Abstract
DNA methylation (5-methylcytosine) in mammalian genomes predominantly occurs at CpG dinucleotides, is maintained by DNA methyltransferase1 (Dnmt1), and is essential for embryo viability. The plant genome also has 5-methylcytosine at CpG dinucleotides, which is maintained by METHYLTRANSFERASE1 (MET1), a homolog of Dnmt1. In addition, plants have DNA methylation at CpNpG and CpNpN sites, maintained, in part, by the CHROMOMETHYLASE3 (CMT3) DNA methyltransferase. Here, we show that Arabidopsis thaliana embryos with loss-of-function mutations in MET1 and CMT3 develop improperly, display altered planes and numbers of cell division, and have reduced viability. Genes that specify embryo cell identity are misexpressed, and auxin hormone gradients are not properly formed in abnormal met1 embryos. Thus, DNA methylation is critical for the regulation of plant embryogenesis and for seed viability.
INTRODUCTION
Early embryogenesis in Arabidopsis thaliana is distinguished by a predictable pattern of cell divisions (Bowman and Mansfield, 1994). The zygote divides asymmetrically to give rise to a small apical cell and a large basal cell, which have distinct developmental fates (Figures 1A to 1F) (Goldberg et al., 1994; Scheres and Benfey, 1999). The apical cell develops into the embryo proper, whereas the basal cell elongates and divides transversely to generate the suspensor, an ephemeral organ that supports the development of the embryo proper (Berleth and Chatfield, 2002). The apical cell undergoes two rounds of longitudinal cell divisions (two- and four-cell stage) and one round of transverse divisions (eight-cell stage). Each of the eight cells derived from the apical cell of the octant embryo divides periclinally to produce a 16-cell embryo. During early embryogenesis, a shoot-root axis of polarity is fixed, shoot and root meristems are formed, cotyledon storage organs are generated, and tissue layers are specified. The embryo passes through a series of stages that are defined morphologically as globular, heart, torpedo, and walking stick stages (Goldberg et al., 1994; Jurgens, 2001). By contrast, the basal cell elongates and divides transversely to form a structure of seven to nine cells. The uppermost cell of the basal lineage, the hypophysis, becomes integrated into the embryo proper and becomes the quiescent center of the root meristem (Berleth and Jurgens, 1993; Hamann et al., 1999). The remaining cells in the basal lineage follow an extraembryonic cell fate and form the suspensor.
Figure 1.
met1-6 Mutation Affects Suspensor and Embryo Development.
(A) to (X) Nomarski photographs of wild-type and homozygous met1-6 mutant embryos at 1 to 6 DAP.
(Y) to (JJ) Histological section photographs of wild-type and met1-6 mutant embryos at 4 to 6 DAP.
Photographs of the wild type (A) to (F) at 1, 2, 3, 4, 5, and 6 DAP, respectively; the met1 mutant embryos at 1 DAP ([G], [M], and [S]), 2 DAP ([H], [N], and [T]), 3 DAP ([I], [O], and [U]), 4 DAP ([J], [P], and [V]), 5 DAP ([K], [Q], and [W]), and 6 DAP ([L], [R], and [X]). Histological sections of wild-type embryos (A) to (C) at 4, 5, and 6 DAP, respectively; the met1 mutant embryos at 4 DAP ([BB], [EE], and [HH]), 5 DAP ([CC], [FF], and [II]), and 6 DAP ([DD], [GG], and [JJ]). (A) to (D), (G) to (J), (M) to (P), (S) to (V), (Y), (BB), (CC), (EE), (HH), and (II) are the same scale, and (E), (F), (K), (L), (Q), (R), (W), (X), (Z), (AA), (DD), (FF), (GG), and (JJ) are the same scale. Bars = 20 μm in (A) and 50 μm in (E). Arrowheads indicate the plane of the first zygotic cell division ([A], [G], [M], and [S]), the boundary between the apical and basal lineage-derived cells ([B] to [L]), the hypophysis (Y), the apical cell nucleus (BB), or cell planes of the first two longitudinal cell divisions of the apical cell ([EE] and [HH]).
Early embryogenesis is regulated by transcription factors, signal transduction pathways mediated by kinases, and proteins that establish and maintain auxin hormone gradients (Willemsen and Scheres, 2004). For example, the YODA (YDA) gene, which encodes a mitogen-activated protein kinase kinase kinase, regulates embryo and suspensor cell identity. In yda mutant plants, the suspensor cells adopt an embryonic cell fate, divide longitudinally, and are integrated into the embryo proper instead of forming the suspensor (Lukowitz et al., 2004). WUSCHEL-related homeobox (WOX) transcription factor genes mark cell fate decisions during early embryogenesis. WOX2 and WOX8 are expressed in the egg cell and zygote, and their expression is limited to the apical and basal cell lineages, respectively. WOX2 is necessary for cell divisions that form the apical embryo domain. Auxin hormone gradients help form the embryonic apical-basal axis, the shoot and root meristems, and the cotyledon organs (Jenik and Barton, 2005; Friml et al., 2006). PIN-formed (PIN) genes encode transporter-like membrane proteins that are important for regulating auxin transport (Friml, 2003; Weijers et al., 2005). PIN proteins display asymmetric subcellular localization at the plasma membrane, regulate polar auxin transport, and establish auxin gradients during embryogenesis. Mutations in PIN1 and PIN7 disrupt the establishment of the embryonic apical-basal axis (Steinmann et al., 1999; Friml et al., 2003). PINOID (PID), encoding a Ser-Thr protein kinase (Christensen et al., 2000), regulates PIN localization and apical-basal axis formation (Friml et al., 2004). Overexpression of PID causes a basal-to-apical shift in PIN localization, resulting in the loss of auxin gradients and defects in embryogenesis.
