Abstract
To gain insight into the processes involved in photosystem II (PSII) biogenesis and maintenance, we characterized the low psii accumulation1 (lpa1) mutant of Arabidopsis thaliana, which generally accumulates lower than wild-type levels of the PSII complex. In vivo protein labeling experiments showed that synthesis of the D1 and D2 proteins was greatly reduced in the lpa1 mutant, while other plastid-encoded proteins were translated at rates similar to the wild type. In addition, turnover rates of the PSII core proteins CP47, CP43, D1, and D2 were higher in lpa1 than in wild-type plants. The newly synthesized PSII proteins were assembled into functional protein complexes, but the assembly was less efficient in the mutant. LPA1 encodes a chloroplast protein that contains two tetratricopeptide repeat domains and is an intrinsic membrane protein but not an integral subunit of PSII. Yeast two-hybrid studies revealed that LPA1 interacts with D1 but not with D2, cytochrome b6, or Alb3. Thus, LPA1 appears to be an integral membrane chaperone that is required for efficient PSII assembly, probably through direct interaction with the PSII reaction center protein D1.
INTRODUCTION
Photosystem II (PSII) is a multisubunit pigment–protein complex that catalyzes the light-driven water oxidation and reduction of plastoquinone. The PSII reaction center is composed of the D1 and D2 proteins, the α- and β-subunits of cytochrome b559 and the PsbI protein. The D1 and D2 heterodimer binds all the essential redox components of PSII required to transfer electrons from the manganese cluster to the plastoquinone pool (Nanba and Satoh, 1987). In addition, oxygen-evolving PSII complexes contain the intrinsic chlorophyll a binding proteins (CP43 and CP47), the oxygen-evolving complex (33-, 23-, and 17-kD proteins), and several low molecular mass proteins (Shi and Schröder, 2004). In recent years, knowledge of the supramolecular organization of PSII has been increasing rapidly, as increasingly high-resolution crystallographic data have been published (Ferreira et al., 2004; Loll et al., 2005).
Although the structure and function of PSII have been extensively examined, our knowledge of the assembly of PSII is still limited. Studies on PSII subunit-specific mutants have shown that the accumulation of D1, D2, and CP47 appears to be coordinated (Jensen et al., 1986; de Vitry et al., 1989; Yu and Vermaas, 1990). In addition, a number of analyses have indicated that an initial complex (probably consisting of D2 and cytochrome b559) serves as a receptor for other PSII core proteins (Adir et al., 1990; van Wijk et al., 1997; Müller and Eichacker, 1999; Zhang et al., 1999). Furthermore, synthesis and integration of the D1 protein appears to involve cotranslational interaction with the D2 protein during the PSII repair process (Zhang et al., 1999; Zhang and Aro, 2002). This process has proved to be highly valuable for analyses of PSII assembly and maintenance due to the high light-dependent turnover rates of the chloroplast proteins associated with it (Chua and Gillham, 1977; Greenberg et al., 1987; Gong and Ohad, 1991; for reviews, see Prasil et al., 1992; Aro et al., 1993; Baena-González and Aro, 2002). Because of the structural complexity of PSII, biogenesis and assembly of PSII require the coordinated synthesis and assembly of both chloroplast- and nuclear-encoded proteins. These are likely to be multistep processes, which are assisted by many nuclear-encoded auxiliary and regulatory proteins. Until recently, the factors involved in regulating the assembly of PSII have remained largely unknown. Protein labeling analyses have shown that plastid-encoded proteins are synthesized in the hcf136 Arabidopsis thaliana mutant, which lacks functional HCF136 protein, but assembly of PSII reaction centers is blocked, and no stable PSII complexes appear to accumulate in it (Meurer et al., 1998; Plücken et al., 2002). These findings strongly indicate that the HCF136 protein is required for the stability and/or assembly of PSII (Meurer et al., 1998; Plücken et al., 2002). In addition, Alb3.1, a homolog of Arabidopsis Alb3, has been found to be required for the efficient assembly of functional PSII in Chlamydomonas reinhardtii, although the integration of D1 into the thylakoid membrane was not affected (Ossenbühl et al., 2004).
Genetic techniques have provided powerful tools for dissecting the molecular mechanisms underlying the biogenesis of thylakoid membrane protein complexes. To identify the genes involved in these processes, several strategies have been applied to isolate photosynthetic mutants, with phenotypic characteristics such as aberrations in leaf pigmentation, seedling lethality, and/or fluorescence kinetics (Bennoun and Delepelaire, 1982; Miles, 1982; Meurer et al., 1996; Niyogi et al., 1997; Shikanai et al., 1999; Varotto et al., 2000; Budziszewski et al., 2001). Screening mutants with altered chlorophyll fluorescence provides a more specific way to obtain photosynthetic mutants. Alterations in chlorophyll fluorescence indicate defects in the photosynthetic electron transport chain due to changes in the structure and function of the thylakoid membrane. For this reason, we have screened mutants with high chlorophyll fluorescence phenotypes from a collection of pSKI015 T-DNA–mutagenized Arabidopsis lines from the Arabidopsis Biological Resource Center. Approximately 90 different recessive mutants have been identified, and those with low PSII accumulation were named lpa mutants. In this study, we characterized the low psii accumulation1 (lpa1) mutant with reduced PSII accumulation and found that LPA1 is involved in efficient assembly of PSII.
RESULTS
Identification and Phenotype of the lpa1 Mutant
The lpa1 mutant was selected by its high chlorophyll fluorescence phenotype (Miles, 1982; Meurer et al., 1996) from the Scheible and Somerville T-DNA Arabidopsis lines (Weigel et al., 2000). The genetic defect in lpa1 segregated as a single recessive mutation. Cosegregation of the phosphinotricin resistance marker of the T-DNA with the mutant phenotype indicated that the mutation was due to the T-DNA insertion (data not shown).
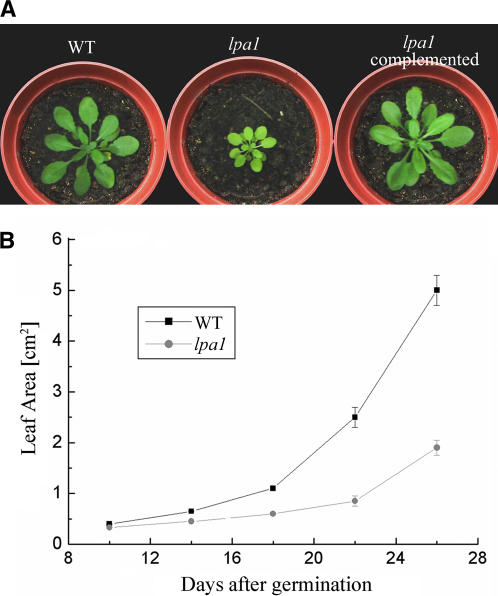
The leaves of the mutant were paler than those of the wild type, and growth was significantly reduced in the mutant (Figure 1). As shown in Figure 1, the leaf area of the mutant was ∼75% lower than that of wild-type plants 26 d after germination.
