophyte mRNA in planta was first estimated by comparing the expression levels of Fl1 β-tubulin with that in axenic culture (Figure 6). This experiment showed that the level of β-tubulin transcript in planta was ∼1/80th the level in mycelia (Figure 6A), a result consistent with estimates of endophyte biomass in perennial ryegrass of between 0.4 and 1.9% (Panaccione et al., 2001; Young et al., 2005). Semiquantitative RT-PCR expression analysis showed that noxA is weakly expressed in axenic culture but strongly expressed in planta (Figure 6A). By contrast, expression of noxB in culture is comparable to that in planta.
Figure 6.
Expression Analysis of Endophyte and Plant Genes.
(A) Expression analysis of E. festucae nox genes. Total RNA was isolated from pseudostems of Fl1-infected perennial ryegrass (in planta) or from mycelia of Fl1 grown in axenic culture on potato dextrose medium (in culture) and used for cDNA synthesis. RT-PCR was performed with primers specific for E. festucae noxA (Ef noxA), noxB (Ef noxB), and β-tubulin (Ef tubB). To compensate for the differences in endophyte biomass in planta versus that in culture, a 1:80 dilution of the Fl1 cDNA was used compared with undiluted pseudostem template.
(B) Expression analysis of L. perenne PR genes in host plant infected with Fl1 or the noxA mutant. Total RNA was isolated from emerging leaf of Fl1-infected wild-type (WT) or noxA mutant–infected (A44) perennial ryegrass. Expression of L. perenne actin (Lp Act1), PR1 (Lp PR1), and PR5 (Lp PR5) and E. festucae noxA (Ef noxA) and β-tubulin (Ef tubB) were analyzed by RT-PCR with specific primers.
Host Plant Response to E. festucae noxA Mutant
To investigate the host response to the noxA mutant, expression of the pathogenesis-related (PR) genes PR1 and PR5 was determined in emerging leaf tissue of perennial ryegrass infected with wild-type Fl1 or the noxA mutant A44. RT-PCR analysis showed that, in contrast with Actin1, both PR1 and PR5 were more highly expressed in plant tissue infected with the noxA mutant than with wild-type Fl1 (Figure 6B). However, the A44–perennial ryegrass association was known to have a greater biomass of endophyte compared with the wild-type association, and this is reflected by the difference in expression levels of the fungal β-tubulin gene (Figure 6B). Therefore, the increased expression of the PR genes in tissue of the A44 association is more likely to reflect the greater number of plant cells responding to the endophyte hyphae rather than an increase in expression at the single-cell level per se.
To investigate whether the noxA mutant elicited a hypersensitive response, plant tissue infected with A44 or with wild-type Fl1 was stained with lactophenol trypan blue to detect localized cell death (Koch and Slusarenko, 1990). Although clusters of dead plant cells indicative of early senescence were found in older plant tissues infected with the noxA mutant, there were no indications of hypersensitive response–induced cell death in young pseudostem tissue for both mutant and wild-type associations (see Supplemental Figure 5 online).
E. festucae noxA Is Involved in ROS Production in Culture and in Host Plants
Mycelial samples of the E. festucae nox mutants grown in axenic culture were examined for their ability to generate superoxide by reduction of nitroblue tetrazolium (NBT), which is reduced by superoxide radicals to a dark-blue, water-insoluble formazan (Munkres, 1990). Within 4 to 5 h after NBT treatment, hyphae of wild-type E. festucae were clearly stained with dark-blue formazan precipitates (Figure 7A). Microscopic observation revealed that only some apical and subapical cells were stained, and in both cases the NBT-reactive blue precipitates were localized to the tip-proximal end of each cell (Figures 7B and 7C). Treatment of wild-type mycelia with diphenylene iodonium, an inhibitor of NADPH oxidase and other FAD-requiring enzymes, prevented reduction of the NBT, confirming that the reduction of NBT was mediated by an NADPH oxidase. Mycelia of both the noxA (A44) and noxB (B7) mutants showed reductions in the intensity of NBT staining compared with the wild type, as did the noxA noxB double mutant A44.B29, which was even further reduced in intensity than the single mutants (Figure 7A). These results suggest that both NoxA and NoxB catalyze ROS production in axenic culture. The weak NBT staining of the double mutant indicates that either there is a third nox gene in E. festucae or the reduction is catalyzed by an alternative oxidase.
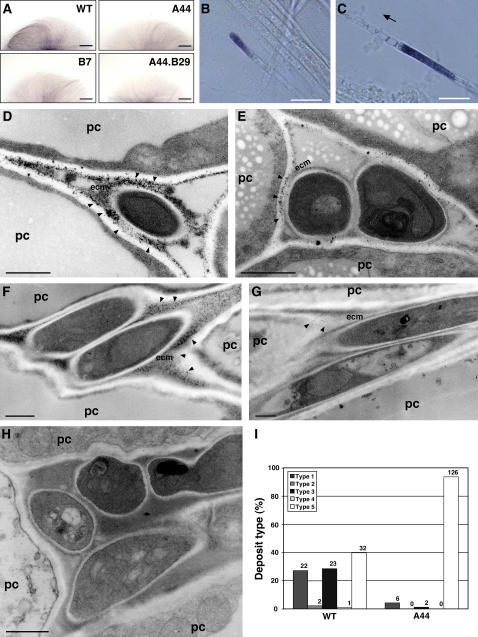
Figure 7.
ROS Production by E. festucae Wild Type and nox Mutants.
(A) Detection of superoxide production by NBT staining. Photographs of colony mycelia of E. festucae wild-type Fl1 (WT), noxA mutant (A44), noxB mutant (B7), and noxA noxB double mutant (A44.B29) grown on potato dextrose agar medium and stained with 0.05% (w/v) NBT solution for 5 h. Bars = 2 mm.