DNA methyltransferases covalently methylate cytosine at the 5-position. DNA methylation is a heritable epigenetic process that regulates developmental processes in animals and plants (Martienssen and Colot, 2001; Reik et al., 2001; Li, 2002). DNA methylation plays an important role in genome stabilization, X chromosome inactivation, silencing of transposons and endogenous retrovirus, gene expression, and imprinting (Bird and Wolffe, 1999; Bestor, 2000; Reik and Walter, 2001; Bender, 2004; Gehring et al., 2004; Chan et al., 2005).
In mammals, 5-methylcytosine is at CpG dinucleotides. During gametogenesis and embryogenesis, DNA methylation is lost and subsequently reestablished by DNA methyltransferase3a (Dnmt3a) and Dnmt3b (Okano et al., 1999). The patterns of DNA methylation are maintained during somatic development by the Dnmt1 DNA methyltransferase (Li et al., 1992). DNA methylation is essential for mammalian embryonic development, and targeted mutations of the Dnmt1 or Dnmt3 genes result in embryonic lethality (Li et al., 1992; Okano et al., 1999). In Arabidopsis, CpG DNA methylation is maintained by METHYLTRANSFERASE1 (MET1), an ortholog of DNA methyltransferase Dnmt1 (Finnegan and Dennis, 1993; Finnegan and Kovac, 2000). In addition to CpG methylation, Arabidopsis has CpNpG and CpNpN methylation. CHROMOMETHYLASE3 (CMT3) and DOMAINS REARRANGED METHYLASE1 (DRM1) and DRM2 (a homolog of Dnmt3 for de novo DNA methylation) maintain patterns of CpNpG and CpNpN methylation (Henikoff and Comai, 1998; Bartee et al., 2001; Lindroth et al., 2001; Cao and Jacobsen, 2002a, 2002b).
Arabidopsis plants with an antisense MET1 transgene, partial-loss-of-function met1 mutations, or cmt3 drm1 drm2 mutations revealed that reduced DNA methylation results in abnormal postembryonic plant development (Finnegan et al., 1996; Kakutani et al., 1996, 2004; Ronemus et al., 1996; Cao, 2003; Kankel et al., 2003; Kato et al., 2003; Saze et al., 2003). Here, we show that a significant fraction of met1 and met1 cmt3 mutant embryos show reduced viability. met1 and met1 cmt3 embryos often have incorrect patterns of cell divisions, polarity, and auxin gradients and misexpress genes that specify embryo cell identity. Thus, DNA methylation is necessary for proper embryo development and viability in Arabidopsis.
RESULTS
Previously, we isolated four met1 mutant alleles (met1-5 to met1-8) (Xiao et al., 2003). The met1-6 allele is likely to be a null allele because, due to a premature translation stop codon, it encodes a truncated DNA methyltransferase that lacks a catalytic domain. The met1-6 mutation leads to late flowering (W. Xiao and R.L. Fischer, unpublished results), results in genomic hypomethylation, and reduces DNA methylation in the promoter of an imprinted gene, MEDEA (Xiao et al., 2003). Plants heterozygous for the met1-6 mutant allele, which were derived from mutagenized plants that were never homozygous for this mutation (Xiao et al., 2003), were used to generate homozygous met1-6 plants used in these studies. We found that siliques from homozygous met1-6 plants contained aborted seeds (12%) at ∼40-fold higher frequency than wild-type controls (0.3%) (Table 1). Observation of embryogenesis using cleared seeds and histological sections revealed that 33% of the met1-6 embryos developed abnormally (Table 1). As described below, these homozygous met1-6 mutant embryos displayed a wide range of developmental abnormalities that were consistently observed.
Table 1.
Effect of Mutations in DNA Methyltransferase Genes on Embryogenesis and Seed Viability
| Genetic Cross
|
Abnormal F1 Embryosa
|
F1 Seed Abortion
|
||||
|---|---|---|---|---|---|---|
| Self-Pollinated | Female | Male | % | n | % | n |
| Wild type | 0 | 967 | 0.3 | 678 | ||
| met1-6/met1-6 | 33 | 568 | 12.2 | 986 | ||
| cmt3-7/cmt3-7 | 3 | 578 | 0.5 | 875 | ||
| MET1/met1-6 | 10 | 562 | 7.8 | 539 | ||
| cmt3-7/cmt3-7 | 23 | 816 | 16.3 | 1096 | ||
| MET1/met1-6 | ||||||
| met1-6/met1-6 | Wild type | 16 | 550 | 7.2 | 690 | |
| Wild type | met1-6/met1-6 | 8 | 822 | 1.2 | 982 | |
| MET1/met1-6 | Wild type | 8 | 420 | 2.2 | 683 | |
| Wild type | MET1/met1-6 | 5 | 403 | 1.0 | 853 | |
Embryos at 1 to 6 d after pollination were examined by whole-mount seed clearing.
Asymmetric First Cell Division
Abnormalities in F2 homozygous met1 embryo development were apparent after the first zygotic division, 1 d after pollination (DAP). We detected met1 mutant zygotes (Figures 1G, 1M, and 1S) that divided more symmetrically than wild-type control embryos (Figure 1A). Approximately 13% (n = 266) of the embryos displayed basal cells that failed to elongate and undergo transverse divisions (Figures 1N and 1T). These results show that genome-wide changes in DNA methylation affect the earliest stages of embryogenesis in Arabidopsis.