Figure 1.
Phenotypes of lpa1 Mutant and Wild-Type Plants.
(A) Five-week-old plants grown in the growth chamber.
(B) Growth kinetics of the lpa1 mutant compared with wild-type plants. Values shown are averages ± se of six replicated experiments.
Reduced PSII Activity in lpa1
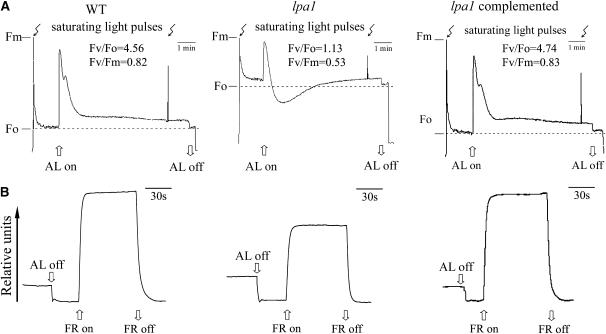
To identify the precise site of the lpa1 mutation, noninvasive fluorometric analyses were performed (Meurer et al., 1996). Fv/Fo decreased from 4.56 in the wild type to 1.13 in the mutant (Figure 2A) because the Fo level was higher in the mutant than in the wild type (0.16 and 0.06, respectively). In turn, this could be due to the mutant having a lower amount of photosystem I (PSI) and PSII reaction centers relative to their antenna complexes. The fluorescence yield in the lpa1 mutant transiently goes below the original and final Fo levels when stimulated by continuous actinic light, which suggests that a nonphotochemical quenching process would transiently develop before being dissipated within 2 to 3 min. The ratio of variable fluorescence to maximum fluorescence (Fv/Fm) reflects the maximum potential capacity of the photochemical reactions of PSII (Krause and Weis, 1991). In the lpa1 mutant, Fv/Fm appeared to be significantly lower (0.53) than in wild-type plants (0.82), implying that the mutants have defects in energy transfer within PSII or a partial loss of PSII capacity. The amplitude of changes in absorbance at 820 nm induced by far-red light (720 nm) was lower in the mutant than in wild-type plants, although the detected changes indicated that P700 can be oxidized in the lpa1 mutant (Figure 2B).
Figure 2.
Chlorophyll Fluorescence Induction and P700 Redox Kinetics of Wild-Type, lpa1, and lpa1 Transformants Complemented with the Open Reading Frame of the At1g02910 Gene.
The minimal level of fluorescence (Fo) of dark-adapted whole plants with all PSII reaction centers open was determined by switching on a pulsed measuring beam of red light. The Fm level with all PSII reaction centers closed was determined by a saturating pulse in the dark-adapted leaves (A). The redox kinetics were investigated by measuring absorbance changes of P700 at 820 nm induced by far-red light (FR; 720 nm) (B). AL, actinic light (120 μmol m−2 s−1).
Ultrastructure of the lpa1 Chloroplast
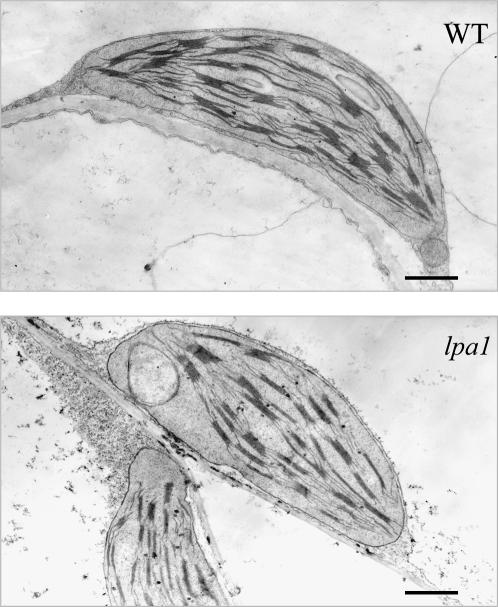
To assess the possibility that the lpa1 mutation causes ultrastructural changes in the chloroplasts, electron micrographs of ultrathin sections from 5-week-old leaves of wild-type and mutant plants were analyzed (Figure 3). Wild-type chloroplasts were found to be larger than those of the mutant (7.4 ± 0.65 and 6.2 ± 0.48 μm, respectively) and had a larger number of discs per grana stack on average (13 ± 2.5 and 7 ± 2, respectively). In addition, the wild-type chloroplasts displayed well-developed membrane systems composed of grana connected by the stroma lamellae, but the thylakoid membrane systems in lpa1 chloroplasts were disturbed, and the membrane spacing was not as clear (Figure 3).
Figure 3.
Electron Micrographs of Chloroplast from the lpa1 and Wild-Type Leaves.
To quantify the differences between wild-type and lpa1 chloroplasts, 48 chloroplast sections were analyzed from 5-week-old plants representing each genotype. Bars = 1 μm.