(B) and (C) Higher magnification light micrographs of hyphae in (A) showing localization of reduced NBT at the hyphal tip (B) and in a subapical cell at the end proximal to the growing tip (C). The direction of the hyphal growing tip in (C) is indicated by the arrow. Bars = 10 μm.
(D) to (H) Transmission electron micrographs of H2O2 localization in meristematic tissue of perennial ryegrass infected with E. festucae wild-type Fl1 (D) to (F) or noxA mutant A44 (G) and (H). Cerium chloride–reactive deposits are indicated by arrowheads. ecm, extracellular matrix; pc, plant cell. Bars = 1 μm.
(I) Distribution of five different types of cerium chloride–reactive deposits showing H2O2 localization in meristematic tissue of perennial ryegrass infected with E. festucae wild-type Fl1 (WT) or noxA mutant (A44). Type 1, cerium chloride–reactive deposits in the extracellular matrix of endophyte; type 2, cerium chloride–reactive deposits in host cell walls; type 3, cerium chloride–reactive deposits in both endophyte extracellular matrix and host cell walls; type 4, cerium chloride–reactive deposits in plasma membrane of endophyte; type 5, no deposit detected. The number of fungal cells of each particular type is given above each column.
The production of ROS, more specifically H2O2, by endophyte in the host plant was detected cytochemically using electron microscopy to locate the deposition of electron-dense cerium perhydroxides, formed by the reaction of cerium ions with H2O2 (Briggs et al., 1975; Bestwick et al., 1997; Shinogi et al., 2003). Cerium-reactive deposits were observed predominantly in the endophyte-produced extracellular matrix (ECM), surrounding the hyphal cell walls, and at the interface between the ECM and the host cell walls of pseudostem tissue infected with wild-type Fl1 and the noxA mutant, but no significant differences were observed in the type or pattern of deposits formed between mutant and wild-type associations (data not shown). By contrast, examination of the meristematic region of the host plant revealed the presence of a range of different types of cerium-reactive deposits in wild-type associations that were rarely found in noxA associations (Figure 7). The deposits found in wild-type cells were localized predominantly in the ECM surrounding the fungal cell walls and at the interface between the ECM and plant cell walls (Figures 7D to 7F). By contrast, reactive deposits were rarely observed in meristematic tissues containing the noxA mutant (Figures 7G and 7H). Analysis of the distribution patterns of cerium-reactive deposits observed in meristematic tissues infected with either wild-type Fl1 or the noxA mutant showed that the majority (95%) of the cells examined in tissue containing the noxA mutant were devoid of deposits, whereas ∼60% of the images examined for the Fl1 wild-type interaction showed some type of cerium-reactive deposit in the ECM and at the interface between the ECM and plant cell walls (Figure 7I). No deposits were detected on the fungal cell walls. These results suggest that endophyte NoxA is largely responsible for the production of ROS in the host meristematic tissue.
DISCUSSION
This work identifies a novel role for ROS in regulating the mutualistic interaction between a fungal endophyte, E. festucae, and its grass host, perennial ryegrass. Plants infected with an E. festucae NADPH oxidase (noxA) mutant lose apical dominance, become severely stunted, show precocious senescence, and eventually die. This antagonistic interaction with the host is accompanied by a dramatic increase in endophyte biomass within the plant compared with that in the wild type. Deletion of noxB, which encodes a second NADPH oxidase isoform, had no effect on the ability of E. festucae to colonize and establish a mutualistic symbiotic association with perennial ryegrass, indicating a specific role for the NoxA isoform in the symbiosis.
NADPH oxidase is an enzyme that catalyzes the production of superoxide by transferring electrons from NADPH to molecular oxygen, with secondary generation of H2O2. Seven mammalian NADPH oxidase enzymes have been identified and characterized. They all contain the core NADPH oxidase domains, including a six-transmembrane region, a FAD binding motif, and four NADPH binding motifs; in addition, however, they also contain unique domains that divide these enzymes into three distinct classes (Lambeth, 2004). The most well-studied member of this family is gp91phox (NOX2), which is responsible for high-level production of superoxide in phagocytic cells in response to microbial invasion. More recently, NOX1, NOX3, and NOX4, close homologs of gp91phox, were shown to be expressed in nonphagocytic cells. These NADPH oxidase enzymes generate low levels of superoxide that activate signal transduction pathways controlling cell proliferation and modification of the ECM (Suh et al., 1999; Lambeth et al., 2000). The NADPH oxidase homologs in plants, designated the Rboh (for respiratory burst oxidase homolog) family of enzymes, are structurally most similar to the mammalian calcium-regulated NADPH oxidase, NOX5, which has an N-terminal calcium binding EF-hand motif (Torres et al., 1998). NADPH-dependent superoxide generation by plants in response to microbial pathogen colonization is a well-known plant defense mechanism (Doke, 1983, 1985; Lamb and Dixon, 1997), and recent studies have shown that activation of particular Rboh isoforms is responsible for ROS accumulation in several plant–microbe interactions (Simon-Plas et al., 2002; Torres et al., 2002, 2005; Yoshioka et al., 2003). Arabidopsis thaliana possesses 10 Rboh isoforms, which have been shown by genetic analysis to be involved in a diverse range of plant cell processes. ROS production by the gene product encoded by ROOT HAIR DEFECTIVE2/RBOHC of Arabidopsis plays a key role in the regulation of root hair growth through the activation of Ca2+ channels (Foreman et al., 2003). Arabidopsis RBOHD and RBOHF have been shown to play a role in ROS production in response to avirulence pathogens (Torres et al., 2002), but a more recent report demonstrated that ROS production by these enzymes suppresses the spread of cell death by antagonizing salicylic acid–dependent pro-death signals (Torres et al., 2005). Arabidopsis RBOHD and RBOHF also regulate stomatal closure, seed germination, and root elongation through abscisic acid signaling (Kwak et al., 2003). These reports demonstrate that ROS production by NADPH oxidases can serve various roles in mammalian and plant systems, including host defense and as a signal for developmental transitions.