Suspensor Development
In wild-type embryos, the basal cell elongates and divides transversely to form a suspensor with seven to nine cells (Figures 1B to 1E). In wild-type suspensors, longitudinal divisions do not occur (Figure 1Y). By contrast, longitudinal cell divisions in the basal cell lineage were detected in ∼27% (n = 266) of homozygous met1-6 embryos at 2 (Figures 1H and 1N) and 3 DAP (Figures 1I and 1O). In wild-type embryos, there is a clear boundary between the spherical proembryo and the linear file of suspensor cells (Figure 1C), and at 4 to 6 DAP, the hypophysis, the uppermost cell of the basal lineage, becomes prominent (Figures 1D to 1F). This clear demarcation between embryo and suspensor is often not detected in met1-6 mutant embryos because of the many longitudinal cell divisions in the suspensor (Figures 1J, 1K, and 1U). Thus, DNA methylation is necessary for proper development of the suspensor during Arabidopsis embryogenesis.
Embryo Development
In wild-type embryos, the apical cell undergoes two rounds of symmetrical divisions with the division planes perpendicular to each other to form a four-cell proembryo (Goldberg et al., 1994). In certain abnormal met1 embryos, the apical cell divided longitudinally, but the subsequent division was not in the correct orientation (Figures 1EE and 1HH). This sometimes generated asymmetric embryos with two cells on one side of the hypophysis and no cells on the other side (Figures 1DD and 1GG). Moreover, these embryos were significantly delayed in their development compared with wild-type control embryos. These results reveal that early planes of cell division in homozygous met1-6 mutant embryos are sometimes not properly specified.
Abnormalities in numbers and planes of cell division persisted throughout met1 embryogenesis. In wild-type Arabidopsis, at 4 to 6 DAP, the embryo passes through a series of stages that are defined morphologically as globular, heart, and torpedo (Figures 1D to 1F and 1AA). Among the abnormal met1 embryos, we observed abnormal embryos without a clear boundary between apical and basal cell lineages due to longitudinal cell divisions in the basal cells of suspensor (Figures 1J, 1K, 1P, and 1U) or embryos that did not display an apical-basal axis and whose basal cell lineage resembled the apical cell lineage (Figures 1Q and 1FF). We also observed abnormal structures in met1-6 embryos (Figures 1V and 1W).
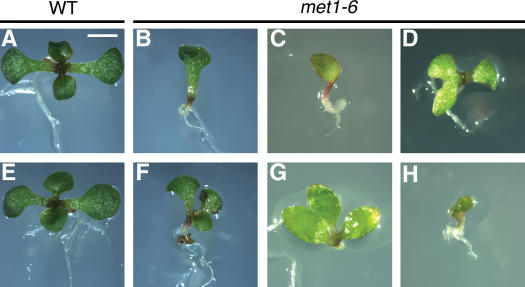
At the transition and early heart stages of wild-type embryogenesis, two symmetrical cotyledons are initiated from lateral domains of the embryo, and an embryonic shoot apical meristem is differentiated from the medial domain between the two cotyledons (Figure 1F) (Berleth and Chatfield, 2002; Prigge et al., 2005). In homozygous met1-6 embryos, the embryo sometimes failed to differentiate two cotyledons (Figures 1L and 1JJ) or initiated three cotyledons (Figure 1R). Mutant embryos having one cotyledon also lacked a medial domain where the embryonic shoot meristem is generated. This phenotype was also observed in F2 homozygous met1-6 seedlings, which had a single cotyledon lacking the apical shoot meristem (Figures 2B and 2C), two abnormal cotyledons (Figure 2D), three cotyledons (Figures 2F and 2G), or four cotyledons (Figure 2H). Taken together, these results show that loss of DNA methylation alters the number and planes of cell division required for generating the embryo proper, apical-basal axis, cotyledons, and meristems.
Figure 2.
Effect of the met1-6 Mutation on Cotyledons and the Shoot Apical Meristem.
Seedlings were photographed at the same magnification at 5 d after germination. Bar = 2 mm.
(A) and (E) Wild-type seedlings.
(B) to (D) and (F) to (H) met1-6 seedlings.
Partial Dominant Effect of met1-6 Mutations on Embryogenesis
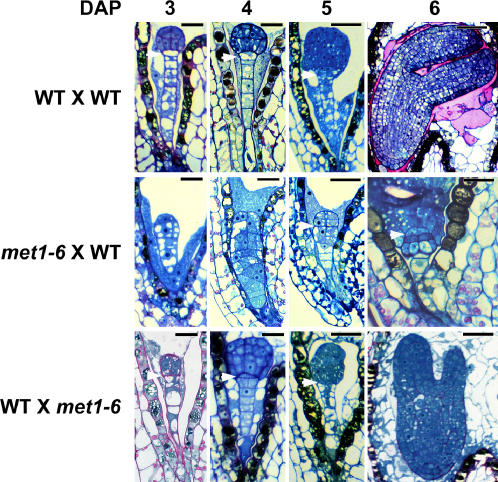
To examine whether a paternal or maternal hypomethylated genome can affect embryogenesis, we reciprocally crossed homozygous met1-6 plants with wild-type plants and examined embryogenesis within the F1 seeds. We found that either maternal- or paternal-derived hypomethylated genomes were sufficient to cause abnormal embryogenesis (Figure 3) and seed abortion (Table 1). Inheritance of a maternal hypomethylated genome resulted in 16% abnormal embryos, whereas 8% of embryos developed improperly when a paternal-derived genome was hypomethylated. These results suggest that hypomethylated genomes have a partial dominant effect on embryogenesis and seed abortion.