Reduced PSII Protein Contents in lpa1
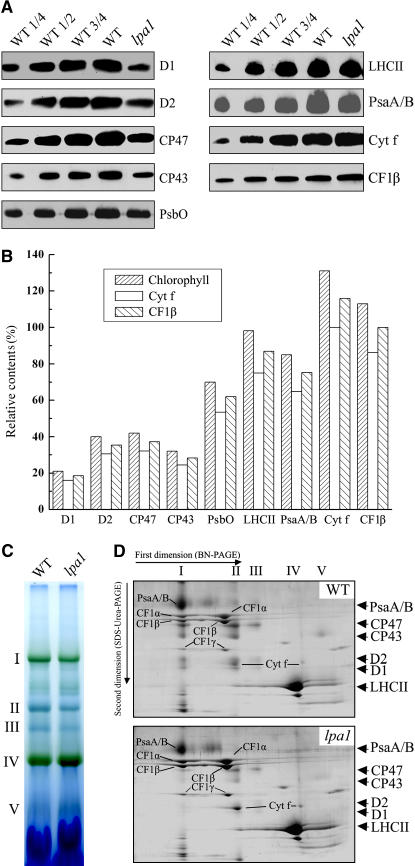
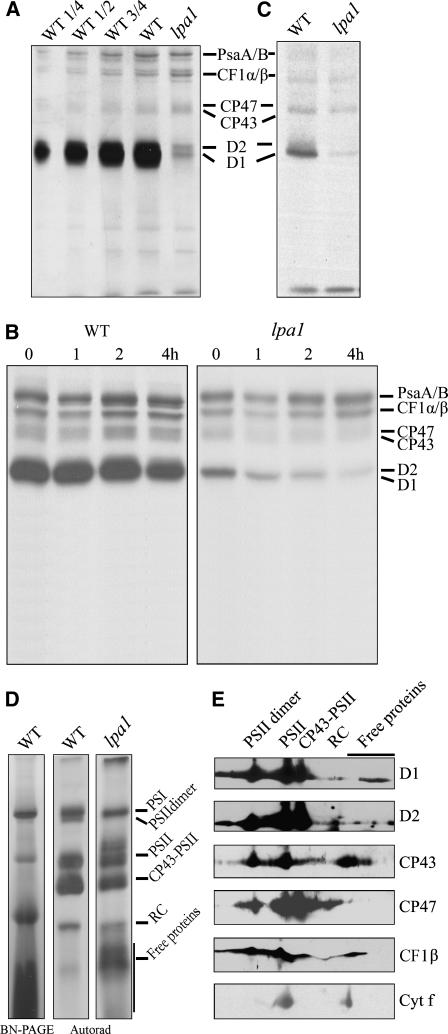
The defect in energy transfer within PSII found in the lpa1 mutant could be due to reduced protein levels of the PSII complex. To examine the steady state levels of the thylakoid membrane proteins, immunoblot analyses were performed with antibodies raised against specific subunits of the photosynthetic thylakoid membrane complexes. Our results showed that levels of the plastid-encoded subunits of the PSII core subunits D1, D2, CP47, and CP43 were reduced to ∼20, 40, 40, and 30% of wild-type levels, respectively (Figure 4A). The nuclear-encoded subunit of the oxygen-evolving complex, the 33-kD protein (PsbO), accumulated to ∼70% of wild-type levels, and light harvesting complex II (LHCII) was decreased slightly. The amounts of the PsaA/B PSI reaction center proteins were reduced to ∼75% of wild-type levels (Figure 4A). The reductions in the contents of these proteins in the lpa1 mutant, relative to those in the wild type, were found to be more pronounced when they were normalized to levels of cytochrome f of the cytochrome b6f complex or β-subunit of the ATP synthase (Figure 4B).
Figure 4.
Analysis of Thylakoid Membrane Proteins from lpa1 and Wild-Type Plants.
(A) Immunodetection of chloroplast proteins. The thylakoid membrane proteins were separated by SDS-urea-PAGE, and the blots were probed with specific anti-D1, anti-D2, anti-CP43, anti-CP47, anti-PsbO, anti-LHCII, anti-PsaA/B, anti-cytochrome f, and anti-CF1β antibodies.
(B) Semiquantitative analysis of chloroplast proteins. X-ray films were scanned and analyzed using an AlphaImager 2200 documentation and analysis system. The protein contents (per unit of chlorophyll) of the thylakoid membrane were determined and compared. The signal intensities of the immunoblot of the mutant, relative to those in the wild type, were also normalized to the cytochrome f and CF1β blot signals.
(C) BN gel analysis of thylakoid membrane protein complexes. Thylakoid membranes (10 μg chlorophyll) from wild-type and lpa1 mutant leaves were solubilized with 1% dodecyl-β-d-maltoside and separated by BN gel electrophoresis. The positions of protein complexes were identified with appropriate antibodies (see Guo et al., 2005).
(D) Two-dimensional separation of protein complexes in the thylakoid membranes. BN-PAGE–separated thylakoid proteins in a single lane from a BN gel were separated in a second dimension by 15% SDS-urea-PAGE and stained with Coomassie blue. Names of the proteins resolved by the second dimension SDS-PAGE, identified by immunodetection and matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry, are indicated by arrows.
To investigate structural alterations of thylakoid proteins, chlorophyll–protein complexes were solubilized from thylakoid membranes using dodecyl-β-d-maltopyranoside (DM) and separated by blue native PAGE (BN-PAGE) (Schägger et al., 1994; Cline and Mori, 2001). After the first-dimensional separation in the presence of Coomassie blue G 250 dye, five major bands were resolved (Figure 4C), apparently representing monomeric PSI and dimeric PSII (band I), monomeric PSII (band II), CP43 minus PSII (band III), trimeric (band IV), and monomeric LHCII (band V) (Guo et al., 2005). The BN-PAGE analysis clearly showed that the amount of PSII per unit of chlorophyll (bands II and III) was significantly lower in thylakoid preparations from the lpa1 mutant than in corresponding preparations from wild-type plants. Analyses of the two-dimensional SDS-urea-PAGE gels after Coomassie blue staining confirmed that the relative amounts of the PSII core subunits CP47, CP43, D1, and D2 were greatly reduced in the mutant, especially the PSII dimer, which was barely detected by this method (Figure 4D). In addition, the amount of PSI was reduced, and the cytochrome b6f and ATP synthase contents were slightly increased per unit of chlorophyll (Figure 4D).
Steady State RNA Levels and Polysome Accumulation in lpa1
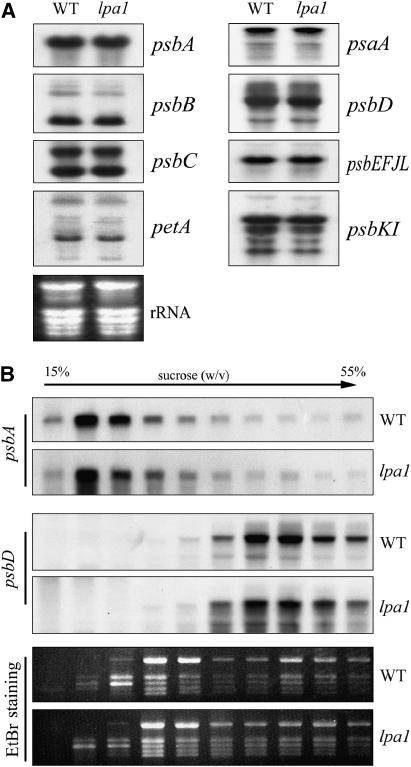
A possible explanation for the dramatic reductions observed in the mutant's PSII contents is that transcription of one or more of the structural PSII proteins is impaired in the mutant. To assess this possibility, the levels of the plastid-encoded PSII transcripts were investigated by RNA gel blot hybridization, which showed that levels of psbA (encoding the D1 subunit of PSII) and psbB (encoding the CP47 subunit of PSII) transcripts were unchanged in the mutant (Figure 5A). The levels of transcripts of other PSII operons (such as the psbD/C, psbE-F-L-J, and psbK-I operons) and transcripts of psaA and petA (which encode the PsaA subunit of PSI and cytochrome f, respectively) were also almost identical in the wild-type and mutant plants (Figure 5A).
