The earth rotates on its axis every 24 h, with the result that any position on the earth's surface alternately faces toward or away from the sun—day and night. That the metabolism, physiology, and behavior of most organisms changes profoundly between day and night is obvious to even the most casual observer. These biological oscillations are apparent as diurnal rhythms. It is less obvious that most organisms have the innate ability to measure time. Indeed, most organisms do not simply respond to sunrise but, rather, anticipate the dawn and adjust their biology accordingly. When deprived of exogenous time cues, many of these diurnal rhythms persist, indicating their generation by an endogenous biological circadian clock. Until recently, the molecular mechanisms by which organisms functioned in this fourth dimension, time, remained mysterious. However, over the last 30 or so years, the powerful approaches of molecular genetics have revealed the molecular underpinnings of a cellular circadian clockwork as complicated and as beautiful as the wonderful chronometers developed in the 18th century. Then, the need to accurately measure time to precisely determine longitude sparked an international competition to claim a prize, the princely sum of 20,000 pounds sterling, offered by the British Crown (Sobel, 1995).
CHARACTERISTICS OF CIRCADIAN RHYTHMS
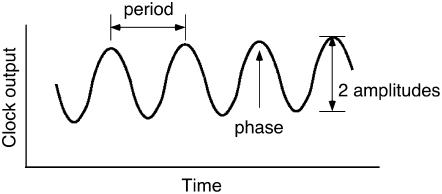
Circadian rhythms are the subset of biological rhythms with period, defined as the time to complete one cycle (Figure 1) of ∼24 h (Dunlap et al., 2004). This defining characteristic inspired Franz Halberg in 1959 to coin the term circadian, from the Latin words “circa” (about) and “dies” (day). A second defining attribute of circadian rhythms is that they are endogenously generated and self-sustaining, so they persist under constant environmental conditions, typically constant light (or dark) and constant temperature. Under these controlled conditions, the organism is deprived of external time cues, and the free-running period of ∼24 h is observed. A third characteristic of all circadian rhythms is temperature compensation; the period remains relatively constant over a range of ambient temperatures (Pittendrigh, 1954). This is thought to be one facet of a general mechanism that buffers the clock against changes in cellular metabolism.
Figure 1.
Critical Terms Used to Describe Circadian Rhythms.
Period is defined as the time to complete one cycle. It is commonly measured from peak to peak but could equally be measured from trough to trough or from any specified phase marker. Phase is the time of day for any given event. For example, if the peak in a rhythm occurred at dawn, the phase of the peak would be defined as 0 h. If a rhythm peaked 6 h after dawn, its phase would be 6 h, and so on. Phase is often defined in zeitgeber time (ZT). Zeitgeber is German for time giver, and any stimulus that imparts time information to the clock is a zeitgeber. The onset of light is a powerful zeitgeber, and dawn is defined as ZT0. The amplitude of the rhythm is defined as one-half the peak-to-trough distance.
Only in exceptional circumstances, such as in the laboratory, is an organism deprived of environmental time cues, such as light/dark cycles or temperature cycles, that derive from the alternation of day and night. These environmental time cues, termed zeitgebers (German for time givers), entrain the endogenous timing system to a period of 24 h, precisely corresponding to the exogenous period of the earth's rotation. The ability of a stimulus to reset the clock is a function of the time of day (phase; see Figure 1) at which the stimulus is administered. A pulse of light given before dawn will advance the phase of the clock, yet the same pulse of light given after dusk will delay the phase. If given at noon, the same pulse of light will have no effect at all. From this it is apparent that the clock regulates its own sensitivity to environmental stimuli. This varying sensitivity can be quantified and displayed as a phase response curve, in which one plots the shift in phase in response to a stimulus applied at different times across the circadian cycle (Dunlap et al., 2004).
THE HISTORY OF CLOCK RESEARCH IN PLANTS
The first writings, at least in the western canon, to recognize diurnal rhythms come from the fourth century BC. Androsthenes described the observation of daily leaf movements of the tamarind tree, Tamarindus indicus, that were observed on the island of Tylos (now Bahrein) in the Persian Gulf during the marches of Alexander the Great (Bretzl, 1903). There was no suggestion that the endogenous origin of these rhythms was suspected at the time, and it took more than two millennia for this to be experimentally tested. The scientific literature on circadian rhythms began in 1729 when the French astronomer de Mairan reported that the daily leaf movements of the sensitive heliotrope plant (probably Mimosa pudica) persisted in constant darkness, demonstrating their endogenous origin (de Mairan, 1729). Presciently, de Mairan suggested that these rhythms were related to the sleep rhythms of bedridden humans. It took 30 years before de Mairan's observations were independently repeated (Hill, 1757; Duhamel duMonceau, 1759; Zinn, 1759). These studies excluded temperature variation as a possible zeitgeber driving the leaf movement rhythms.
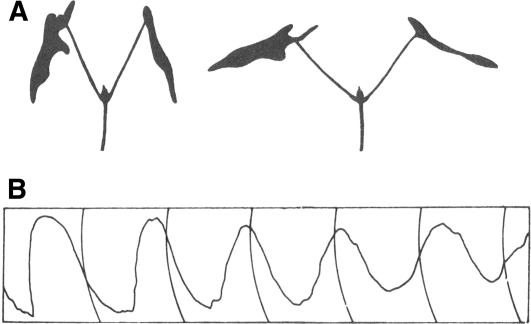
Nearly a century passed before period length of these leaf movements was accurately measured and it was realized that these rhythms were only ∼24 h, making the rhythms circadian and suggesting that these rhythms were endogenous and not simply responses to environmental time cues. de Candolle (1832) determined that the free running period of M. pudica was 22 to 23 h, discernably shorter than 24 h. He further showed that the rhythm could be inverted by reversing the alternation of light and dark. A number of authors repeated and expanded these observations through the 19th and early 20th centuries, in each case exploiting plant leaf movements (Figure 2), the only known circadian rhythm (for a more complete historical account, see Bünning, 1960; Cumming and Wagner, 1968). As an aside, animal circadian rhythms were not scientifically described until much later, with pigment rhythms in arthropods (Kiesel, 1894) and daily activity in rats (Richter, 1922) being among the first in the literature.
Figure 2.
Leaf Movements of a Representative Species.
(A) Sleep movements of Phaseolus coccineus. The position of the primary leaves of a seedling at night is at the left and during the day is at the right.
(B) Circadian rhythm of leaf movements of P. coccineus entrained to light/dark cycles and monitored in continuous light. As can be inferred from the leaf positions in (A), the peaks of the curve represent the nighttime leaf position. The vertical lines indicate 24-h intervals. The period for this trace is ∼27 h.