In contrast with plants and mammals, filamentous fungi generally have just two NADPH oxidase isoforms (Lalucque and Silar, 2003). The fungal NoxA and NoxB have all of the core structural domains found in the animal gp91phox but appear to lack any additional fungus-specific domains. Two plant pathogenic fungi, Fusarium graminearum and Magnaporthe grisea, possess a third nox gene, noxC, which encodes a protein with an extra N-terminal calcium binding motif, similar to that found in human NOX5 and plant Rbohs (Aguirre et al., 2005). Functional analysis of A. nidulans noxA and the P. anserina homolog, Nox1, showed that the NADPH oxidase encoded by this gene is essential in the differentiation of sexual fruiting bodies from their progenitor cells (Lara-Ortíz et al., 2003; Malagnac et al., 2004). In addition to the block in sexual development, the P. anserina Nox1 mutants were defective in aerial hyphal formation and mycelial pigmentation, additional differentiations that characterize the vegetative-to-stationary phase transition in this organism. Disruption of noxA in A. nidulans had no effect on asexual hyphal growth (Lara-Ortíz et al., 2003).
Inactivation of E. festucae noxA changes the interaction of endophyte with its host from mutualistic to antagonistic. Results to date suggest that E. festucae grows predominantly by tip growth within the leaf and axillary bud primordia (Tan et al., 2001), but after colonization of the leaf expansion zone, the hyphae further elongate by intercalary extension (M.J. Christensen, unpublished data). This proposed pattern of growth would allow the endophyte to recapitulate the growth pattern of the plant and avoid mechanical shear as the leaf cells are displaced away from the leaf expansion zone. Based on this model, hyphal cells that leave the leaf expansion zone will be fully mature. In young emerging leaves of perennial ryegrass infected with the E. festucae noxA mutant, hyphal growth and morphology were very similar to those of the wild type, although the fungal biomass was much greater. In older leaves, most of the hyphae had unusual branching and the cells were highly vacuolated, a morphology indicative of tip growth (Grove et al., 1970; Barelle et al., 2003), but some hyphae were similar in growth pattern and morphology to those found in younger leaves. Using cerium chloride to detect H2O2 accumulation, reactive deposits were found in the endophyte ECM and at the interface of the ECM and the host cell walls of meristematic tissue infected with the wild-type gene but were absent from tissue infected with the noxA mutant. By contrast, no difference was observed in the frequency or type of cerium-reactive deposits found between the wild type and the mutant in pseudostem tissue, indicating that there is compensatory production of ROS from either the endophyte or the plant in the mutant associations.
These results highlight a fundamental difference in the steady state levels of ROS in meristematic tissue versus pseudostem that is NoxA-dependent. In axenic culture, in which fungal hyphae grow by tip growth, we detected ROS production in some apical and subapical cells at the leading edge of the colony. The intensity of the reduced NBT formazan deposits was greatest on the end of the cell proximal to the hyphal tip. On the distal side of the growing colony edge, the cells were highly vacuolated, a morphology characteristic of colony-polarized tip growth. Based on these observations, we propose that the ROS produced by E. festucae NoxA in planta negatively regulates hyphal tip growth, thereby preventing excessive colonization of the leaf and axillary bud primordial tissue in the basal meristem as well as inhibiting tip growth of the hypha in mature leaf tissue that has ceased expanding. By contrast, NoxA positively regulates the differentiation of aerial hyphae in axenic culture, as loss of this activity results in hyphal colonies that are less fluffy than wild-type colonies, a phenotype similar to that observed for the Nox1 mutant of P. anserina (Malagnac et al., 2004). Despite these contrasting phenotypes, there may be some consistency in the underlying regulatory mechanisms if the primary role of NoxA is the regulation of developmental transitions, with the type of growth transition dependent on the environment in which the fungus is growing (Aguirre et al., 2005).
E. festucae, like most other filamentous fungi, has a second nox gene, noxB, the gene product of which has an extended N terminus compared with NoxA. Disruption of E. festucae noxB had no distinguishable effect on intercellular growth, biomass, and branching in planta and on mycelial growth in axenic culture. Moreover, the noxB mutation had no additional or modifying effect on the phenotype conferred by noxA in planta. These results indicate that NoxA and NoxB have distinct functional roles in E. festucae, as has been shown for P. anserina, in which Nox1 is required for sexual development and Nox2 is required for ascospore germination (Malagnac et al., 2004). We have yet to test whether E. festucae noxB also functions in ascospore germination, because the sexual cycle in this fungus is only initiated under field conditions, in which an external stromata forms on the flag leaf surrounding the developing inflorescence during reproductive growth of the host (Leuchtmann et al., 1994).
Expression of E. festucae noxA in planta was greater than that in axenic culture, a result consistent with the critical role established for NoxA in the symbiotic interaction. By contrast, noxB expression in planta was similar to that in axenic culture, and at a level comparable to that of noxA in the host plant. However, disruption of noxB had no detectable effect on either symbiotic or vegetative growth, a result not unlike that for Nox2 in P. anserina, in which expression was shown to be constitutive in vegetative hyphae yet disruption of the gene had no effect on mycelial growth (Malagnac et al., 2004). Direct detection of ROS by NBT staining of noxB mycelia showed a reduction in superoxide production over the wild type that was comparable to that of the noxA mutant, despite the higher levels of expression of the noxB gene in culture compared with noxA. These results suggest that production of ROS by NADPH oxidase proteins is regulated at the posttranscriptional level, a result consistent with known posttranslational mechanisms for the activation of mammalian NADPH oxidases (Lambeth, 2004).