Figure 3.
A Hypomethylated Maternal or Paternal Genome Influences Embryo Development.
Histological sections of wild-type embryos and the F1 seeds of reciprocal crosses with wild-type and met1-6 plants at 3 to 6 DAP. Bars = 20 and 50 μm at 3 and 5 DAP, respectively.
DNA Hypomethylation during Gametogenesis Affects Embryogenesis
Plants heterozygous for met1 mutations produce gametes that are hypomethylated during meiosis (Kankel et al., 2003; Saze et al., 2003). To determine if loss of DNA methylation during gametogenesis is sufficient to influence embryo and seed development, we self-pollinated heterozygous met1-6/MET1 plants and analyzed the F1 seed. Approximately 10% of the F1 embryos displayed developmental abnormalities (e.g., unusual numbers and planes of cell division) similar to the embryos shown in Figure 1, and ∼8% of the F1 seed aborted (Table 1). To determine if the loss of DNA methylation in the female or male gametophyte is sufficient to perturb seed development, we performed reciprocal crosses with met1-6/MET1 heterozygotes and wild-type plants. When the maternal parent was heterozygous, ∼8% of F1 embryos displayed abnormalities in the number and planes of cell division, and 2% of the F1 embryos aborted (Table 1). When the paternal parent was heterozygous, the effect was diminished, with 5 and 1% of the F1 embryos showing developmental abnormalities and aborting, respectively (Table 1). In control crosses with wild-type plants, we did not detect any abnormal F1 embryos, and only 0.3% aborted (Table 1). These results show that loss of DNA methylation during female or male gametogenesis is sufficient to influence embryogenesis and seed viability.
Synergy between Mutations in the MET1 and CMT3 DNA Methyltransferase Genes
One hypothesis to explain how met1-6 plants produce viable seeds is that both MET1 and CMT3 are biologically redundant; mutating one methyltransferase does not cause 100% lethality because the other methyltransferase is still present. To test this hypothesis, we self-pollinated MET1/met1-6 cmt3-7/cmt3-7 plants and analyzed the F1 progeny. In these experiments, the parent plants were never homozygous for the met1-6 mutation, so that the CpG hypomethylation was initiated in the female and male gametophytes. We found that 23% of the F1 embryos developed abnormally compared with 10% of self-pollinated MET1/met1-6 and 4% of cmt3-7/cmt3-7 controls (Table 1). Based upon visual inspection of seeds, ∼16% of the F1 progeny from self-pollinated MET1/met1-6 cmt3-7/cmt3-7 plants had aborted (Table 1). We determined the genotype of 276 seedlings from a self-pollinated MET1/met1-6 cmt3-7/cmt3-7 plant. All progeny were homozygous for cmt3-7, 36% were homozygous for MET1, 60% were heterozygous for MET1/met1-6, and 4% were homozygous for met1-6. The 2:1 ratio of homozygous MET1 and heterozygous MET1/met1-6 progeny (χ2 = 2.8, P > 0.09) demonstrates Mendelian segregation of the met1-6 allele during meiosis and is consistent with most of the homozygous met1-6 seeds not producing viable seedlings. Rare met1-6 cmt3-7 homozygous plants showed a dramatic reduction in stature compared with met1-6 and cmt3-7 control plants (Figure 4) and were sterile (data not shown). Thus, reduction of both CpG and non-CpG DNA methylation caused by met1-6 and cmt3-7 mutations results in a synergistic decrease in seed viability and plant robustness.
Figure 4.
met1-6 and cmt3-7 Mutations Have a Synergistic Effect on Arabidopsis Growth and Development.
Representative 45-d-old plants were photographed. Bars = 2.5 cm in (A) to (D) and 0.4 cm in (E).
(A) Wild type.
(B) Homozygous cmt3-7.
(C) Homozygous met1-6.
(D) and (E) The same homozygous cmt3-7 met1-6 plant.
Specification of Cell Identity
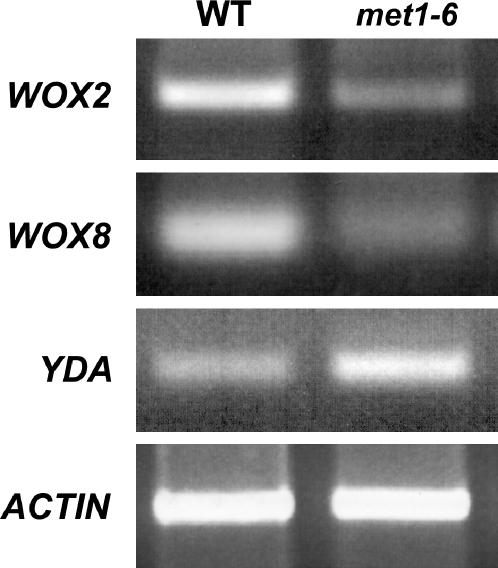
Longitudinal cell divisions in the suspensor (Figures 1H, 1I, 1N, and 1O) suggest that the suspensor cells are adopting an embryonic fate (Lukowitz et al., 2004). To investigate whether DNA methylation might influence cell fate decision during early embryogenesis, we analyzed expression of genes important for cell fate specification. As shown in Figure 5, expression of YDA, a mitogen-activated protein kinase kinase kinase gene (Lukowitz et al., 2004), is elevated in homozygous met1-6 seeds at 4 DAP. By contrast, expression of the WOX2 and WOX8 homeodomain transcription factor genes is reduced in homozygous met1-6 seeds at 4 DAP. Thus, DNA methylation, either directly or indirectly, influences transcription of genes that regulate cell identity during early embryogenesis.