(A) was originally published as Figure 14 and (B) as Figure 4 in Chapter 2 in Bünning (1973). Both are reproduced with kind permission of Springer Science and Business Media.
Not surprisingly, that these circadian rhythms in leaf movements were truly endogenous was disputed. Pfeffer (1873), for example, suspected that light leaking into the darkrooms (and wine cellars and caves) employed in these studies foiled the attempts to provide constant conditions and invalidated the claims that these rhythms had endogenous origins. However, the critics ultimately were persuaded by the accumulating mass of evidence. Pfeffer himself extensively studied leaf movements and provided many examples of the free-running periods of leaf movement rhythms differing from 24 h (Pfeffer, 1915). That the rhythms were circadian and not exactly 24 h was an extremely important point because it was the best evidence, until experiments on the fungus Neurospora crassa were conducted in space (Sulzman et al., 1984), that these rhythms were truly endogenous and not driven by some subtle and undetected geophysical cue associated with the rotation of the earth on its axis.
The third key criterion of circadian rhythms is temperature compensation, and it took much longer for this attribute to become appreciated. The rationale for examining temperature dependence of the period length emerged from the expectation that the clock mechanism was based on alternating chemical processes. Thus, it was anticipated that, like chemical processes, the clock should exhibit marked temperature dependence. The rate of a typical chemical reaction doubles with a 10° increase in temperature (Q10 = 2). However, the period of leaf movement in P. coccineus exhibited a Q10 of only 1.2 (Bünning, 1931). By the 1960s, this observation had been extended to many other plants as well as to animals (Sweeney and Hastings, 1960). That the clocks were not temperature independent but, instead, exhibited less than expected temperature dependence strongly supported the concept of a temperature compensation mechanism that was imperfect. Consistent with this view was the observation of Q10 values of <1.0: an imperfect compensation mechanism could lengthen the period either insufficiently or too greatly at higher temperatures or, conversely, shorten the period too little or too much at lower temperatures.
As early as 1880, Charles and Francis Darwin suggested the heritability of circadian rhythms (Darwin and Darwin, 1880), as opposed to the imprinting of a 24-h period by exposure to diurnal cycles during development. This was initially explored in the 1930s by two strategies. In one, plants or animals were raised in constant conditions for multiple generations. One of the most grueling among such studies demonstrated the retention of stable rhythms among fruit flies reared in constant conditions for 700 generations (reviewed in Johnson, 2005). In a second strategy, seedlings or animals were exposed to cycles that differed from 24 h in an effort to imprint novel periods; such studies could sometimes impose the novel period length during the novel cycles, but upon release into continuous conditions, the endogenous circadian period was restored (Bünning, 1973). The inheritance of period length among progeny from crosses of parents with distinct period lengths was first reported in Phaseolus; hybrids had period length intermediates between those of the parents (Bünning, 1932, 1935).
Forward genetic analysis to identify components of circadian clocks began in the 1970s. Although now it seems axiomatic that circadian clocks are composed of the products of genes, just how this might be so was the source of considerable controversy. It was argued that forward genetic efforts would be fruitless because clocks were sufficiently complex to reasonably be expected to exhibit polygenic inheritance (Bünning, 1935) and would not yield easily to standard genetic approaches. However, mutations conferring altered period length were identified and characterized in the fruitfly Drosophila melanogaster (Konopka and Benzer, 1971), the green alga Chlamydomonas reinhardtii (Bruce, 1972), and the filamentous fungus N. crassa (Feldman and Hoyle, 1973). It took more than a decade to clone the first clock gene, the Drosophila period (per) gene (Bargiello and Young, 1984; Zehring et al., 1984), and another 5 years to clone the second, the Neurospora frequency gene (McClung et al., 1989). However, the decade of the 1990s saw rapid progress toward the identification of clock components and the elucidation of oscillator mechanisms central to the circadian clock in a number of organisms, most notably Drosophila, Neurospora, and mice (Dunlap, 1999).
In plants, it was realized that the leaf movement rhythm was only one among many rhythms that included germination, growth, enzyme activity, stomatal movement and gas exchange, photosynthetic activity, flower opening, and fragrance emission (Cumming and Wagner, 1968). However, genetic studies of plant clocks languished after Bünning's first experiments. Two critical discoveries changed this. First, Kloppstech (1985) described a circadian rhythm in pea in the abundance of three nuclear-encoded transcripts encoding the light-harvesting chlorophyll a/b binding protein (LHCB; also called CAB), the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, and an early light-induced protein. This observation was replicated and extended in wheat, where it was shown that the transcription rate for the Cab-1 gene was under circadian control (Nagy et al., 1988). Neither pea nor wheat was particularly suitable for positional gene cloning, but Arabidopsis thaliana was emerging as a powerful system in which to combine forward genetic analysis with molecular gene cloning techniques (Somerville and Koornneef, 2002). It was soon established that the transcription rate and transcript accumulation of Arabidopsis LHCB (Millar and Kay, 1991) and a number of other genes (McClung and Kay, 1994) were also under circadian control.
These initial Arabidopsis experiments were quite labor intensive, as tissues for RNA extraction, RNA gel blotting, and nuclear run-on analyses had to be harvested at frequent intervals over fairly lengthy time courses. Such experiments inevitably became exercises in sleep deprivation for the experimenters and provided considerable disincentive to the recruitment of graduate students into the field. Moreover, forward genetic analysis required a sensitive, reliable, and nondestructive assay that could score the circadian activity of individual seedlings without killing them. The luciferases offered a versatile class of noninvasive reporter genes. Firefly luciferase (LUC) catalyzes the ATP-dependent oxidative decarboxylation of luciferin with the concomitant release of a photon at 560 nm; this light emission can be quantified with luminometers or with sensitive charge-coupled device cameras (Welsh et al., 2005). Millar et al. (1992) demonstrated that a short fragment of the Arabidopsis LHCB1*3 (CAB2) promoter would drive rhythmic transcription and mRNA accumulation of LUC mRNA detectable as rhythmic light emission from individual Arabidopsis seedlings bearing the LHCB:LUC transgene. There was an element of luck in this, as it turned out that the LUC protein itself was quite stable, and bulk LUC protein failed to oscillate in abundance. However, LUC protein loses catalytic activity after only a few enzymatic cycles, with the net result that light production requires de novo LUC synthesis that is limited by transcript abundance. The LUC mRNA is sufficiently unstable that its accumulation tracks the transcription rate, which, when driven by the LHCB promoter, is rhythmic. After this initial demonstration in Arabidopsis, LUC use in circadian studies spread to other organisms, including Drosophila and mammals (Welsh et al., 2005).