As discussed above, the phenotype observed for the E. festucae noxA mutant in planta is exceptional, compared with the culture phenotypes observed for nox mutants from other fungi, because development is enhanced rather than impaired. Because plant-generated superoxide in response to pathogen attack is considered to be a crucial signaling mechanism in plant defense, it is also possible that ROS produced by the endophyte induces a moderate plant defense response, resulting in the suppression of endophyte growth in the host plant. However, in the E. festucae–ryegrass interaction, we did not find evidence for an endophyte-induced hypersensitive response in the host tissue. Although expression levels of perennial ryegrass PR1 and PR5 were greater in noxA associations, this most likely reflected the increase in hyphal biomass rather than any specific increase in expression at the level of a single cell. Plant cell death was not detected in young plant tissue infected with wild-type E. festucae or the noxA mutant. Together, these results indicate that there is no difference between the noxA mutant and the wild type in the localized defense response with the host. Therefore, the plant phenotype observed for the noxA mutant interaction is more likely to be a consequence of either the increased growth of the endophyte in the host plant or an alteration in host signaling that is not expressed as a localized host defense response.
Perennial ryegrass infected with the noxA mutant showed severe stunting and increased tiller number. This phenotype could result from an imbalance in hormone levels in the shoot apical meristem leading to decreased apical dominance (Shimizu-Sato and Mori, 2001). Physiological studies have shown that two phytohormones, cytokinin and auxin, are mainly responsible for the regulation of apical dominance through positive and negative control of axillary bud growth, respectively (Taiz and Zeiger, 1998). One explanation for the increase in tiller number observed in this study could be a loss of apical dominance from higher levels of cytokinins in plant meristematic tissue as a consequence of the greater fungal biomass.
We propose that the primary role of ROS produced by E. festucae NoxA is to control fungal growth in planta. This could be achieved either by regulation of a fungal signaling pathway that controls hyphal growth or by a direct effect of the ROS, such as cross-linking of fungal cell walls and/or linking of fungal cell walls to plant cell walls. Whatever the mechanism, the regulation of NoxA is clearly under the control of signal(s) from the host plant. In mammalian phagocytic cells, activation of gp91phox (NOX2) is triggered by the recognition of microorganism or inflammatory mediators, and subsequent signaling events through protein phosphorylation and lipid metabolism result in the recruitment and assembly of cytosolic p47phox, p40phox, p67phox, and Rac-GTP with the membrane-associated gp91phox and p22phox to form a ROS-producing active complex (Vignais, 2002; Lambeth, 2004). Some of these regulatory proteins are shared with the NOX1 and NOX3 complexes (Cheng et al., 2004; Lambeth, 2004). Because fungal NoxA and NoxB proteins are structurally similar to human NOX1 to NOX4, it is very likely that the E. festucae NADPH oxidase proteins will have regulatory components to control NADPH oxidase activity, even though homologs of the phox regulatory proteins have yet to be reported in filamentous fungi. The small GTP binding protein Rac has been identified in several fungi (Weinzierl et al., 2002; Boyce et al., 2003; Chen and Dickman, 2004), and the Rac homolog from Colletotrichum trifolii was shown to be a regulator of ROS generation and hyphal growth and development (Chen and Dickman, 2004). Rac homologs also play a key regulatory role in ROS production in plant systems (Kawasaki et al., 1999). Therefore, the E. festucae Rac homolog would be a good target in which to further investigate the regulation of Nox.
In summary, we describe here a novel role for ROS in regulating the growth of a mutualistic endophyte within its plant host. It will be of considerable interest to determine whether ROS production is a mechanism used by other symbiotic fungi, and possibly biotrophic pathogens, to control their growth and development in their host plants and whether this mechanism differs in the necrotrophic pathogens.
METHODS
Fungal Strains and Growth Conditions
Cultures of Epichloë festucae (see Supplemental Table 1 online) were grown on 2.4% potato dextrose agar or complete medium (0.1% yeast extract, 0.1% peptone, 1% KNO3, 0.5% KH2PO4, 0.25% MgSO4·7H2O, 0.002% FeCl3, 1% glucose, and 1.5% agar) (Sanderson and Srb, 1965) under conditions described previously (Moon et al., 1999, 2000). Cultures of Fl1 used for RT-PCR analysis were grown in 2.4% potato dextrose broth. YPS medium contained 0.1% yeast extract, 0.1% peptone, and 1 M sucrose. The naming of E. festucae and perennial ryegrass (Lolium perenne) genes and gene products follows the Aspergillus nidulans (Bennett and Lasure, 1985) and Oryza sativa (Wu et al., 1991) conventions, respectively. The names of genes from other organisms follow the conventions for those organisms. The term “epichloe” is used throughout as a general reference to endophytes of the Epichloë (sexual) and Neotyphodium (asexual) species in accordance with the proposal by Schardl et al. (2004).
Plant Growth and Endophyte Inoculation Conditions
Inoculation of endophyte-free seedlings of perennial ryegrass, tall fescue (Festuca arundinacea), and meadow fescue (Festuca pratensis) with E. festucae was performed by the method of Latch and Christensen (1985). Plants were grown and tested for the presence of endophyte as described previously (Tanaka et al., 2005).
DNA Preparations, Hybridizations, and PCR
Fungal genomic DNA was isolated from freeze-dried mycelium by the method of Byrd et al. (1990) or using the Extract-N-Amp plant PCR kit (Sigma-Aldrich) according to the manufacturer's instructions. Plasmid and cosmid DNA was isolated by alkaline lysis and purified by polyethylene glycol precipitation (Sambrook et al., 1989). Genomic digests were transferred to positively charged nylon membranes (Roche) by capillary transfer and fixed by UV light cross-linking in a Cex-800 UV light cross-linker (Ultra-Lum) at 254 nm for 2 min. Filters were probed with digoxigenin-11-dUTP–labeled probes (Roche). DNA was labeled by PCR incorporation of digoxigenin-11-dUTP. Hybridizations and light detection were performed according to the manufacturer's instructions.