Figure 5.
mRNA Expression in Chloroplasts and Association of Chloroplast mRNA with Polysomes.
(A) RNA gel blot analysis of transcripts in wild-type and lpa1 mutant plants. Transcripts of the genes psbA, psbB, psbC, psbD, psbEFJL, psbKI, petA, and psaA were detected by probing the filter with the appropriate gene-specific probes. Twenty micrograms of total RNA from wild-type and lpa1 mutant plants were size-fractionated by agarose gel electrophoresis, transferred to a nylon membrane, and probed with 32P-labeled cDNA probes.
(B) Association of psbA and psbD mRNAs with polysomes. Total extracts from wild-type and lpa1 leaves were fractionated on sucrose gradients. Ten fractions of equal volume were collected from the top to the bottom of the sucrose gradients, and equal proportions of the RNA purified from each fraction were analyzed by gel blot hybridization. rRNAs were detected by ethydium bromide (EtBr) staining.
The effects of the lpa1 mutation on protein synthesis in the chloroplast were further examined by analyzing the association of chloroplast mRNA with ribosomes by sucrose density gradient fractionation (Figure 5B). Total leaf lysates were fractionated, and RNA purified from the gradient fractions was analyzed with probes specific for psbA and psbD transcripts. No significant difference in the association with polysomes between the wild type and the mutant was detected for the psbA and psbD genes (Figure 5B).
Synthesis of the Thylakoid Membrane Proteins in lpa1
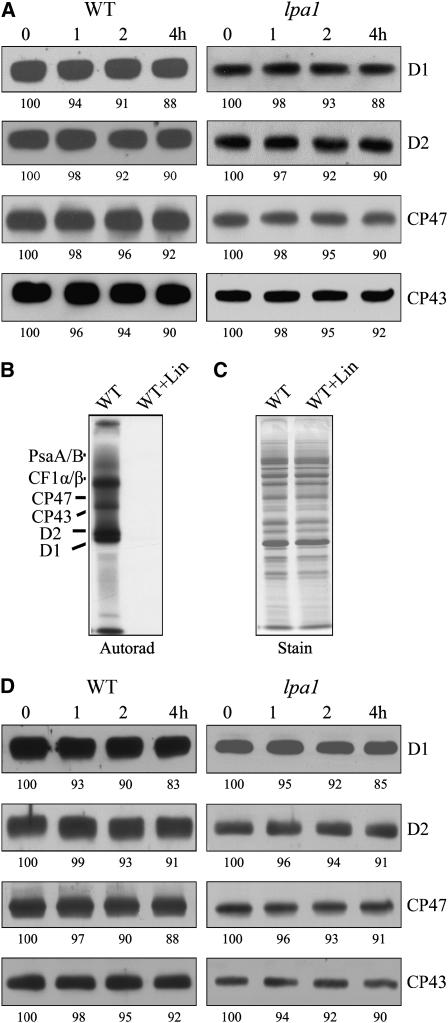
To investigate whether the diminished accumulation of PSII is due to either impaired translation or accelerated degradation of its component proteins, the synthesis of thylakoid membrane proteins was investigated by in vivo pulse-chase labeling experiments. For this purpose, the leaf proteins were pulse labeled with [35S]Met in the presence of cycloheximide, which blocks the synthesis of the nuclear-encoded proteins. As shown in Figure 6A, the rates of synthesis of the PSII subunits CP47 and CP43, PSI reaction center PsaA/B proteins, and the α- and β-subunits of the chloroplast ATP synthase (CF1-α/β) remained almost unchanged in the mutant. However, the incorporation of [35S]Met into the D1 and D2 proteins was dramatically reduced to <10% of wild-type levels (Figure 6A). Pulse labeling for 20 min was followed by a chase with unlabeled Met to monitor the turnover rates of plastid-encoded proteins. Our results showed that turnover rates of four major PSII subunits (CP47, CP43, D1, and D2) were higher in the lpa1 mutant than in wild-type plants (Figure 6B). Analysis of the protein labeling in mature leaves, in which net PSII biogenesis had ceased, showed that the incorporation of [35S]Met into the D1 and D2 proteins was also significantly reduced in the mutant (Figure 6C).
Figure 6.
In Vivo Protein Synthesis of Plastid-Encoded Membrane Proteins and Immunoblot Analysis after Second Dimension BN-PAGE.
(A) Incorporation of [35S]Met into thylakoid membrane proteins in young seedlings. After a 20-min pulse in Arabidopsis young seedlings in the presence of cycloheximide, the thylakoid membranes were isolated, and the proteins were separated by SDS-urea-PAGE and visualized autoradiographically.
(B) Pulse and chase of thylakoid membrane proteins. The 20-min pulse in Arabidopsis young seedlings was followed by a chase of cold Met for 1, 2, and 4 h. The thylakoid membranes were then isolated, separated by SDS-urea-PAGE, and visualized autoradiographically.
(C) Incorporation of [35S]Met into thylakoid membrane proteins in mature leaves. After a 30-min pulse in Arabidopsis mature leaves in the presence of cycloheximide, the thylakoid membranes were isolated, and the proteins were separated by SDS-urea-PAGE and visualized by autoradiogram.
(D) Autoradiogram of thylakoid membrane protein complexes of young seedlings solubilized and separated by BN-PAGE after pulse labeling for 20 min. A lane of BN gel separated thylakoid membrane proteins is shown at the left to indicate the locations of the identified complexes. RC, reaction center.
(E) Representative immunoblots from a second dimension SDS-PAGE separation of the thylakoid membrane. Horizontal strips of immunoblots with anti-D1, anti-D2, anti-CP47, anti-CP43, anti-CF1β, and anti-cytochrome f antibodies are shown.
Assembly of PSII and Stability of PSII Proteins in lpa1
After 20 min of pulse labeling, most of the radioactivity in the thylakoid membrane preparations was found to be associated with various PSII complexes, subcomplexes, and constituents (Figure 6D). The unassembled proteins were barely detectable in preparations from wild-type plants, indicating that assembly of PSII is very efficient in the wild-type plants, in accordance with our previous studies of isolated chloroplasts (Zhang et al., 2000). In the lpa1 mutant, PSII complexes were also detected, but there were higher levels of the free proteins than in the wild type (Figure 6D). Thus, assembly of PSII is less efficient in the lpa1 mutant than in the wild type. Since synthesis of D1 and D2 was greatly reduced in the mutant, longer exposure times were required to detect the bands in the lpa1 than in the wild-type autoradiograms. A very intensely labeled band above the one corresponding to PSII in the mutant was possibly due to CF1 ATP synthase by immunodetection (Figure 6E) and MALDI-TOF mass spectrometry analyses (data not shown). Immunoblot analysis showed streaking of PSII core proteins in this region (Figure 6E). Thus, it is likely that some other forms of PSII subcomplexes were also present in this radiolabeled band.