Figure 5.
Effect of the met1-6 Mutation on Expression of Genes That Regulate Embryo Cell Identity.
RNA was isolated from wild-type and homozygous met1-6 seeds. WOX2, WOX8, YDA, and ACTIN RNAs were amplified by RT-PCR as described in Methods.
Auxin Gradients
Abnormal met1-6 mutant embryos (Figure 1) resembled those with defects in establishing auxin gradients (Friml et al., 2003). To understand the relationship between DNA methylation and auxin gradients during embryogenesis, we compared DR5:green fluorescent protein (GFP) transgene expression in wild-type embryos and homozygous met1-6 aborted embryos. DR5:GFP is a synthetic auxin-responsive promoter (DR5) ligated to the GFP that has been used to reveal auxin gradients in the early stages of wild-type embryogenesis (Sabatini et al., 1999; Friml et al., 2002a, 2002b). As has been reported previously (Friml et al., 2003), DR5:GFP expression is primarily in the basal lineage cells 3 to 4 DAP, especially in the hypophysis and upper suspensor cells (Figure 6). By 5 to 6 DAP, maximum DR5:GFP activity is detected in the quiescent center of the root meristem, the future columella and its initials, and weak expression in the tips of the developing cotyledons and provascular strands (Figure 5). We found that auxin gradients, as revealed by the pattern of DR5:GFP promoter activity, were not properly specified in abnormal homozygous met1-6 embryos (Figure 6). DR5:GFP expression was relatively evenly distributed in cells derived from either the apical or basal cells in abnormal embryos at 4, 5, and 6 DAP. This result reveals that normal DNA methylation patterns, either directly or indirectly, are required for generating auxin gradients consistently during embryogenesis.
Figure 6.
Expression of the DR5:GFP Transgene in Abnormal met1-6 Mutant Embryos.
Micrographs of DR5:GFP transgene expression in wild-type and abnormal met1-6 embryos at 3, 4, 5, and 6 DAP. For each embryo, two micrographs were taken, one in bright field (top) and the other using epifluorescence with blue light excitation (bottom). Arrowheads point to the boundary between the apical and basal lineage-derived cells.
PIN1 encodes an auxin efflux carrier and is responsible for establishing auxin gradients in early embryogenesis (Friml, 2003; Weijers et al., 2005). To determine whether PIN1 promoter activity, either directly or indirectly, is affected by DNA methylation, we introduced the PIN1:GFP transgene into met1-6/MET1 heterozygous plants. As shown in Figure 7, in control wild-type plants at 3 and 4 DAP, the PIN1:GFP transgene was primarily expressed in the globular embryo proper, especially within the top half of the embryo proper (Friml et al., 2003). However, in abnormal met1-6 embryos, PIN1:GFP was expressed throughout the entire embryo proper and was evenly distributed in both apical- and basal-derived cells (Figure 7). This result suggests that DNA methylation, either directly or indirectly, regulates PIN1 gene expression that, in turn, is necessary for establishing auxin gradients.
Figure 7.
Expression of the PIN1:GFP Transgene in Abnormal met1-6 Mutant Embryos.
Micrographs of PIN1:GFP transgene expression in wild-type and abnormal met1-6 embryos at 3 and 4 DAP. For each embryo, two micrographs were taken, one in bright field (top) and the other using epifluorescence with blue light excitation (bottom). Arrowheads point to the boundary between the apical and basal lineage-derived cells taken in bright field, and the plane between the top half of the embryo proper where PIN1:GFP was mainly expressed and the bottom half in wild-type embryos.
DNA Methylation Status of Genes That Regulate Embryogenesis
DNA methylation usually represses gene expression (Bender, 2004). Embryogenesis may be affected in met1-6 mutants because genes are misexpressed or overexpressed due to the absence of repressive DNA methylation. Alternatively, embryogenesis in met1-6 mutants may be defective because of ectopic de novo methylation and gene silencing, a process that has been previously documented in DNA methylation mutant backgrounds (Chan et al., 2005).
To investigate whether DNA methylation could directly influence embryonic regulatory genes, we performed gel blot analyses on DNA isolated from wild-type or homozygous met1-6 seedlings. Genomic DNA was digested with endonucleases HpaII (CCGG) and HpyCH4 IV (ACGT), both of which are inhibited by the presence of 5-methylcytosine within their recognition sequences. Digested DNAs were blotted and hybridized to labeled probes complementary to the 5′-region, coding region, and 3′-region of PIN1 and YDA. These genes were examined because of their integral roles in embryogenesis and possibility of being directly regulated by MET1.
We detected no DNA methylation at PIN1. Both wild-type and met1 mutant DNA had the same size HpaII and HpyCH4 IV restriction fragments, indicating that 5-methylcytosine was not present at these restriction enzyme sites (data not shown). Thus, it is likely that MET1 indirectly affects the expression of the PIN1 gene. By contrast, we found the methylation status for the YDA gene was affected in the met1-6 mutant background. In the 5′-region, coding region, and 3′-region of YDA, HpaII and HpyCH4 IV sites were not digested in wild-type DNA, whereas these sites were digested in met1-6 DNA (Figure 8). This indicates that these sites are methylated in wild-type plants, and this methylation is dependent on MET1 activity. Thus, the met1-6 mutation directly affects DNA methylation at the YDA locus. This may account for the higher YDA expression detected in met1-6 mutant seeds (Figure 5).