The development of the LUC assay system permitted the first screen for Arabidopsis clock mutants. Arabidopsis seeds bearing the LHCB:LUC transgene were mutagenized, and M2 seedlings were screened to yield the first plant clock mutant, timing of cab expression1 (toc1-1; Millar et al., 1995b). The LHCB:LUC transgene also was introduced into various genetic backgrounds to provide a sensitive assay system to test mutants for effects on circadian function (Millar et al., 1995a).
ARABIDOPSIS DISPLAYS MANY CIRCADIAN RHYTHMS
Arabidopsis exhibits myriad rhythmic outputs or “hands” of the clock (McClung, 2001; McClung et al., 2002; Staiger, 2002). Like many plants, Arabidopsis displays rhythmic cotyledon and leaf movement, although this rhythm in Arabidopsis is based on differential growth and thus differs from the rhythmic turgor-driven expansion and contraction of the pulvinus that underlies rhythmic leaf movement in legumes, including Tamarindus and Mimosa (Kim et al., 1993). In Arabidopsis, there is a circadian rhythm in the elongation rate of the abaxial and adaxial cells of the petiole that confers an oscillation in position of cotyledons and leaves (Engelmann and Johnsson, 1998). Arabidopsis also exhibits a circadian rhythm in the rate of hypocotyl elongation (Dowson-Day and Millar, 1999) and in the elongation rate of inflorescence stem (Jouve et al., 1998).
Circadian control of transcription is widespread (Dunlap, 1999), and the list of plant genes regulated by the circadian clock is extensive. Microarray analyses suggest that ∼10% of all Arabidopsis genes regulated at the level of mRNA abundance and have identified multiple metabolic pathways under circadian control (Harmer et al., 2000; Schaffer et al., 2001). Circadian-regulated transcripts are enriched in the subset of transcripts with short half-lives (Gutierrez et al., 2002); it may be that high transcript stability may obscure some transcriptional oscillations when one simply monitors steady state transcript abundance. Indeed, enhancer trapping suggests that up to 35% of the transcriptome may show clock regulation (Michael and McClung, 2003).
Although the study of circadian rhythms has focused on constant conditions, it is important to remember that plants in nature grow in a changing world. In plants grown in diurnal cycles, there is an important interaction with sugar metabolism that strongly influences cycling gene expression (Bläsing et al., 2005). In addition, recent data make it clear that the circadian clock modulates the ability to respond to abiotic stresses, such as cold (Fowler et al., 2005). Clock modulation of response to biotic stresses has been the subject of speculation but remains to be established.
THE CURRENT CLOCK PARADIGM: INTERLOCKED FEEDBACK LOOPS
With the cloning of the Drosophila per gene, which encodes a novel protein of unknown function, the central question in clock research immediately became, “how can this gene product generate a circadian rhythm?” Negative feedback loops had been suspected to underlie the circadian clock, and several observations on per suggested that it might fit into such a loop. per mRNA abundance showed a circadian oscillation that was followed, with a lag of ∼4 h, by oscillations in PER protein (Hardin et al., 1990). As PER protein accumulated, per mRNA declined in abundance. This suggested a simple autoregulatory negative feedback loop: the clock gene is transcribed and the transcript is translated into a protein that accumulates in the nucleus to inhibit further transcription. Degradation of both mRNA and protein relieves this inhibition, and the cycle renews. This simple model has largely withstood the test of time, although it has increased in complexity. PER protein complexes with a second clock protein, TIMELESS (TIM), to inhibit the transcriptional activation of the per and tim promoters by a heterodimer of the dCLOCK (dCLK) and CYCLE (CYC) transcription factors. The dCLK/CYC heterodimer also activates the transcription of vrille and Pdp1ɛ, which encode negative and positive regulators, respectively, of clk transcription (Hardin, 2004). Thus, there are at least two interlocked feedback loops that include both positive and negative feedback. Positive components promote the transcription of negative components, and negative components play a dual role, blocking their own expression as well as increasing the expression of positive components, which interlocks the loops to create a robust sustained oscillation. In addition, a variety of posttranslational mechanisms, including nucleocytoplasmic localization, phosphorylation, and regulated protein degradation, affect clock function (Harms et al., 2004).
This paradigm of interlocked transcriptional/translational feedback loops underpins the molecular mechanisms of the circadian clock in all eukaryotes studied to date (Dunlap et al., 2004). However, the combination of components recruited to form the clock varies among organisms; the fungal clock is quite distinct from the animal clock, although fly and mouse clocks are fairly similar. It is also clear that cyanobacteria provide a stunning exception to the essential ubiquity of transcriptional regulation in clock function, as a temperature-compensated circadian rhythm can be reconstituted in vitro with three Synechococcus proteins and ATP (Nakajima et al., 2005; Tomita et al., 2005).
THE CURRENT PARADIGM APPLIED TO PLANTS: A MODEL OF THE PLANT OSCILLATOR
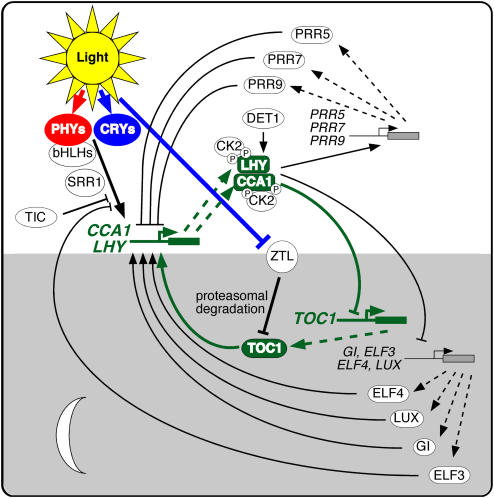
The sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) identified no obvious orthologs to most known clock proteins, which means that the Arabidopsis clock mechanism is novel, at least in terms of its composition. Nonetheless, the paradigm of interlocked feedback loops seems to be conserved. A number of recent reviews discuss the increasingly complex picture of the Arabidopsis clock (Salomé and McClung, 2004, 2005b; Harmon et al., 2005; Mizuno and Nakamichi, 2005). A simplified version of the Arabidopsis circadian clock is illustrated in Figure 3 (see also Table 1). It comprises three interlocked feedback loops, with two single Myb domain transcription factors, CIRCADIAN AND CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), playing roles in each loop. TOC1, the founding member of a family of pseudo-response regulators (PRRs), closes one loop, while three TOC1 paralogs, PRR5, PRR7, and PRR9, close a second loop. A third loop includes a Myb transcription factor, LUX ARRHYTHMO (LUX).
Figure 3.
A Molecular Model of the Arabidopsis thaliana Circadian Oscillator.