Standard PCR amplifications of genomic, plasmid, and cosmid DNA templates were performed as described previously (Tanaka et al., 2005). Sequences of PCR primers used in this study are provided in Supplemental Table 2 online.
Real-time quantitative PCR determination of endophyte DNA biomass in perennial ryegrass associations infected with Fl1 wild type or the A44 ΔnoxA mutant was performed using primers endo1 and endo2, as described previously (Young et al., 2005).
Cosmids pPN70 (noxA) and pPN71 (noxB) were isolated from an Fl1 cosmid library (Tanaka et al., 2005) by colony hybridization using probes amplified with primers noxAf and noxAr (344 bp, from pPN72) and nox2a and nox2d (1.2 kb, from Fl1 genomic DNA). The latter were unique primers designed to conserved DNA regions identified from an alignment of noxB sequences from Fusarium graminearum (accession number EAA75161), Magnaporthe grisea (EAA56588), Neurospora crassa (XP_331064), and Podospora anserina (AAQ74977).
Preparation of RNA, and RT-PCR Analysis
Total RNA was isolated from frozen mycelium or perennial ryegrass pseudostem tissue using TRIzol reagent (Invitrogen) and reverse-transcribed as described previously (Tanaka et al., 2005). Gene-specific amplifications of cDNA were performed under the same conditions as for standard PCR described above except that the cycle number was 29. The gene-specific primer (see Supplemental Table 2 online) combinations used for expression analysis were as follows: T1.1 and T1.2 (β-tubulin), noxAr2 and noxAr (noxA), nox2e and nox2j (noxB), Lp-actin-F and Lp-actin-R (Actin1), LpPR1-F and LpPR1-R (PR1), and LpPR5-F and LpPR5-R (PR5). For intron analysis, the cDNA was amplified with the following primer pairs (see Supplemental Table 2 online): for noxA, noxAf2 and efnox1b (introns 1 and 2); for noxB, nox2e and efnox2u (introns 1 and 2).
Preparation of Enhanced GFP Vector
Plasmid pPN82 (9.7 kb), for the constitutive expression of GFP, was prepared by sequentially ligating into pBluescriptII KS+ a 0.7-kb BamHI/SalI fragment of enhanced GFP (EGFP) from pEGFP (Clontech), a 1.4-kb HindIII fragment of hph under the control of the trpC promoter from pCB1004 (Carroll et al., 1994), a 2.3-kb EcoRI/NcoI fragment of gpd promoter from pAN7-1 (Punt et al., 1987), and a 0.6-kb NotI/SacII fragment of trpC terminator from pII99 (Namiki et al., 2001). The gpd promoter fragment was prepared by digesting a PCR product amplified with primer set M13-reverse and pgpdNc using pAN7-1 as template. The fragment of the trpC terminator was prepared by digesting a PCR product amplified with primer set TnotI and Tsc2 using pII99 as template.
Preparation of Deletion and Complementation Constructs
Plasmid pPN74 was prepared by cloning a 5.1-kb XbaI/XhoI fragment containing the full-length noxA, together with 2.5 kb 5′ and 0.8 kb 3′ of noxA, from pPN73 into pII99 (Figure 5A). Plasmid pPN75 was prepared by replacing a 0.1-kb HindIII fragment in pPN73 with a 1.4-kb HindIII fragment of hph under the control of the trpC promoter from pPN1686, as shown in Figure 5A. Plasmid pPN78 was prepared by sequentially ligating into pII99 a 1.8-kb BamHI/EcoRI fragment 5′ of noxB from pPN76 and a 1.2-kb SpeI/XhoI fragment 3′ of noxB, as shown in Figure 5A. The SpeI/XhoI fragment was prepared by digesting a PCR product amplified with primer set Xh2.8Spe and Sm1.2hr using pPN71 as template. The primers Xh2.8Spe and Sm1.2hr contained mismatches relative to the genomic sequence to introduce the appropriate enzyme recognition site (see Supplemental Table 2 online). PCR amplification used for preparing pPN78 was performed in a 50-μL reaction volume containing 1 ng of the template DNA, 5 μL of 10× reaction buffer (Roche), each primer at 300 nM, each deoxynucleotide triphosphate at 200 μM, and 1.75 units of Expand Hi Fidelity polymerase (Roche). The thermocycle conditions used were as follows: one cycle at 94°C for 2 min; 35 cycles at 94°C for 15 s, 60°C for 30 s, and 68°C for 4 min; and 68°C for 7 min.
The 6.4- and 6.3-kb linear products of pPN75 and pPN78 used for transformation were amplified using Expand Long Template polymerase (Roche) as described above except that each deoxynucleotide triphosphate was at 350 μM, each primer (M13F and M13R) was at 400 nM, and the thermocycle conditions were as follows: one cycle at 93°C for 1 min; 30 cycles at 93°C for 10 s, 60°C for 30 s (with a 20-s incremental increase per cycle after cycle 11), and 68°C for 4.5 min; and 68°C for 10 min.
E. festucae Transformation
Protoplasts of E. festucae were prepared as described previously (Young et al., 1998, 2005). Protoplasts were transformed with 5 μg of either circular (pPN74 and pPN82) or linear PCR-amplified product (primers M13F and M13R for pPN75, pII99-2 and pII99-3 for pPN78) or cotransformed with 5 μg of circular pII99 and 10 μg of pPN83 using the method described previously (Itoh et al., 1994). Putative pPN75 (noxA) replacements were initially screened by PCR using primers noxAf2 and noxAr2 (890 bp wild type, 2.1 kb pPN75), which are within the noxA region deleted. Putative pPN78 (noxB) replacements were screened by PCR using primers that flank nptII (nox2e and nox2j; 1.2 kb wild-type, 3.9 kb replacement) and the 5′ (nox2l and pII99-1; 2.0 kb) and 3′ (pII99-4 and nox2k; 1.6 kb) flanking regions of the replacement.