Figure 8.
DNA Methylation of the YDA Gene.
Wild-type and met1-6 DNAs were digested with HpaII or HpyCH4 IV, blotted, and hybridized to probes that hybridize to the YDA 5′-region, gene, and 3′-region that were prepared as described in Methods.
(A) YDA 5′-region.
(B) YDA gene.
(C) YDA 3′-region
DISCUSSION
DNA Methylation Is Critical for Arabidopsis Embryo Development
Arabidopsis embryo development involves a predictable pattern of planes and numbers of cell division (Bowman and Mansfield, 1994). We found this pattern was not maintained in a significant fraction of embryos with mutations in the MET1 and CMT3 DNA methyltransferase genes (Table 1, Figure 1). We observed defects in the plane of the first asymmetric division that produces the apical and basal cell lineages, as well as those divisions in the embryo proper that form distinct cell layers and partition the globular stage embryo (Mayer et al., 1993). Later-stage embryos sometimes displayed massive cell proliferation of the basal cell lineage and thereby lost their apical-basal axis of polarity (Figure 1). This may be attributed to a failure of the embryo to suppress the embryonic potential of the suspensor, which has been previously observed (Yeung and Meinke, 1993; Yadegari et al., 1994). Many of the abnormal embryos likely abort their development, resulting in nonviable seed (Figure 1, Table 1).
Pleiotropic phenotypes were observed in both met1-6 and met1-6 cmt3-7 developing embryos. This fact makes it difficult to pinpoint the developmental processes and genes directly affected in these backgrounds. We were however able to determine that embryonic auxin gradients (Figure 6) and PIN1 promoter activity (Figure 7) were highly perturbed in abnormal met1-6 embryos. Mutants defective in auxin transport and signaling also exhibit pleiotropic phenotypes (Friml, 2003), some of which are similar to ones described here (Figure 1).
Mechanism of DNA Methylation in Regulating Embryogenesis
How does DNA hypomethylation affect embryogenesis and reduce seed viability in Arabidopsis? Loss of DNA methylation derepresses silenced transposons (Kakutani et al., 2004), and these could insert into genes necessary for early embryogenesis; however, the low frequency of transposition events in a single generation (Miura et al., 2001) cannot account for the high level of abnormal embryos in met1-6 and met1-6 cmt3-7 mutants (Table 1). Hypomethylation in general is more likely to cause phenotypic defects due to improper gene expression (Bender, 2004), such as the case of ectopic FWA expression and delayed flowering in met1 mutant backgrounds (Soppe et al., 2000). It is also possible that ectopic hypermethylation and gene silencing, a phenomenon that occurs at the SUPERMAN and AGAMOUS loci in methylation mutants (Jacobsen et al., 2000), may be responsible for some met1-6 embryonic phenotypes. Thus, met1-6 embryogenesis may be perturbed because hypomethylation and ectopic hypermethylation cause changes in gene transcription.
We found subtle yet reproducible differences in the mRNA levels of WOX2, WOX8, and YDA between wild-type and met1-6 developing seeds (Figure 5). We also found that PIN1:GFP is improperly expressed in abnormal met1-6 embryos (Figure 7). If DNA methylation directly affects the establishment of auxin gradients, it is likely not through the regulation of PIN1 gene transcription, as there was no DNA methylation detected at the HpyCH4 IV and HpaII sites of the PIN1 gene in wild-type or met1-6 plants. In contrast with PIN1, we found that the YDA locus was methylated in a wild-type genome and that this methylation was dependent on MET1 (Figure 8). This loss of DNA methylation correlates with an increase in YDA expression in met1-6 developing seeds (Figure 5). Whether this methylation directly influences YDA expression is unknown. YDA regulates extraembryonic cell fates in the basal cells (Lukowitz et al., 2004). It is possible that some of the met1-6 embryonic phenotypes are attributable to ectopic expression of YDA. However, we do not know the number or identity of all the genes that are directly regulated by DNA methylation during embryogenesis. It is likely that they encode both regulatory proteins and enzymes involved in cell metabolism.
Parent-of-Origin Effects of DNA Hypomethylation on Embryogenesis, Viability, and Seed Size
Reciprocal crosses between a wild-type parent and a hypomethylated parent due to expression of an antisense MET1 transgene result in F1 seeds with altered embryo and endosperm size (Adams et al., 2000). Inheritance of a hypomethylated maternal genome produced larger embryo and endosperm, whereas inheritance of a hypomethylated paternal genome produced smaller embryo and endosperm. It is thought that hypomethylation of one parental genome allows for expression of normally silenced, imprinted alleles that influence seed size (Adams et al., 2000). We observed parent-of-origin effects on seed size in progeny from reciprocal crosses between wild-type and homozygous met1-6 plants (W. Xiao and R.L. Fischer, unpublished results) similar to those observed with MET1 antisense plants (Adams et al., 2000). Reciprocal crosses between homozygous met1-6 and wild-type parents also produced a significant fraction of F1 aborted embryos (Figure 3, Table 1). Thus, a hypomethylated maternal or paternal genome is sufficient to have an adverse influence on embryogenesis. This partial dominant effect is probably due to the fact that regions that have lost their DNA methylation due to mutations in DNA methyltransferases are very inefficiently remethylated (Chan et al., 2004), allowing the hypomethylated state of the maternal- or paternal-derived genome to persist in the F1 heterozygous progeny.