To study the stability of the assembled PSII in the lpa1 mutant, we treated leaves of mutant and wild-type plants with lincomycin to block chloroplast-encoded protein synthesis and measured the contents of PSII proteins by immunoblot analysis. As shown in Figure 7A, levels of D1, D2, CP47, and CP43 were quite stable during the treatment. In the lpa1 mutant, PSII proteins were decreased by the same extent as in the wild type (Figure 7A). To examine whether lincomycin effectively inhibited translation, we treated the leaves with lincomycin for 30 min and then performed in vivo protein labeling. The results confirmed that preincubating the leaves with lincomycin for 30 min completely blocked the synthesis of chloroplast proteins (Figures 7B and 7C).
Figure 7.
Stability of PSII Proteins.
(A) Immunoblot analysis of thylakoid membrane proteins. The Arabidopsis leaves were incubated with lincomycin for 30 min and illuminated for various times. After this treatment, the thylakoid membranes were isolated, and the contents of PSII proteins were determined through immunoblot analysis. X-ray films were scanned and analyzed using an AlphaImager 2200 documentation and analysis system. The percentages of protein levels shown below the lanes were estimated by comparison with levels found in corresponding samples taken at time 0.
(B) and (C) Effects of lincomycin (Lin) on protein synthesis. After incubation in the presence of 20 μg/mL cycloheximide and 100 μg/mL lincomycin for 30 min, the Arabidopsis leaves were labeled for 20 min. After labeling, the thylakoid membrane proteins were isolated, separated by SDS-urea-PAGE, and visualized autoradiographically (B). The Coomassie blue–stained gel (C) is presented to show that equal amounts of proteins were loaded.
(D) Immunoblot analysis of thylakoid membrane proteins. The Arabidopsis leaves were incubated with cycloheximide for 30 min and illuminated for various times. After the treatments, the thylakoid membranes were isolated, and the contents of PSII proteins were determined through immunoblot analysis. The percentages of protein levels shown below the lanes were estimated by comparison with levels found in corresponding samples taken at time 0.
The above findings showed that the steady state protein levels of PSII are fairly stable (Figure 7A). By contrast, the newly synthesized PSII proteins displayed very short half-lives in the mutant (Figure 6B). It is possible that some critical short-lived and nuclear-encoded factor(s) is (are) required to protect newly synthesized subunits from degradation in the mutant. A control experiment in the presence of cycloheximide was performed to assess this possibility. The results showed that PSII core subunits were stable in the wild type as well as in the mutant in the presence of cycloheximide (Figure 7D).
Cloning of the LPA1 Gene
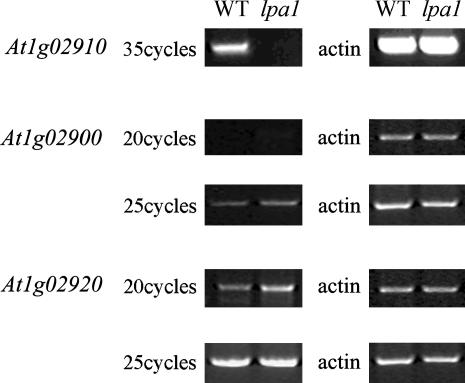
Cosegregation analysis of the lpa1 mutant phenotype and the phosphinotricin resistance marker indicated that the mutated LPA1 gene was tagged by the T-DNA. To determine the genetic basis of the lpa1 phenotypes, inverse PCR (Li et al., 2001) was performed, and the genomic region flanking the right border of the T-DNA was isolated. Sequence analysis showed that the T-DNA was inserted in the first exon of At1g02910, at position +7 relative to the ATG codon. To study the effect of the T-DNA insertion on At1g02910 expression, the At1g02910 cDNA was used as a probe in RNA gel blot hybridization. No expression of At1g02910 was detected in the wild type when 35 μg of total RNA was loaded, probably because of its very low level of expression. RT-PCR analysis showed that expression of the At1g02910 gene was undetectable after 35 cycles, at which point strong signals were obtained from wild-type samples (Figure 8). In addition, two annotated genes close to the insertion site (At1g02900 and At1g02920) were slightly activated, as shown by RT-PCR analysis (Figure 8).
Figure 8.
RT-PCR Analysis of Mutant Plants.
RT-PCR was performed with specific primers for At1g02900, At1g02910, and At1g02920 (left gel) and actin-specific primers (right gel).
To confirm that the LPA1 gene disruption was responsible for the mutant phenotype, complementation was performed with the isolated At1g02910 cDNA. The resulting clone containing the cDNA sequence under the control of the cauliflower mosaic virus 35S promoter was introduced into a homozygous lpa1 mutant using the floral dip method (Clough and Bent, 1998). Ten independent transgenic plants were obtained, and their chlorophyll fluorescence induction kinetics showed normal wild-type behavior (Figure 2). Thus, it can be concluded that the lpa1 mutant phenotype is due to loss of the function of the At1g02910 gene.
LPA1 Contains Two Tetratricopeptide Repeat Domains
BLAST searches of the complete Arabidopsis sequence confirmed that only one copy of the LPA1 gene is present in the nuclear genome. The open reading frame of LPA1 encodes a polypeptide of 453 amino acids with a calculated molecular mass of 50 kD. Database searches revealed that the N-terminal half of the protein contains two tetratricopeptide repeat (TPR) motifs that are arranged tandemly, each consisting of a 34-residue degenerate consensus sequence (Goebl and Yanagida, 1991). The N-terminal sequence is rich in positive and hydroxylated amino acid residues, which is characteristic of chloroplast transit peptides. The TMHMM program revealed that LPA1 has two transmembrane domains in the regions 202 to 224 and 239 to 258, suggesting that LPA1 is probably a membrane protein (Figure 9).
Figure 9.
Amino Acid Sequence Alignment of LPA1.
The amino acid sequence of the At1g02910 protein was compared with homologous sequences from O. sativa, C. reinhardtii, and Arabidopsis. Black boxes indicate strictly conserved amino acids, and gray boxes indicate closely related amino acids. Bars above the sequences indicate the functional TPR domains. Two possible transmembrane domains are shown by A and B. The sequences were aligned with ClustalW (Thompson et al., 1994).
Database searches and protein sequence alignments revealed that LPA1 shares significant sequence identity with an Oryza sativa protein (BAD52962; 71% identity, 85% similarity). Another protein from Chlamydomonas reinhardtii (C_130158; 21% identity) has one TPR domain and higher identity (32 to 45%) to LPA1 at its C terminus. Two hypothetical proteins, At4g28740 from Arabidopsis (29% identity, 47% similarity) and XP_473105 from O. sativa (33% identity, 51% similarity), were also found to have higher similarity to the C terminus of LPA1. However, these two proteins do not have chloroplast transit peptides, transmembrane domains, or TPR-like motifs (Figure 9).