Genes are indicated by solid boxes with the gene names indicated at the left. Proteins are indicated by oval and oblong shapes, with the protein name indicated within the shape. Transcription and translation are indicated by dashed lines. Protein activity is indicated with solid lines, with lines ending in arrowheads indicating positive action and lines ending in perpendicular dashes indicating negative action. The core CCA1/LHY/TOC1 feedback loop is highlighted in green with thick lines and closed shapes. Phosphorylation of LHY and CCA1 by CK2 is indicated with circled Ps. Shaded area indicates activities peaking in the subjective night, and white area indicates activities peaking during the subjective day.
Table 1.
Known Arabidopsis Genes with Clock Functions
| Circadian Clock Phenotype
|
||||
|---|---|---|---|---|
| Gene | Locus ID | Function | Loss of Function | Overexpression |
| CCA1 | At2g46830 | Single Myb domain transcription factor | Short period | Arrhythmic |
| CKB3 | At3g60250 | Casein kinase II regulatory subunit | Not known (gene family) | Short period |
| CRY1 | At4g08920 | Blue light photoreceptor | Long period in blue light | Short period in blue light |
| CRY2 | At1g04400 | Blue light photoreceptor | Long period in blue light | Short period in blue light |
| DET1 | At4g10180 | Repressor of photomorphogenesis | Short period | Not known |
| ELF3 | At2g25930 | Unknown | Arrhythmic in continuous light | Long period |
| ELF4 | At2g40080 | Unknown | Arrhythmic | Not known |
| GI | At1g22770 | Unknown | Short period, low amplitude | Short period, low amplitude |
| LHY | At1g01060 | Single Myb domain transcription factor | Short period | Arrhythmic |
| LUX | At3g46640 | Myb transcription factor | Arrhythmic | Arrhythmic |
| PHYA | At1g09570 | Red light photoreceptor | Long period in far-red light | Short period in far-red light |
| PHYB | At2g18790 | Red light photoreceptor | Long period in red light, leading phase in white light | Short period in red light, lagging phase in white light |
| PIF3 | At1g09530 | Basic helix-loop-helix transcription factor | Wild type | Wild type |
| PRR3 | At5g60100 | Pseudo-response regulator | Short period | Wild type |
| PRR5 | At5g24470 | Pseudo-response regulator | Short period | Low amplitude, long period |
| PRR7 | At5g02810 | Pseudo-response regulator | Long period | Not known |
| PRR9 | At2g46790 | Pseudo-response regulator | Long period | Short period |
| SRR1 | At5g59560 | Unknown | Leading phase, low amplitude | Not known |
| TIC | Gene not yet identified | Short period, low amplitude | Not known | |
| TOC1 | At5g61380 | Pseudo-response regulator | Short period | Arrhythmic |
| ZTL | At5g57360 | F-box protein | Long period | Arrhythmic |
How do we conclude this? A toc1 loss-of-function mutant was identified as a short period mutant through a forward genetic screen, as described above. If the oscillating mRNA and protein abundance of a clock component, such as TOC1, is necessary for oscillator function but becomes pegged at a constant high level through overexpression, arrhythmicity should result. Indeed, TOC1 overexpressors are arrhythmic (Makino et al., 2002; Más et al., 2003b). CCA1 was identified initially as binding to the LHCB1*3 promoter. Loss of CCA1 function causes short period (Green and Tobin, 1999), but its overexpression causes arrhythmicity (Wang and Tobin, 1998), suggesting that it too is a core clock component. LHY, CCA1's closest paralog in Arabidopsis, was identified in a screen for late-flowering mutants. The allele identified in a screen of transposon-tagged mutants turned out to be overexpressed, which conferred arrhythmicity (Schaffer et al., 1998; Wang and Tobin, 1998). lhy loss of function, like that of cca1, confers short period, but the cca1 lhy double mutant is arrhythmic, suggesting that they are core clock components that function redundantly (Alabadí et al., 2002; Mizoguchi et al., 2002).
How these genes form an oscillator loop is not completely understood. CCA1 and LHY bind to the TOC1 promoter, and overexpression of either results in low levels of TOC1 expression, consistent with their roles as negative regulators of TOC1. TOC1 is inferred to be a positive regulator because expression of CCA1 and LHY is greatly reduced in a severe toc1-2 mutant (Alabadí et al., 2001, 2002; Matsushika et al., 2002b; Mizoguchi et al., 2002; Harmer and Kay, 2005; Hazen et al., 2005). Although TOC1 overexpression results in arrhythmicity, neither CCA1 nor LHY expression levels are dramatically elevated (Makino et al., 2002). TOC1 contains a CCT (for CONSTANS, CONSTANS-LIKE, TOC1) domain thought to be involved in transcription (Strayer et al., 2000) but has not been shown to bind to either CCA1 or LHY promoters. It seems that TOC1 on its own is insufficient for expression of CCA1 and LHY. Several other genes, including GIGANTEA (GI), EARLY FLOWERING3 (ELF3), ELF4, and LUX, are required for CCA1 and LHY expression (Park et al., 1999; Doyle et al., 2002; Mizoguchi et al., 2002; Hazen et al., 2005).
In other systems, the oscillator has been shown to include multiple interlocked feedback loops. Consistent with this paradigm, modeling studies show that available data cannot be accounted for within a single feedback loop (Locke et al., 2005). At least two other loops are thought to interlock with the TOC1/CCA1/LHY loop. Locke et al. (2005) proposed a second loop in which TOC1 is activated by a hypothetical evening-expressed protein that itself is repressed by TOC1 and demonstrated that GI behavior was consistent with that predicted for this hypothetical component. A number of investigators have proposed a third loop. CCA1 and LHY are positive regulators of three TOC1 relatives, PRR5, PRR7, and PRR9 (Farré et al., 2005; Harmer and Kay, 2005; Mizuno and Nakamichi, 2005). PRR5/7/9 are negative regulators of CCA1/LHY because CCA1 and LHY transcripts accumulate in prr7 and prr7 prr9 mutants (Farré et al., 2005), and CCA1 is constitutively transcribed in the arrhythmic prr5 prr7 prr9 triple mutant (Nakamichi et al., 2005b). PRR5/7/9 and TOC1 are thought to be mutually repressive (Mizuno and Nakamichi, 2005). Loss of function of prr7 or prr9 causes period lengthening, while loss of function of prr5 causes period shortening (Kaczorowski and Quail, 2003; Michael et al., 2003). The circadian phenotypes of the single prr mutants are small (period changes of 1 to 1.5 h) compared with the period shortening (3 to 4 h) seen in toc1-2 mutants, but redundancy among the PRRs may partially account for this. The phenotype of the prr7 prr9 double mutant is more than additive; the period lengthening is dramatically increased, and the double mutant is arrhythmic in the dark (Farré et al., 2005; Nakamichi et al., 2005a; Salomé and McClung, 2005a). Emphasizing the centrality of the PRRs to clock function, the triple prr5 prr7 prr9 mutant is essentially arrhythmic under all conditions tested (Nakamichi et al., 2005b). However, overexpression of PRR3, PRR5, or PRR9 has only small period effects (Matsushika et al., 2002a; Sato et al., 2002; Murakami et al., 2004), suggesting that additional factors are required for full PRR function.