For restriction enzyme–mediated integration mutagenesis, protoplasts were transformed with 5 μg of HindIII-linearized pAN7-1, plus 20 units of HindIII added to the plasmid/protoplast mixture, using the method described previously (Itoh et al., 1994). Transformants were selected on YPS medium containing either hygromycin (150 μg/mL) or geneticin (200 μg/mL). The resulting transformants were nuclear-purified by two rounds of subculturing of mycelium. Transformants expressing EGFP were selected under UV light using a BX51 stereomicroscope (Olympus).
DNA Sequencing and Bioinformatics
DNA fragments were sequenced by the dideoxynucleotide chain-termination method using Big-Dye (version 3) chemistry (Applied BioSystems). Products were separated on an ABI 3730 analyzer (Applied Biosystems). Sequence data were assembled into contigs with SEQUENCHER version 4.1 (Gene Codes) and analyzed and annotated as described previously (Tanaka et al., 2005).
Microscopy
Small leaf pieces, 1- to 2-mm thick, from endophyte-infected plant tissues were fixed in 3% glutaraldehyde and 2% formaldehyde in 0.1 M phosphate buffer, pH 7.2, and then prepared for light microscopy and transmission electron microscopy, as described by Spiers and Hopcroft (1993). For light microscopy, the sections were stained with toluidine blue, as described by Christensen et al. (2002). A Philips 201C transmission electron microscope was used to examine hyphal structure.
For H2O2 detection, a modified cytochemical method was used (Briggs et al., 1975; Bestwick et al., 1997). Meristematic or pseudostem tissue (2- to 3-mm thick) was vacuum-infiltrated in 5 mM CeCl3 solution buffered with 50 mM 3-(N-morpholino)-propanesulfonic acid (MOPS), pH 7.2, or MOPS at room temperature for 1 min, and incubated at room temperature for 1 h. The samples were then prefixed in 2.5% glutaraldehyde buffered with 0.1 M cacodylate buffer, pH 7.2, at 4°C overnight. After prefixation, the specimens were prepared for transmission electron microscopy, as described by Shinogi et al. (2001), and observed with a Hitachi 7100 transmission electron microscope.
Production of superoxide was detected with NBT using a method modified from that described by Shinogi et al. (2003). Cultures of E. festucae were grown on slides covered with a thin layer of potato dextrose agar for 10 d. Mycelia were incubated in 0.05 M sodium phosphate buffer, pH 7.5, containing 0.05% (w/v) NBT (Sigma-Aldrich) and where necessary preincubated for 20 min with 50 μM diphenylene iodinium (Sigma-Aldrich). After 5 h of incubation with NBT, the cultures were fixed in ethanol to stop the reaction. The stained samples were either photographed or examined with a compound microscope at ×100 and ×400 magnification.
Confocal laser scanning fluorescence images were recorded on a TCS 4D confocal system (Leica Microsystems) with a ×40 numerical aperture 1.0 oil-immersion lens. A krypton–argon laser was used as the excitation source at 488 nm, and GFP fluorescence was recorded between 515 and 545 nm. Images of GFP fluorescence shown in the figures are from a depth series of 28 optical sections taken at 1.5 μm. The images were stored as TIF files and processed with Canvas 10 software (ACD Systems International).
Accession Numbers
Sequence data from this article can be found in the DDBJ/EMBL/GenBank databases under the accession numbers AB236860 (E. festucae noxA), AB236861 (E. festucae noxB), AB239929 (L. perenne PR1 cDNA partial sequence), and AB239930 (L. perenne PR5 cDNA partial sequence).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Growth of Endophyte-Infected Perennial Ryegrass.
Supplemental Figure 2. Growth of Epiphyllous Hyphae on the Surface of Leaf Blades.
Supplemental Figure 3. Symbiotic Phenotype of E. festucae FR2.
Supplemental Figure 4. Relative Biomass of Wild-Type E. festucae and the noxA Mutant in Perennial Ryegrass.
Supplemental Figure 5. Lactophenol Trypan Blue–Stained Perennial Ryegrass Tissue.
Supplemental Table 1. Biological Materials.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
This research was supported by Grants MAU103 and MAU0403 from the Royal Society of New Zealand Marsden Fund. We thank Andrea Bryant and Elizabeth Nickless (Massey University) and Douglas Hopcroft and Raymond Bennett (HortResearch) for technical assistance, Geoff Jameson (Massey University) and Christine Foyer (Rothamsted) for technical advice, and David Jones (Australian National University) and Christine Foyer (Rothamsted) for discussion of the research and comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Barry Scott (d.b.scott@massey.ac.nz).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039263.
References
- Aguirre, J., Ríos-Momberg, M., Hewitt, D., and Hansberg, W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13 111–118. [DOI] [PubMed] [Google Scholar]
- Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827. [DOI] [PubMed] [Google Scholar]
- Arnold, A.E., Mejía, L.C., Kyllo, D., Rojas, E.I., Maynard, Z., Robbins, N., and Herre, E.A. (2003). Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 100 15649–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle, C.J., Bohula, E.A., Kron, S.J., Wessels, D., Soll, D.R., Schäfer, A., Brown, A.J., and Gow, N.A.R. (2003). Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot. Cell 2 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J.W., and Lasure, L.L. (1985). Conventions for gene symbols. In Gene Manipulation in Fungi, J.W. Bennett and L.L. Lasure, eds (London: Academic Press), pp. 537–544.