LPA1 Is an Intrinsic Chloroplast Membrane Protein
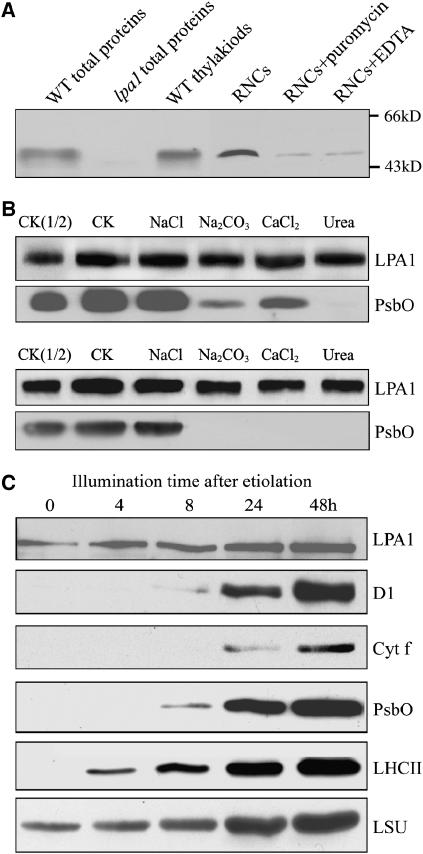
To determine the localization of LPA1, polyclonal antiserum was raised against recombinant LPA1 protein (amino acids 307 to 417). A 47-kD protein was detected in total protein and membrane fractions prepared from the wild-type plants, but no signal was detected in total protein preparations of the lpa1 plants (Figure 10A). These findings indicated that the T-DNA insertion leads to the loss of LPA1 protein accumulation. Immunoblot analysis also showed that LPA1 is associated with ribosome nascent chain (RNC) fractions (Figure 10A). The association of LPA1 with RNC fractions was greatly reduced in the presence of either puromycin or EDTA (Figure 10A).
Figure 10.
Immunolocalization of LPA1 and Light-Induced Accumulation of LPA1 and Plastid Proteins.
(A) Immunoblot analysis of LPA1. Samples from wild-type and lpa1 plants consisting of total leaf proteins, the thylakoids (equivalent to 5 μg chlorophyll), RNCs and RNCs treated with 1 mM puromycin or 10 mM EDTA (equivalent to 50 μg chlorophyll) were separated by SDS-PAGE and immunodetected with the antibody raised against LPA1.
(B) Salt washing of the membranes. The membrane preparations were incubated with 1 M NaCl, 200 mM Na2CO3, 1 M CaCl2, and 6 M urea for 30 min at 0°C (top panel) or sonicated in the presence of these salts (bottom panel) and incubated for another 30 min at 0°C. PsbO, the 33-kD luminal protein of PSII, was used as a marker. CK, the membrane preparations without treatments were used as a control.
(C) Immunoblot analysis of protein accumulation during light-induced greening of etiolated Arabidopsis seedlings. After growth in the dark for 5 d, the etiolated seedlings were illuminated for 4, 8, 24, and 48 h. The samples were harvested and processed for immunoblot analysis. Protein loadings were based on equal amounts of seedling fresh weight. The antibodies used for the analysis are indicated at the right.
To further investigate whether LPA1 is a transmembrane protein, the membrane fractions were isolated, and the proteins were subjected to immunoblot analysis. Washing the membranes with 0.25 M NaCl, 0.2 M Na2CO3, 1 M CaCl2, or 6 M urea did not release LPA1 from the membranes. LPA1 was still retained in the membranes, even when membrane preparations were sonicated in the presence of the listed salts (Figure 10B). During these treatments, PsbO, the 33-kD luminal protein of PSII, was used as a control.
Since LPA1 is required for PSII accumulation, we investigated the accumulation of LPA1 and PSII during light-induced greening. The PSII reaction center proteins D1, PsbO, LHCII, and cytochrome f were not detected in etiolated seedlings (Figure 10C), but LHCII appeared after illumination for 4 h, and the D1 and cytochrome f proteins were detected after illumination for 24 h. The large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase was present in the etiolated seedlings, and PsbO was detected after illumination for 8 h. The greening study showed that LPA1 accumulates even in dark-grown seedlings, and increases preceded accumulation of the D1 protein of PSII subunits (Figure 10C).
LPA1 Interacts with PSII Reaction Center Protein D1
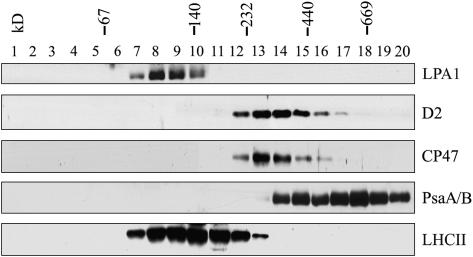
To test whether LPA1 is part of a multiprotein complex or an integral subunit of PSII, isolated thylakoid membranes were solubilized by DM and separated by sucrose gradient sedimentation (Zhang et al., 1999). After centrifugation, 20 fractions were collected from the gradients and subjected to immunoblot analysis using specific antibodies (Figure 11). The immunoblots indicated that LPA1 was present in fractions at ∼120 kD based on the migration of molecular standards.
Figure 11.
Sucrose Density Gradient Analysis of LPA1 in Thylakoids.
Thylakoids (0.5 mg/mL) were solubilized with 1% DM and separated in a linear 0.1 to 1 M sucrose gradient. Twenty fractions were collected from the gradients, and the proteins from each fraction were separated by SDS-PAGE. After electrophoresis, the separated proteins were immunodetected with the antibody directed against LPA1. To identify the positions of the major photosynthetic complexes, blots were also probed with anti-D2, anti-CP47, anti-LHCII, and anti-PsaA/B antibodies. The positions of 669-kD (thyroglobulin), 440-kD (ferritin), 232-kD (catalase), 140-kD (lactate dehydrogenase), and 66-kD (BSA) molecular size markers are indicated.
The above results show that LPA1 is required for the accumulation of PSII and that it does not comigrate with PSII. However, if LPA1 is involved in the assembly of PSII, one would expect it to interact with at least some subunits of the PSII complex. To test this possibility, we examined the interaction of LPA1 with the subunits of PSII in vivo using a modified split ubiquitin system to assess interactions of membrane proteins (Stagljar et al., 1998; Pasch et al., 2005; Yan et al., 2005). In this system, interactions between membrane-bound fusion proteins can be detected by monitoring the release of an artificial transcription factor consisting of protein A, LexA, and VP16 (PLV). To ensure that the interactions detected by the split ubiquitin system were specific for each individual protein, an unrelated membrane protein (Alg5) or the soluble ubiquitin domains Cub or NubG were used as bait or prey proteins in the control experiments.