Transcriptional regulation is important in clock function, but it is clear that posttranscriptional regulation is an essential constituent of the clock mechanism. While incompletely understood, casein kinase II (CK2) phosphorylates CCA1 and LHY, and overexpression of the regulatory β3 subunit (CKB3) confers short period (Sugano et al., 1998, 1999). CK2-mediated phosphorylation of CCA1 is necessary for in vivo function (Daniel et al., 2004). LHY is degraded via the proteasome, and this is accelerated in det1-1, providing a molecular explanation of the period-shortening effect of this mutation (Song and Carré, 2005), although a role for phosphorylation in degradation remains possible. Recently, a second type of posttranslational modification has been implicated in clock function. SPINDLY is an N-acetylglucosamine transferase that decorates GI, among other targets, and spy mutants exhibit altered rhythms in leaf movement (Tseng et al., 2004).
The identification of a novel family of proteins, ZEITLUPE (ZTL), LOV KELCH PROTEIN2 (LKP2), and FLAVIN binding KELCH REPEAT F-BOX (FKF), with PAS/LOV domains, Kelch repeats, and F-boxes (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001), has contributed to our understanding of the role of protein degradation in the Arabidopsis circadian clock. The LOV domains are similar to those of phototropins, and the LOV domain of FKF is photoactive (Imaizumi et al., 2003). FKF seems restricted to photoperiodism (Nelson et al., 2000), but ZTL and LKP2 affect the clock (Somers et al., 2000; Jarillo et al., 2001; Schultz et al., 2001). ztl-1 and ztl-2 mutants are affected in the period length of numerous rhythms (Somers et al., 2000, 2004; Dodd et al., 2004). ZTL mRNA abundance is not clock regulated, but ZTL protein levels peak around dusk, while trough levels are reached around dawn (Kim et al., 2003). The rate of proteasome-mediated degradation of ZTL varies during the course of the day: ZTL is more stable at dusk, around its peak value, and is more rapidly degraded at dawn when it reaches its trough. F-box proteins provide specificity to proteasomal degradation pathways by specific interaction with and polyubiquitination of targets for degradation. In this case, ZTL is a component of an SCF complex that recruits TOC1 for proteasomal degradation (Somers et al., 2000; Más et al., 2003a; Han et al., 2004). In the ztl mutant, protein levels of TOC1 are elevated and only weakly rhythmic, demonstrating that ZTL is critical for degradation of TOC1. Increasing expression of ZTL confers corresponding dosage-dependent period shortening (Han et al., 2004). Collectively, these data argue that the level of TOC1 activity, as regulated through transcriptional repression by CCA1 and LHY and via protein degradation by ZTL, is a key determinant of circadian period.
ENTRAINMENT
Although chronobiologists commonly study rhythms in constant conditions, organisms live in the cycling world of day and night. The two chief entraining stimuli that synchronize the endogenous clock with the exogenous temporal environment are light and temperature (Millar, 2004; Salomé and McClung, 2005b). Both phytochromes and cryptochromes provide light input to the clock, although the signal transduction pathways are incompletely defined. Interestingly, photoreceptor expression is itself rhythmic, indicating that the clock gates its sensitivity to light (e.g., Tóth et al., 2001), although bulk phytochrome protein levels do not oscillate (Sharrock and Clack, 2002). Light input is negatively regulated by ELF3; loss-of-function alleles of elf3 yield conditional arrhythmicity in continuous light but remain rhythmic in the dark (Hicks et al., 1996, 2001; McWatters et al., 2000; Covington et al., 2001; Liu et al., 2001). TIME FOR COFFEE (TIC) may have a similar effect on gating light input, although during a distinct phase; the tic elf3 double mutant is fully arrhythmic in light or dark (Hall et al., 2003). The period alteration of ztl mutants shows fluence rate dependence, suggesting a role for ZTL in light input (Somers et al., 2000).
It seems reasonable that both dawn and dusk provide important entraining cues. Possibly, the dusk signal involves relief from light repression of TOC1 degradation mediated by SCFZTL. The dawn cue likely involves induction of CCA1 and LHY and other clock components, although we lack a detailed mechanistic understanding of this signaling. Light input is positively regulated by SENSITIVITY TO RED LIGHT REDUCED1 (SRR1), which also plays an as yet undefined role in the core oscillator (Staiger et al., 2003). The basic helix-loop-helix transcription regulator, PHYTOCHROME-INTERACTING FACTOR3 (PIF3) induces CCA1 and LHY expression via binding to the G-box (Martínez-García et al., 2000). Phytochrome B (PHYB) in its active form binds specifically and reversibly to DNA-bound PIF3, suggesting a direct link from light perception to modification of the negative limb of the circadian clock (Martínez-García et al., 2000). However, loss of function of PIF3 does not affect period length of rhythmic gene expression (Monte et al., 2004). It is also important to note that the conclusive resetting of the clock by transient expression of CCA1 or LHY has not been demonstrated nor has it been definitively shown that levels of CCA1 or LHY set phase (Salomé and McClung, 2005b).
Temperature signaling to the clock is much less well defined. Abundant evidence supports the importance of temperature cycles in clock entrainment. Temperature steps as small as 0.5°C can entrain the Kalanchoë clock, showing the exquisite sensitivity of the system (Rensing and Ruoff, 2002). In Arabidopsis, gene expression and cotyledon movement can be entrained by temperature cycles (Michael and McClung, 2002; Salomé et al., 2002; Salomé and McClung, 2005a), but the mechanism of action is currently unknown. It has been established that PRR7 and PRR9 are important as the prr7 prr9 double mutant fails to entrain to temperature cycles that effectively entrain the wild type (Salomé and McClung, 2005a).
Considerable natural variation in temperature compensation has been described, and GI has been identified as a quantitative trait locus responsible for a substantial portion of that variation (Edwards et al., 2005). It seems likely that this clock property will prove amenable to forward and reverse genetic approaches.