Reciprocal crosses with met1 and wild-type plants revealed a higher percentage of abnormal F1 embryos among progeny with a hypomethylated maternal genome than those with a paternal hypomethylated genome (Table 1). These results suggest that embryogenesis is particularly sensitive to hypomethylation of the maternally derived genome and support the hypothesis that the maternal genome plays the predominant role in controlling early seed development (Vielle-Calzada et al., 2000).
Redundancy of DNA Methylation in the Plant Genome
We found that mutations in both the MET1 and CMT3 genes had a much more dramatic effect on embryogenesis, seed viability (Table 1), and plant development (Figure 4) compared with mutations in just one of these genes. This suggests that the level of DNA methylation must be reduced below a critical threshold level before its role in seed viability and plant development is evident. In mammals, 5-methylcytosine is mainly present at a single sequence context, CpG dinucleotides, that is maintained by the DNA methyltransferase Dnmt1. Loss-of-function mutations in the Dnmt1 gene remove most DNA methylation and result in embryo lethality (Li et al., 1992). By contrast, plant genomes have 5-methylcytosine in multiple sequence contexts (CpG, CpNpG, and CpNpN) that is maintained by multiple DNA methyltransferases (Bender, 2004; Chan et al., 2005). Our results suggest that the plant genome is epigenetically modified by MET1 and CMT3 in a partially redundant fashion.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in greenhouses under continuous light at 23°C. cmt3-7 mutant plants were crossed with MET1/met1-6 heterozygous plants, then MET1/met1-6 CMT3/cmt3-7 plants were selected in the F1 progeny, and MET1/met1-6 cmt3-7/cmt3-7 plants were obtained in the F2 progeny. Genotyping plants for met1-6 and cmt3-7 was performed as described (Lindroth et al., 2001; Xiao et al., 2003).
Whole-Mount Seed Clearing, Histology, and Microscopy
Whole-mount immature seeds 1 to 6 DAP were cleared in Hoyer's fluid (70% chloral hydrate, 4% glycerol, and 5% gum arabic) and observed with a Zeiss Axioskop 50 microscope using Nomarski optics. Thin section studies of seeds were performed using methods as described (Brown et al., 1999). Briefly, seeds were fixed in 4% glutaraldehyde in 0.1 M phosphate buffer, pH 6.9, postfixed in osmium ferricyanide, dehydrated through a graded acetone series, and infiltrated with Spurr's resin (EMS). Ovules were sectioned sagittally in the plane of the micropyle and stalk with an LKB historange microtome equipped with glass knives. The 2.0- to 5.0-μm sections were mounted on glass slides (Brown and Lemmon, 1995) and stained with polychrome stain (Fox, 1997). GFP fluorescence microscopy was conducted as described (Yadegari et al., 2000).
RT-PCR Analysis
RT-PCR analysis was performed as described (Kinoshita et al., 1999). Total RNA was isolated from wild-type and met1-6 mutant seeds at 4 DAP. Primers used in the experiment were as follows: for WOX2, WOX2F (5′-CGTTTCTTCTACCCCCCTCC-3′) and WOX2R (5′-ATCACGGAGGGCAAATCTGT-3′); for WOX8, WOX8F (5′-CCTATCATCTTCCTTTTCCTCA-3′) and WOX8R (5′-TTGTGATGAACACGAAGCTTG-3′); for YDA, YDA-F (5′-ATACCGGTGCTGAGCCTGAT-3′) and YDA-R (5′-GTCCAGATCCAAGCAAGGAA-3′). All primer pairs spanned intron sequences so that amplification of RNA could be distinguished from amplification of any contaminating DNA.
DNA Gel Blot Analysis
Genomic DNA was isolated from 10-d-old wild-type (Columbia gl1) and met1-6 seedlings grown in culture (Tai and Tanksley, 1990). DNAs were cleaved by methylation-sensitive restriction endonucleases HpaII and HpyCH4 IV for 4 h at 37°C, run on 1.2% agarose gels, and blotted to a positively charged nylon membrane (Amersham Pharmacia Biotech). Membranes were hybridized with probes randomly labeled using Prime-It II (random primer labeling kit) from Stratagene. Primers used to amplify DNA for radioactive labeling were as follows: for the PIN1 promoter region, PIN1m_F1 (5′-CAAGGCGGCACGAATTTTAGT-3′) and PIN1m_R4 (5′-ATAGCTACGTATAACGGAACC-3′); for the PIN1 gene, PIN1m_F4 (5′-CGAGCGATTTTGTTAACTAGTG-3′) and PIN1m_R2 (5′-TGAAGGAAATGAGGGACCAG-3′); for the PIN1 3′-intergenic region, PIN1m3-F1 (5′-GAGATATTACCAAAACACAGGG-3′) and PIN1m3-R4 (5′-AAGAATCGGTAAAAGGATACAC-3′); for the YDA promoter region, pYDA-f1 (5′-TTTTTCACTTTTTAAATATTTTGC-3′) and pYDA-r (5′-GATCTTCTTCCCACAAACCA-3′); for the YDA gene, YDAm-F1 (5′-ATGCCTTGGTGGAGTAAATCA-3′) and YDAm-R1 (5′-GGGTCCTCTGTTTGTTGATC-3′); for the YDA 3′-intergenic region, YDAm-F2 (5′-CCCGTTCGAGTCAAATGATTC-3′) and YDAm-R2 (5′-GTTGTTCTCACTTGCTCGATT-3′).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G73590 (PIN1) and AT1G63700 (YDA).