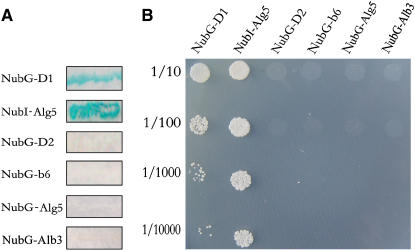
A series of plasmids in which the NubG moiety was fused to the N terminus of X (where X represents D1, D2, or cytochrome b6) were prepared. They were subsequently transformed into the NWY32 strain in which LPA1-Cub-LexA-VP16 was expressed. The resulting transformants were analyzed for β-galactosidase (β-Gal) activity and for growth on plates lacking His, Leu, and Trp (SD-His-Leu-Trp). As shown in Figure 12, coexpression of LPA1-Cub-LexA-VP16 with NubG-D1 resulted in positive β-Gal activity (Figure 12A) and growth on SD-His-Leu-Trp plates (Figure 12B). However, coexpression of LPA1-Cub-LexA-VP16 with NubG-D2 and NubG-cytochrome b6 yielded transformants that showed no β-Gal activity (Figure 12A) and growth on SD-His-Leu-Trp plates (Figure 12B).
Figure 12.
Interaction of LPA1 with the Reaction Center D1 Protein of PSII.
(A) β-Gal activity of the transformants expressing LPA1-Cub-LexA-VP16 together with the NubG-X (where X represents D1, D2, cytochrome b6, or Alb3) fusion proteins. Cells were grown on SD-His-Leu-Trp plates, transferred to Whatman filters, permeabilized, and incubated in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
(B) Growth of the cells expressing LPA1-Cub-LexA-VP16 with various NubG-X constructs on SD-His-Leu-Trp plates. Cells were grown to logarithmic phase, and 5-μL portions of 1:10 series dilutions were spotted on the SD-His-Leu-Trp plates and incubated at 30°C for 2 d. As a positive control, NMY32 was transformed with the NubI-Alg5 expression plasmid, and as a negative control, NMY32 was transformed with the plasmid expressing NubG-Alg5.
Alb3.1 in Chlamydomonas has been shown to be involved in the efficient integration of D1 into the reaction center of PSII (Ossenbühl et al., 2004). Therefore, we next examined whether LPA1 interacts with Alb3 in Arabidopsis. As shown in Figure 12, coexpression of LPA1-Cub-LexA-VP16 with NubG-Alb3 yielded transformants that showed no β-Gal activity (Figure 12A) and growth on SD-His-Leu-Trp plates (Figure 12B). These results strongly suggest that LPA1 interacts with D1 but not with D2, cytochrome b6, or Alb3.
DISCUSSION
The biogenesis and assembly of PSII complexes requires the coordinated synthesis and assembly of both plastid- and nuclear-encoded proteins. Due to the limited coding capacity of the chloroplast genome, the biogenesis and assembly of PSII is mainly regulated by the nuclear-encoded proteins (Goldschmidt-Clermont, 1998; Wollman et al., 1999; Barkan and Goldschmidt-Clermont, 2000; Rochaix, 2001; Leister, 2003). Genetic and biochemical studies have identified a number of nuclear-encoded factors that are involved in various posttranscriptional processes, such as mRNA stability, mRNA processing, splicing, translation, and assembly. Nac2, Mbb1 in Chlamydomonas, and HCF107 in Arabidopsis have functions related to mRNA stability, processing, and/or translation (Boudreau et al., 2000; Vaistij et al., 2000; Felder et al., 2001). Several mutants have also been isolated that have specific deficiencies in translation of psbA and psbC mRNA. Tba1 of Chlamydomonas encodes a putative oxidoreductase that functions as a redox regulator of cPAB1 RNA binding activity and, thus, indirectly promotes psbA mRNA translation initiation (Somanchi et al., 2005). The Tbc2 gene product, together with Tbc1 and Tbc3, is required for translation of psbC mRNA in Chlamydomonas (Auchincloss et al., 2002). HCF136 has been identified as an essential factor for the stable assembly of PSII in Arabidopsis (Meurer et al., 1998). Alb3.1 in Chlamydomonas probably acts as a membrane-integral chaperone in an early step of PSII assembly (Ossenbühl et al., 2004). Here, we describe a nuclear high chlorophyll fluorescence mutant of Arabidopsis with greatly reduced PSII levels and report the isolation of the mutated gene, LPA1, which appears to be required for efficient PSII assembly. Evidence regarding its probable functions during the assembly of PSII is also presented and discussed.
PSII Is Greatly Reduced in the lpa1 Mutant
In the Arabidopsis lpa1 mutant, amounts of the PSII reaction center proteins were reduced to ∼20% of wild-type levels (Figure 4). The contents of thylakoid membrane protein complexes, cytochrome b6f and ATPase, were slightly elevated per unit of chlorophyll, since levels of PSII proteins were greatly reduced in the mutant (Figure 4). Levels of the PsaA/B subunits of PSI were also reduced in the mutant, in accordance with spectroscopic data we acquired indicating that reduced levels of functional PSI were present in the mutant (Figure 2B). Reductions in the PSI protein levels have been observed in several PSII mutants, indicating that the reduced PSI levels in PSII mutants are mostly due to secondary effects of their mutations (Meurer et al., 1998; Plücken et al., 2002). Thus, this lpa1 mutant can be classified as a PSII mutant.
No alterations in the abundance or patterns of PSII, PSI, and cytochrome b6f gene transcripts were detected in the lpa1 mutant (Figure 5A). These results demonstrate that the reduced PSII contents are not due to the absence of transcripts encoding one of these structural PSII proteins but may be due to impaired translation or, alternatively, accelerated degradation of the PSII subunits once they have been synthesized. In vivo protein labeling showed that the incorporation of [35S]Met into the D1 and D2 proteins was dramatically reduced in the mutant, although the rate of synthesis of other plastid-encoded proteins was not affected (Figure 6A). It is not clear whether the primary effect of the mutation is on D1, with secondary effects on D2, or whether both proteins are affected directly. Analysis of the association of psbA and psbD transcripts with polysomes showed that initiation of the translation of psbA and psbD mRNA was not altered in the mutant (Figure 5B).