CIRCADIAN CLOCKS AND PHOTOPERIODISM
The role of photoperiod (daylength) in controlling seasonal responses was noted early in the 20th century (Tournois, 1912; Klebs, 1913). Garner and Allard (1920) demonstrated that many plants flower in response to changes in daylength. The connection between photoperiodism and the circadian clock was first noted by Bünning (1936) and was developed into the external coincidence model, in which a rhythmic process that controls the photoperiodic response is sensitive to light at certain times of day (Pittindrigh and Minis, 1964).
Flowering of Arabidopsis is accelerated in long days, and the mechanism by which this occurs is becoming clear (for review, see Corbesier and Coupland, 2005). Briefly, a key promoter of flowering is CONSTANS (CO). The transcription of the CO gene is clock regulated so that CO mRNA only accumulates late in the day. At least in part, this is because the clock-regulated F-box protein FKF controls the stability of a cycling Dof transcription factor, CDF1, which is a repressor of CO transcription (Imaizumi et al., 2005). FKF has a photoactive LOV domain and likely serves as a photoperiodic blue-light receptor (Imaizumi et al., 2003). Once CDF1 is degraded, CO transcription ensues. However, CO protein is unstable and fails to accumulate in the dark. Light perception via CRY2 and PHYA stabilizes CO (Valverde et al., 2004) in long days when CO mRNA accumulates and is translated in the light but not in short days when CO mRNA only accumulates and is translated after dusk. Thus, CO protein accumulates to activate its target, FLOWERING LOCUS T, in long but not in short days (Suárez-López et al., 2001; Yanovsky and Kay, 2002). Flowering is only promoted in Arabidopsis when there is the proper coincidence of the internal oscillation in CO transcription and subsequent translation with the external oscillation in light. Excitingly, this model applies to rice, a short-day plant—the salient difference seems to be that CO serves as a floral repressor in that species (Hayama and Coupland, 2004).
ADAPTIVE FITNESS CONFERRED BY CIRCADIAN CLOCKS
It has long been presumed that the ability to anticipate light/dark cycles gives organisms a fitness advantage. One long-standing idea, termed the escape from light hypothesis, posits that organisms would accrue advantage from phasing light-sensitive processes, such as DNA replication, to the dark portion of the daily cycle (Pittendrigh, 1993). In cyanobacteria, competitive ability depends on the correspondence between a strain's free-running period and ambient daylength; wild-type strains outcompete either long- or short-period mutants when grown in 24-h days (12 h light/12 h dark). This does not reflect a competitive advantage to the wild type under all conditions because long-period (30-h period) mutants outcompete the wild type (25-h period) when grown in long cycles (15 h light/15 h dark) (Johnson, 2005).
Early studies in tomato showed that growth improved on light/dark cycles of 24 h rather than short (6 h light/6 h dark) or long (24 h light/24 h dark) cycles or continuous light (Withrow and Withrow, 1949; Highkin and Hanson, 1954; Hillman, 1956), although this work only indirectly implicates the circadian clock in the growth response. More direct testing has come in recent years. Arabidopsis clock mutants with longer than normal periods (28 h) have lower biomass than those with short periods (20 h) when grown under short cycles (10 h light/10 h dark), and these differences in size are largely attributable to impaired physiological function, including lower rates of chlorophyll production and carbon fixation (Green et al., 2002; Dodd et al., 2005).
In Arabidopsis, there is considerable circadian variation among natural genotypes (for examples, see Swarup et al., 1999; Michael et al., 2003; Edwards et al., 2005). There is a positive correlation between period length of a set of natural accessions and daylength encountered at latitude of origin of the accessions (Michael et al., 2003), which may indicate a selection of altered clock function under differing environmental conditions (temperature and daylength covary with latitude). In addition to the effects of period length on carbon fixation, biomass, and survival described above (Dodd et al., 2005), period length may also affect the flowering timing response (for example, see Yanovsky and Kay, 2002). Under the entraining conditions of a light/dark cycle, the period is 24 h. The effects of a long endogenous circadian period are seen as a lagging phase under entraining conditions. Thus, lengthening the period would delay the accumulation of CO mRNA, which would increase the critical daylength required for the accumulation of CO protein. This would delay flowering until the longer days encountered later in the season, which could be advantageous at higher latitudes where daylength increases rapidly and precedes the cessation of freezing weather. Altered temperature compensation might also underlie this latitudinal cline; there is a similar latitudinal cline in a polymorphism in the Drosophila per gene that is thought to be related to altered temperature compensation properties conferred by the per alleles (Costa et al., 1992; Sawyer et al., 1997).
CRITICAL QUESTIONS THAT REMAIN
The progress achieved in the last 15 years toward unraveling the plant circadian clock mechanism is remarkable, but much remains unfinished. An outline of the oscillator mechanism has emerged but remains incomplete. Although we can safely conclude that the paradigm of interlocked feedback loops constituting a circadian oscillator is conserved in plants, not all the components have yet been identified, and the mechanistic details of almost every step are only incompletely understood. It is humbling that, after so much effort and progress, almost all questions remain only incompletely answered and, effectively, all questions remain! Moreover, the field is now expanding its view from the purely reductionist goal of identifying the oscillator itself to a consideration of the evolutionary and ecological consequences of variation in clock function, so a host of new questions are being considered. It is exhilarating to consider what a retrospective view a decade from now will reveal.
Acknowledgments
I thank Patrice Salomé (Dartmouth College, Hanover, NH) for much data, for innumerable thought-provoking conversations, and for critical reading of this manuscript. I also thank Ann Lavanway (Dartmouth College) for help with Figure 2. I apologize to all authors whose work was omitted due to the length limitations. Work in my laboratory on circadian rhythms is supported by a grant from the National Science Foundation (MCB-0343887).
References
- Alabadí, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Más, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadí, D., Yanovsky, M.J., Más, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bargiello, T.A., and Young, M.W. (1984). Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. USA 81 2142–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing, O.E., Gibon, Y., Günther, M., Höhne, M., Morcuende, R., Osuna, D., Thimm, O., Usadel, B., Scheible, W.-R., and Stitt, M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretzl, H. (1903). Botanische Forschungen des Alexanderzuges. (Leipzig, Germany: B.G. Teubner).
- Bruce, V.G. (1972). Mutants of the biological clock in Chlamydomonas reinhardi. Genetics 70 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünning, E. (1931). Untersuchungen über die autonomen tagesperiodischen Bewungen der Primärblätter von Phaseolus multiflorus. Jahrb. Wiss. Bot. 75 439–480. [Google Scholar]
- Bünning, E. (1932). Über die Erblichket der Tagesperiodizitat bei den Phaseolus-Blättern. Jahrb. Wiss. Bot. 77 283–320. [Google Scholar]
- Bünning, E. (1935). Zur Kenntnis der erblichen Tagesperioizität bei den Primärblätter von Phaseolus multiflorus. Jahrb. Wiss. Bot. 81 411–418. [Google Scholar]
- Bünning, E. (1936). Die endogene Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber. Dtsch. Bot. Ges. 54 590–607. [Google Scholar]
- Bünning, E. (1960). Opening address. Biological Clocks. Cold Spring Harb. Symp. Quant. Biol. 25 1–9. [Google Scholar]
- Bünning, E. (1973). The Physiological Clock. 3rd ed. (New York: Springer-Verlag).