- Bestwick, C.S., Brown, I.R., Bennett, M.H., and Mansfield, J.W. (1997). Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce, K.J., Hynes, M.J., and Andrianopoulos, A. (2003). Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 116 1249–1260. [DOI] [PubMed] [Google Scholar]
- Briggs, R.T., Drath, D.B., Karnovsky, M.L., and Karnovsky, M.J. (1975). Localization of NADH oxidase on the surface of human polymorphonuclear leukocytes by a new cytochemical method. J. Cell Biol. 67 566–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, L.P., Wilkinson, H.H., and Schardl, C.L. (1997). Bioprotective alkaloids of grass-fungal endophyte symbioses. Plant Physiol. 114 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, A.D., Schardl, C.L., Songlin, P.J., Mogen, K.L., and Siegel, M.R. (1990). The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 18 347–354. [DOI] [PubMed] [Google Scholar]
- Carroll, A.M., Sweigard, J.A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41 22. [Google Scholar]
- Chen, C., and Dickman, M.B. (2004). Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 51 1493–1507. [DOI] [PubMed] [Google Scholar]
- Cheng, G., Ritsick, D., and Lambeth, J.D. (2004). Nox3 regulation by NOXO1, p47phox, and p67phox. J. Biol. Chem. 279 34250–34255. [DOI] [PubMed] [Google Scholar]
- Christensen, M.J. (1995). Variation in the ability of Acremonium endophytes of Lolium perenne, Festuca arundinacea and F. pratensis to form compatible associations in the three grasses. Mycol. Res. 99 466–470. [Google Scholar]
- Christensen, M.J., Ball, O.J.-P., Bennett, R.J., and Schardl, C.L. (1997). Fungal and host genotype effects on compatibility and vascular colonization by Epichloë festucae. Mycol. Res. 101 493–501. [Google Scholar]
- Christensen, M.J., Bennett, R.J., and Schmid, J. (2002). Growth of Epichloë/Neotyphodium and p-endophytes in leaves of Lolium and Festuca grasses. Mycol. Res. 106 93–106. [Google Scholar]
- Clay, K. (2004). Fungi and the food of the gods. Nature 427 401–402. [DOI] [PubMed] [Google Scholar]
- Clay, K., and Schardl, C. (2002). Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160 (suppl.), S99–S127. [DOI] [PubMed] [Google Scholar]
- DeLeo, F.R., Yu, L., Burritt, J.B., Loetterle, L.R., Bond, C.W., Jesaitis, A.J., and Quinn, M.T. (1995). Mapping sites of interaction of p47-phox and flavocytochrome b with random-sequence peptide phage display libraries. Proc. Natl. Acad. Sci. USA 92 7110–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke, N. (1983). Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 23 345–357. [Google Scholar]
- Doke, N. (1985). NADPH-dependent O2− generation in membrane fractions isolated from wounded potato tubers inoculated with Phytophthora infestans. Physiol. Plant Pathol. 27 311–322. [Google Scholar]
- Finegold, A.A., Shatwell, K.P., Segal, A.W., Klausner, R.D., and Dancis, A. (1996). Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271 31021–31024. [DOI] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H.F., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D.G., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Grove, S.N., Bracker, C.E., and Morre, D.J. (1970). An ultrastructural basis for hyphal tip growth in Pythium ultimum. Am. J. Bot. 57 245–266. [Google Scholar]
- Harper, A.M., Chaplin, M.F., and Segal, A.W. (1985). Cytochrome b-245 from human neutrophils is a glycoprotein. Biochem. J. 227 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, Y., Johnson, R., and Scott, B. (1994). Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli. Curr. Genet. 25 508–513. [DOI] [PubMed] [Google Scholar]
- Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10 22–29. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, H., Christensen, M.J., and Bennett, R.J. (1993). Incompatibility of some grass/Acremonium endophyte associations. Mycol. Res. 97 1237–1244. [Google Scholar]
- Kornfeld, R., and Kornfeld, S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54 631–664. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.-M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D.G., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucque, H., and Silar, P. (2003). NADPH oxidase: An enzyme for multicellularity? Trends Microbiol. 11 9–12. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4 181–189. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D., Cheng, G., Arnold, R.S., and Edens, W.A. (2000). Novel homologs of gp91phox. Trends Biochem. Sci. 25 459–461. [DOI] [PubMed] [Google Scholar]
- Lane, G.A., Christensen, M.J., and Miles, C.O. (2000). Coevolution of fungal endophytes with grasses: The significance of secondary metabolites. In Microbial Endophytes, C.W. Bacon and J.F.J. White, eds (New York: Marcel Dekker), pp. 341–388.
- Lara-Ortíz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50 1241–1255. [DOI] [PubMed] [Google Scholar]
- Latch, G.C.M., and Christensen, M.J. (1985). Artificial infection of grasses with endophytes. Ann. Appl. Biol. 107 17–24. [Google Scholar]
- Leuchtmann, A., Schardl, C.L., and Siegel, M.R. (1994). Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue grasses. Mycologia 86 802–812. [Google Scholar]
- Lodwig, E., and Poole, A. (2003). Metabolism of Rhizobium bacteroids. CRC Crit. Rev. Plant Sci. 22 37–78. [Google Scholar]
- Malagnac, F., Lalucque, H., Lepère, G., and Silar, P. (2004). Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41 982–997. [DOI] [PubMed] [Google Scholar]
- Moon, C.D., Scott, B., Schardl, C.L., and Christensen, M.J. (2000). The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia 92 1103–1118. [Google Scholar]
- Moon, C.D., Tapper, B.A., and Scott, B. (1999). Identification of Epichloë endophytes in planta by a microsatellite-based PCR fingerprinting assay with automated analysis. Appl. Environ. Microbiol. 65 1268–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkres, K.D. (1990). Histochemical detection of superoxide radicals and hydrogen peroxide by Age-1 mutants of Neurospora. Fungal Genet. Newsl. 37 24–25. [Google Scholar]
- Namiki, F., Matsunaga, M., Okuda, M., Inoue, I., Nishi, K., Fujita, Y., and Tsuge, T. (2001). Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant Microbe Interact. 14 580–584. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5 566–576. [DOI] [PubMed] [Google Scholar]