Acknowledgments
We thank J. Penterman for stimulating discussions and for help with the preparation of this manuscript. We also thank M. Gehring, T.-F. Hsieh, J.H. Huh, and D. Michaeli for critically reading this manuscript and J. Friml and the ABRC (Ohio State University, Columbus, OH) for providing the DR5:GFP and PIN1:GFP transgenic seeds. This work was supported by National Institutes of Health Grant GM069415 and USDA Grant 2005-02355 to R.L.F.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robert L. Fischer (rfischer@berkeley.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038836.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502. [DOI] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55 41–68. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Chatfield, B. (2002). Embryogenesis: Pattern formation from a single cell. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0054, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Berleth, T., and Jurgens, G. (1993). The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118 575–587. [Google Scholar]
- Bestor, T.H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet. 14 2395–2402. [DOI] [PubMed] [Google Scholar]
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression–Belts, braces, and chromatin. Cell 99 451–454. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Mansfield, S.G. (1994). Embryogenesis. In Arabidopsis: An Atlas of Morphology and Development, J. Bowman, ed (New York: Springer-Verlag), pp. 351–361.
- Brown, R.C., and Lemmon, B.E. (1995). Methods in plant immunolight microscopy. Methods Cell Biol. 49 85–107. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.-A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 32–42. [Google Scholar]
- Cao, X. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13 2212–2217. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002. a). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002. b). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Henderson, I.R., and Jacobsen, S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Chan, S.W.-L., Zilberman, D., Xia, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1136. [DOI] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., and Dennis, E.S. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., and Kovac, K.A. (2000). Plant DNA methyltransferases. Plant Mol. Biol. 43 189–201. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, L.M. (1997). Microscopy 101. Polychrome stain for epoxy sections. Microsc. Today 97 21. [Google Scholar]
- Friml, J. (2003). Auxin transport - Shaping the plant. Curr. Opin. Plant Biol. 6 7–12. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002. b). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benkova, E., Mendgen, K., and Palme, K. (2002. a). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2006). Apical-basal polarity: Why plant cells don't stand on their heads. Trends Plant Sci. 11 12–14. [DOI] [PubMed] [Google Scholar]
- Gehring, M., Choi, Y., and Fischer, R.L. (2004). Imprinting and seed development. Plant Cell 16 (suppl.), S203–S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., de Paiva, G., and Yadegari, R. (1994). Plant embryogenesis: Zygote to seed. Science 266 605–614. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jurgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126 1387–1395. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Comai, L. (1998). A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Sakai, H., Finnegan, E.J., Cao, X., and Meyerowitz, E.M. (2000). Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr. Biol. 10 179–186. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., and Barton, M.K. (2005). Surge and destroy: The role of auxin in plant embryogenesis. Development 132 3577–3585. [DOI] [PubMed] [Google Scholar]
- Jurgens, G. (2001). Apical/basal pattern formation in Arabidopsis embryogenesis. EMBO J. 20 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K., and Richards, E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Kato, M., Kinoshita, T., and Miura, A. (2004). Control of development and transposon movement by DNA methylation in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 69 139–143. [DOI] [PubMed] [Google Scholar]
- Kankel, M.W., Ramsey, D.E., Stokes, T.L., Flowers, S.K., Haag, J.R., Jeddeloh, J.A., Riddle, N.C., Verbsky, M.L., and Richards, E.J. (2003). MET1 cytosine methyltransferase mutants. Genetics 163 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Miura, A., Bender, J., Jacobsen, S.E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13 421–426. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E. (2002). Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3 662–673. [DOI] [PubMed] [Google Scholar]
- Li, E., Bestor, T.H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 915–926. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Roeder, A., Parmenter, D., and Somerville, C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 109–119. [DOI] [PubMed] [Google Scholar]
- Martienssen, R.A., and Colot, V. (2001). DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293 1070–1074. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Buttner, G., and Jurgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 117 149–162. [Google Scholar]
- Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411 212–214. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D.W., Haber, D.A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 247 247–257. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W., Dean, W., and Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science 293 1089–1093. [DOI] [PubMed] [Google Scholar]
- Reik, W., and Walter, J. (2001). Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2 21–32. [DOI] [PubMed] [Google Scholar]
- Ronemus, M.J., Galbiati, M., Ticknor, C., Chen, J., and Dellaporta, S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273 654–657. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Saze, H., Scheid, O.M., and Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34 65–69. [DOI] [PubMed] [Google Scholar]
- Scheres, B., and Benfey, P.N. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 505–537. [DOI] [PubMed] [Google Scholar]
- Soppe, W.J.J., Jacobsen, S.E., Alonso-Blanco, C., Jackson, J.P., Kakutani, T., Koornneef, M., and Peeters, A.J.M. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6 791–802. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Tai, T.H., and Tanksley, S.D. (1990). A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol. Biol. Rep. 8 297–303. [Google Scholar]
- Vielle-Calzada, J.-P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404 91–94. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Sauer, M., Meurette, O., Friml, J., Ljung, K., Sandberg, G., Hooykaas, P., and Offringa, R. (2005). Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., and Scheres, B. (2004). Mechanisms of pattern formation in plant embryogenesis. Annu. Rev. Genet. 38 587–614. [DOI] [PubMed] [Google Scholar]
- Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J.J., Goldberg, R.B., Pennell, R.I., and Fischer, R.L. (2003). Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5 891–901. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., de Paiva, G.R., Laux, T., Koltunow, A.M., Apuya, N., Zimmerman, L., Fischer, R., Harada, J.J., and Goldberg, R.B. (1994). Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 7 1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Katz, A., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, E.C., and Meinke, D.W. (1993). Embryogenesis in angiosperms: Development of the suspensor. Plant Cell 10 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]