- Corbesier, L., and Coupland, G. (2005). Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 28 54–66. [Google Scholar]
- Costa, R., Peixoto, A.A., Barbujani, G., and Kyriacou, C.P. (1992). A latitudinal cline in a Drosophila clock gene. Proc. R. Soc. Lond. B. Biol. Sci. 250 43–49. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming, B.G., and Wagner, E. (1968). Rhythmic processes in plants. Annu. Rev. Plant Physiol. 19 381–416. [Google Scholar]
- Daniel, X., Sugano, S., and Tobin, E.M. (2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C., and Darwin, F. (1880). The Power of Movement in Plants. (London: J. Murray).
- de Candolle, A.P. (1832). Physiologie Végétale. (Paris: Bechet Jeune).
- de Mairan, J. (1729). Observation botanique. Hist. Acad. Roy. Sci. 35–36.
- Dodd, A.N., Parkinson, K., and Webb, A.A.R. (2004). Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol. 162 63–70. [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., Hibberd, J.M., Millar, A.J., and Webb, A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17 63–71. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77. [DOI] [PubMed] [Google Scholar]
- Duhamel duMonceau, H.L. (1759). La Physique des Arbres. (Paris: H.L. Guerin and L.F. Delatour).
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96 271–290. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C., Loros, J.J., and DeCoursey, P. (2004). Chronobiology: Biological Timekeeping. (Sunderland, MA: Sinauer Associates).
- Edwards, K.D., Lynn, J.R., Gyula, P., Nagy, F., and Millar, A.J. (2005). Natural allelic variation in the temperature compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, W., and Johnsson, A. (1998). Rhythms in organ movement. In Biological Rhythms and Photoperiodism in Plants, P.J. Lumsden and A.J. Millar, eds (Oxford: BIOS Scientific Publishers), pp. 35–50.
- Farré, E.M., Harmer, S.L., Harmon, F.G., Yanovsky, M.J., and Kay, S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15 47–54. [DOI] [PubMed] [Google Scholar]
- Feldman, J.F., and Hoyle, M. (1973). Isolation of circadian clock mutants of Neurospora crassa. Genetics 75 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S.G., Cook, D., and Thomashow, M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, W.W., and Allard, H.A. (1920). Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J. Agric. Res. 18 553–606. [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.-Y., and Tobin, E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., and Tobin, E.M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, R.A., Ewing, R.M., Cherry, J.M., and Green, P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L., Mason, M., Risseeuw, E.P., Crosby, W.L., and Somers, D.E. (2004). Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 40 291–301. [DOI] [PubMed] [Google Scholar]
- Hardin, P.E. (2004). Transcription regulation within the circadian clock: The E-box and beyond. J. Biol. Rhythms 19 348–360. [DOI] [PubMed] [Google Scholar]
- Hardin, P.E., Hall, J.C., and Rosbash, M. (1990). Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343 536–540. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., and Kay, S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, F., Imaizumi, T., and Kay, S. (2005). The plant circadian clock: Review of a clockwork Arabidopsis. In Endogenous Plant Rhythms, A. Hall and H. McWatters, eds (Oxford: Blackwell Scientific Publications), pp. 1–23.
- Harms, E., Kivimae, S., Young, M.W., and Saez, L. (2004). Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19 361–373. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Albertson, T.M., and Wagner, D.R. (2001). EARLY FLOWERING 3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Highkin, H.R., and Hanson, J.B. (1954). Possible interactions between light-dark cycles and endogenous daily rhythms on the growth of tomato plants. Plant Physiol. 29 301–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, H.D. (1757). The Sleep of Plants. (London: R. Baldwin).
- Hillman, W.S. (1956). Injury of tomato plants by continuous light and unfavorable photoperiodic cycles. Am. J. Bot. 43 89–96. [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Tang, R.-H., Yang, H.-Q., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and PHYB. Nature 410 487–490. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H. (2005). Testing the adaptive value of circadian systems. Methods Enzymol. 393 818–837. [DOI] [PubMed] [Google Scholar]
- Jouve, L., Greppin, H., and Agosti, R.D. (1998). Arabidopsis thaliana floral stem elongation: Evidence for an endogenous circadian rhythm. Plant Physiol. Biochem. 36 469–472. [Google Scholar]
- Kaczorowski, K.A., and Quail, P.H. (2003). Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15 2654–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesel, A. (1894). Untersuchungen zur Physiologie des facettierten Auges. Sitzungsber. Akad. Wiss. Wien 103 97–139.
- Kim, H.Y., Coté, G.G., and Crain, R.C. (1993). Potassium channels in Samanea saman protoplasts controlled by phytochrome and the biological clock. Science 260 960–962. [DOI] [PubMed] [Google Scholar]
- Kim, W.-Y., Geng, R., and Somers, D.E. (2003). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. USA 100 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebs, G. (1913). Über das Verhåltnis der Aussenwelt zur Entwicklung der Pflanze. Heidelb. Acad. Wiss. 5 1–47. [Google Scholar]
- Kloppstech, K. (1985). Diurnal and circadian rhythmicity in the expression of light-induced nuclear messenger RNAs. Planta 165 502–506. [DOI] [PubMed] [Google Scholar]
- Konopka, R., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock–regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C.W., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (June 28, 2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1 (online), doi/10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- Makino, S., Matsushika, A., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43 58–69. [DOI] [PubMed] [Google Scholar]
- Martínez-García, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863. [DOI] [PubMed] [Google Scholar]
- Más, P., Alabadí, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003. b). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más, P., Kim, W.-Y., Somers, D.E., and Kay, S.A. (2003. a). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Imamura, A., Yamashino, T., and Mizuno, T. (2002. a). Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol. 43 833–843. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., Yamashino, T., and Mizuno, T. (2002. b). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 43 118–122. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2001). Circadian rhythms in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 139–162. [DOI] [PubMed] [Google Scholar]
- McClung, C.R., Fox, B.A., and Dunlap, J.C. (1989). The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature 339 558–562. [DOI] [PubMed] [Google Scholar]
- McClung, C.R., and Kay, S.A. (1994). Circadian rhythms in Arabidopsis thaliana. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 615–637.