- Panaccione, D.G., Johnson, R.D., Wang, J., Young, C.A., Damrongkool, P., Scott, B., and Schardl, C.L. (2001). Elimination of ergovaline from a grass-Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. USA 98 12820–12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M. (2004). Molecular genetics of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 7 414–421. [DOI] [PubMed] [Google Scholar]
- Punt, P.J., Oliver, R.P., Dingemanse, M.A., Pouwels, P.H., and van den Hondel, C.A.M.J.J. (1987). Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56 117–124. [DOI] [PubMed] [Google Scholar]
- Royer-Pokora, B., Kunkel, L.M., Monaco, A.P., Goff, S.C., Newburger, P.E., Baehner, R.L., Cole, F.S., Curnutte, J.T., and Orkin, S.H. (1986). Cloning the gene for an inherited human disorder—chronic granulomatous disease—on the basis of its chromosomal location. Nature 322 32–38. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sánchez, O., Navarro, R.E., and Aguirre, J. (1998). Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 258 89–94. [DOI] [PubMed] [Google Scholar]
- Sanderson, K.E., and Srb, A.M. (1965). Heterokaryosis and parasexuality in the fungus Ascochta imperfecta. Am. J. Bot. 52 72–81. [PubMed] [Google Scholar]
- Schardl, C.L. (2001). Epichloë festucae and related mutualistic symbionts of grasses. Fungal Genet. Biol. 33 69–82. [DOI] [PubMed] [Google Scholar]
- Schardl, C.L., Leuchtmann, A., and Spiering, M.J. (2004). Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 55 315–340. [DOI] [PubMed] [Google Scholar]
- Schiestl, R.H., and Petes, T.D. (1991). Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88 7585–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, B. (2001). Epichloë endophytes: Symbionts of grasses. Curr. Opin. Microbiol. 4 393–398. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato, S., and Mori, H. (2001). Control of outgrowth and dormancy in axillary buds. Plant Physiol. 127 1405–1413. [PMC free article] [PubMed] [Google Scholar]
- Shinogi, T., Suzuki, T., Kurihara, T., Narusaka, Y., and Park, P. (2003). Microscopic detection of reactive oxygen species generation in the compatible and incompatible interactions of Alternaria alternata Japanese pear pathotype and host plants. J. Gen. Plant Pathol. 69 7–16. [Google Scholar]
- Shinogi, T., Suzuki, T., Tagashira, M., Yamane, K., Yao, N., Uwo, M., Kawakami, S., Narusaka, Y., and Park, P. (2001). A low viscosity epoxy resin “Quetol-651” as a substitute of Spurr's resin for hard biological materials in transmission electron microscopy. J. Electr. Microsc. Technol. Med. Biol. 16 1–10. [Google Scholar]
- Simon-Plas, F., Elmayan, T., and Blein, J.-P. (2002). The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 31 137–147. [DOI] [PubMed] [Google Scholar]
- Spiering, M.J., Moon, C.D., Wilkinson, H.H., and Schardl, C.L. (2005). Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering, M.J., Wilkinson, H.H., Blankenship, J.D., and Schardl, C.L. (2002). Expressed sequence tags and genes associated with loline alkaloid expression by the fungal endophyte Neotyphodium uncinatum. Fungal Genet. Biol. 36 242–254. [DOI] [PubMed] [Google Scholar]
- Spiers, A.G., and Hopcroft, D.H. (1993). Black canker and leaf spot of Salix in New Zealand caused by Glomerella miyabeana (Colletotrichum gloeosporioides). Eur. J. For. Pathol. 23 92–102. [Google Scholar]
- Suh, Y.-A., Arnold, R.S., Lassegue, B., Shi, J., Xu, X., Sorescu, D., Chung, A.B., Griendling, K.K., and Lambeth, J.D. (1999). Cell transformation by the superoxide-generating oxidase Mox1. Nature 401 79–82. [DOI] [PubMed] [Google Scholar]
- Taiz, L., and Zeiger, E. (1998). Plant Physiology. (Sunderland, MA: Sinauer Associates).
- Tan, Y.Y., Spiering, M.J., Scott, V., Lane, G.A., Christensen, M.J., and Schmid, J. (2001). In planta regulation of extension of an endophytic fungus and maintenance of high metabolic rates in its mycelium in the absence of apical extension. Appl. Environ. Microbiol. 67 5377–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Tapper, B.A., Popay, A., Parker, E.J., and Scott, B. (2005). A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57 1036–1050. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D.G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Jones, J.D.G., and Dangl, J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37 1130–1134. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K.E., and Jones, J.D.G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14 365–370. [DOI] [PubMed] [Google Scholar]
- Vignais, P.V. (2002). The superoxide-generating NADPH oxidase: Structural aspects and activation mechanism. CMLS Cell. Mol. Life Sci. 59 1428–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach, T.M., and Segal, A.W. (1997). Analysis of glycosylation sites on gp91phox, the flavocytochrome of the NADPH oxidase, by site-directed mutagenesis and translation in vitro. Biochem. J. 321 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Machado, C., Panaccione, D.G., Tsai, H.-F., and Schardl, C.L. (2004). The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 41 189–198. [DOI] [PubMed] [Google Scholar]
- Weinzierl, G., Leveleki, L., Hassel, A., Kost, G., Wanner, G., and Bolker, M. (2002). Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 45 219–231. [DOI] [PubMed] [Google Scholar]
- Wu, R., Hirai, A., Mundy, J., Nelson, R., and Rodriguez, R. (1991). Guidelines for nomenclature of cloned genes or DNA fragments in rice. Rice Genet. Newsl. 8 51–53. [Google Scholar]
- Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., Jones, J.D.G., and Doke, N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, C., Itoh, Y., Johnson, R., Garthwaite, I., Miles, C.O., Munday-Finch, S.C., and Scott, B. (1998). Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 33 368–377. [DOI] [PubMed] [Google Scholar]
- Young, C.A., Bryant, M.K., Christensen, M.J., Tapper, B.A., Bryan, G.T., and Scott, B. (2005). Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genomics 274 13–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.