- McClung, C.R., Salomé, P.A., and Michael, T.P. (2002). The Arabidopsis circadian system. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0044, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408 716–720. [DOI] [PubMed] [Google Scholar]
- Michael, T.P., and McClung, C.R. (2002). Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol. 130 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., and McClung, C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis thaliana. Plant Physiol. 132 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., Salomé, P.A., Yu, H.J., Spencer, T.R., Sharp, E.L., Alonso, J.M., Ecker, J.R., and McClung, C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053. [DOI] [PubMed] [Google Scholar]
- Millar, A.J. (2004). Input signals to the plant circadian clock. J. Exp. Bot. 55 277–283. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carré, I.A., Strayer, C.A., Chua, N.-H., and Kay, S.A. (1995. b). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., and Kay, S.A. (1991). Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Short, S.R., Chua, N.-H., and Kay, S.A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Straume, M., Chory, J., Chua, N.-H., and Kay, S.A. (1995. a). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267 1163–1166. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.-R., Carré, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641. [DOI] [PubMed] [Google Scholar]
- Mizuno, T., and Nakamichi, N. (2005). Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 46 677–685. [DOI] [PubMed] [Google Scholar]
- Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., and Quail, P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, M., Yamashino, T., and Mizuno, T. (2004). Characterization of circadian-associated APRR3 Pseudo-Response Regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 45 645–650. [DOI] [PubMed] [Google Scholar]
- Nagy, F., Kay, S.A., and Chua, N.-H. (1988). A circadian clock regulates transcription of the wheat Cab-1 gene. Genes Dev. 2 376–382. [Google Scholar]
- Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., and Kondo, T. (2005). Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308 414–415. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005. a). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46 609–619. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005. b). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46 686–698. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340. [DOI] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Pfeffer, W. (1873). Physiologische Untersuchungen. (Leipzig, Germany: W. Engelmann).
- Pfeffer, W. (1915). Beiträge zur Kenntnis der Entstehung der Schlafbewegungen. Abh. Sächs. Akad. Wiss. Math. Physiol. Kl. 34 1–154. [Google Scholar]
- Pittendrigh, C.S. (1954). On the temperature independence in the clock system controlling emergence time in Drosophila. Proc. Natl. Acad. Sci. USA 40 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C.S. (1993). Temporal organization: Reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55 17–54. [DOI] [PubMed] [Google Scholar]
- Pittindrigh, C.S., and Minis, D.H. (1964). The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am. Nat. 98 261–322. [Google Scholar]
- Rensing, L., and Ruoff, P. (2002). Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol. Int. 19 807–864. [DOI] [PubMed] [Google Scholar]
- Richter, C.P. (1922). A behavioristic study of the activity of the rat. Comp. Psychol. Monogr. 1 1–55. [Google Scholar]
- Salomé, P.A., and McClung, C.R. (2004). The Arabidopsis thaliana clock. J. Biol. Rhythms 19 425–435. [DOI] [PubMed] [Google Scholar]
- Salomé, P.A., and McClung, C.R. (2005. a). PRR7 and PRR9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé, P.A., and McClung, C.R. (2005. b). What makes Arabidopsis tick: Light and temperature entrainment of the circadian clock. Plant Cell Environ. 28 21–38. [Google Scholar]
- Salomé, P.A., Michael, T.P., Kearns, E.V., Fett-Neto, A.G., Sharrock, R.A., and McClung, C.R. (2002). The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, E., Nakamichi, N., Yamashino, T., and Mizuno, T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43 1374–1385. [DOI] [PubMed] [Google Scholar]
- Sawyer, L.A., Hennessy, J.M., Peixoto, A.A., Rosato, E., Parkinson, H., Costa, R., and Kyriacou, C.P. (1997). Natural variation in a Drosophila clock gene and temperature compensation. Science 278 2117–2120. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schultz, T.F., Kiyosue, T., Yanovsky, M., Wada, M., and Kay, S.A. (2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock, R.A., and Clack, T. (2002). Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel, D. (1995). Longitude: The True Story of a Lone Genius Who Solved the Greatest Scientific Problem of His Time. (New York: Walker Publishing Company).
- Somers, D.E., Kim, W.Y., and Geng, R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Somerville, C., and Koornneef, M. (2002). A fortunate choice: The history of Arabidopsis as a model plant. Nat. Rev. Genet. 3 883–889. [DOI] [PubMed] [Google Scholar]
- Song, H.-R., and Carré, I.A. (2005). DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 57 761–771. [DOI] [PubMed] [Google Scholar]
- Staiger, D. (2002). Circadian rhythms in Arabidopsis: Time for nuclear proteins. Planta 214 334–344. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sugano, S., Andronis, C., Green, R.M., Wang, Z.-Y., and Tobin, E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 95 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S., Andronis, C., Ong, M.S., Green, R.M., and Tobin, E.M. (1999). The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 12362–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzman, F.M., Ellman, D., Fuller, C.A., Moore-Ede, M.C., and Wassmer, G. (1984). Neurospora circadian rhythms in space: A reexamination of the endogenous-exogenous question. Science 225 232–234. [DOI] [PubMed] [Google Scholar]
- Swarup, K., Alonso-Blanco, C., Lynn, J.R., Michaels, S.D., Amasino, R.M., Koornneef, M., and Millar, A.J. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20 67–77. [DOI] [PubMed] [Google Scholar]
- Sweeney, B., and Hastings, J.W. (1960). Effects of temperature upon diurnal rhythms. Cold Spring Harb. Symp. Quant. Biol. 25 87–104. [DOI] [PubMed] [Google Scholar]
- Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307 251–253. [DOI] [PubMed] [Google Scholar]
- Tóth, R., Kevei, É., Hall, A., Millar, A.J., Nagy, F., and Kozma-Bognár, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournois, J. (1912). Influence de la lumière sur la floraison du houblon japonais et du chanvre déterminées par des semis haitifs. C. R. Acad. Sci. Paris 155 297–300. [Google Scholar]
- Tseng, T.-S., Salomé, P.A., McClung, C.R., and Olszewski, N.E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering and rhythms in leaf movements. Plant Cell 16 1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Welsh, D.K., Imaizumi, T., and Kay, S.A. (2005). Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 393 269–288. [DOI] [PubMed] [Google Scholar]
- Withrow, A.P., and Withrow, R.B. (1949). Photoperiodic chlorosis in tomato. Plant Physiol. 24 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Zehring, W., Wheeler, D., Reddy, P., Rosbash, M., and Hall, J. (1984). P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39 369–376. [DOI] [PubMed] [Google Scholar]
- Zinn, J.G. (1759). Von dem Schlafe der Pflanzen. Hamburg. Mag. 22 40–50. [Google Scholar]