A possible explanation for the decreased synthesis of the D1 and D2 proteins in the lpa1 mutant is that their translation may be dependent on the assembly of photosynthetic complexes, as suggested by several authors (Choquet et al., 1998; Choquet and Vallon, 2002; Choquet and Wollman, 2002). In lpa1, assembly of PSII complexes is retarded, and the presence of unassembled proteins in the thylakoid membrane (Figure 6D) could downregulate the translation of corresponding transcripts. Similar phenomena have been observed in the alb3.1p mutant in Chlamydomonas, in which synthesis of the D1 protein appears to be greatly reduced by the accumulation of unassembled D1 protein in the membrane (Ossenbühl et al., 2004). The processes involved may be similar to the epistatic synthesis of cytochrome f, which is regulated by the assembly state of cytochrome b6f (unassembled cytochrome f accumulated in the membrane downregulates initiation of translation of further petA transcripts through interactions with their 5′ untranslated regions; Choquet et al., 1998). This mechanism has been found to operate in the biogenesis of the subunits of PSI and PSII (Wostrikoff et al., 2004; Minai et al., 2005). Similar assembly-dependent regulation of translation may have occurred at the translation elongation level rather than the initiation level in our experiments. In support of this hypothesis, we previously found the translation elongation of D1 to be severely inhibited and the accumulation of full-length D1 protein to be greatly reduced in an organelle translation/assembly system when normal interactions between the D1 and D2 proteins were hampered (Zhang et al., 2000). Another possibility is that labeling of D1 and D2 proteins was reduced in the mutant because the degradation rates of these proteins were higher than in the wild type. Inactivation of single, central PSII genes in cyanobacteria does not appear to have severe effects on the translation of their respective proteins, but instead appears to accelerate the degradation of close assembly partners (Yu and Vermaas, 1990, 1993). Thus, it is possible that the accumulation of unassembled proteins may increase turnover rates of protein synthesized de novo in the mutant.
Interestingly, PSII reaction center proteins still accumulated to up to ∼20% of wild-type levels in the mutant, despite severe reductions in the labeling (at least 10-fold) of the D1 and D2 proteins and in the half-life (to ∼1 h) of newly synthesized PSII subunits (Figure 6B). During the pulse labeling, PSII subunits, especially the D1 and D2 reaction center proteins, were intensively labeled both in young seedlings and in mature leaves (Figures 6A and 6C). Thus, active PSII repair continually occurs, in parallel with the de novo synthesis of PSII complexes, even in very young seedlings' leaves, in accordance with evidence presented by Rokka et al. (2005). Furthermore, it is possible that PSII repair rather than biogenesis is affected in the lpa1 mutant. If so, the immunoblot data could reflect rates of biogenesis, while pulse labeling data may reflect rates at which the various subunits are replaced.
LPA1 Is Required for Efficient PSII Assembly
In vivo labeling experiments in the wild type and lpa1 mutant revealed the presence of PSII complexes with identical molecular masses (Figure 6D), suggesting that functional assembly takes place in the mutant. Measurement of the PSII oxygen evolution activity further confirmed that functional PSII complexes are formed in the lpa1 mutant, and the activity retained in the mutant was consistent with the amount of PSII subunits (data not shown). The ability of the mutant to grow photoautotrophically provides further support for the above conclusion. However, assembly of PSII in lpa1 was less efficient as a result of the loss of LPA1 (Figure 6D), although immunoblot analysis showed that the subunits in the assembled PSII complexes of the mutant are as stable as those in the wild-type plants (Figure 7). These results suggest that LPA1 is involved in the assembly of the PSII complex rather than in maintaining its stability.
Cell fractionation and immunoblot analysis demonstrated that LPA1 is an intrinsic membrane protein (Figure 10B), in accordance with the presence of a plastidal transit peptide and two transmembrane domains in it (Figure 9). Sucrose gradient fractionation analysis further confirmed that LPA1 is not an integral subunit of PSII but may form complexes with itself or, more likely, with other proteins (Figure 11). The N terminus of LPA1 also contains two TPR domains (Figure 9), which have been shown to mediate protein–protein interactions and the assembly of multiprotein complexes (Goebl and Yanagida, 1991). Although LPA1 is not a constituent of PSII complexes, its demonstrated requirement for PSII accumulation implied that it probably interacts with one or more subunits of PSII. This implication was confirmed by yeast two-hybrid analysis showing that LPA1 specifically interacts with the D1 protein but not with the D2 protein or cytochrome b6 of the cytochrome b6f complex (Figure 12). The PSII assembly factor Alb3.1 in Chlamydomonas has also been found to interact with the D1 protein (Ossenbühl et al., 2004). Furthermore, protein–protein interaction studies have revealed that Alb3 not only binds to the PSII core proteins D1, D2, and CP43, but also to the PSI reaction center protein PsaA and the ATPase subunit CFoIII, suggesting that Alb3 plays an essential role in the assembly of photosynthetic protein complexes (Pasch et al., 2005). However, no interaction between LPA1 and Alb3 was detected in our yeast two-hybrid analysis (Figure 12). Thus, the functions of LPA1 and Alb3 in PSII assembly probably involve different mechanisms.
Another TPR domain–containing factor, PratA, has been shown to be involved in PSII biogenesis in cyanobacteria (Klinkert et al., 2004). Targeted inactivation of PratA in synechocystis, a periplasmic TPR repeat protein, resulted in severe reductions in PSII levels (Klinkert et al., 2004). Further analysis indicated that PratA may assist the C-terminal processing of the D1 protein, most likely through its TPR domains (Klinkert et al., 2004). In addition, TPR domains are known to be functionally important in Ycf3, which interacts with at least two PSI subunits, PsaA and PsaD (Boudreau et al., 1997; Naver et al., 2001), and several TPR cochaperones have been shown to participate transiently in intermediate assembly stages before the functional maturation of steroid receptors (Smith, 2004). Thus, the flexible and mutable TPR domain appears to have a general capacity to facilitate specific protein–protein interactions by holding them together and thus providing essential contact between them (Goebl and Yanagida, 1991; Das et al., 1998; Smith, 2004).
Assembly of PSII occurs in a sequential manner via a series of intermediates. The first of these intermediates, the complex formed by D2 and cytochrome b559, appears to serve as a receptor for newly synthesized D1 (Adir et al., 1990; van Wijk et al., 1997; Müller and Eichacker, 1999; Zhang et al., 1999). LPA1 appears to be essential for the efficient assembly of PSII, probably through direct interaction with D1, which is integrated into PSII cotranslationally during the insertion of nascent D1 chains into the thylakoid membrane (Zhang et al., 1999). The association of LPA1 with RNC fractions suggests that a key function may be assisting the correct assembly of the D1 protein into functional PSII during its translation. Thus, LPA1 may act as a chaperone, assisting the proper folding and integration of D1 into the thylakoid membrane and promoting the subsequent interaction between D1 and another key PSII reaction center protein D2.
METHODS
Plant Materials and Growth Conditions
The lpa1 mutant was isolated in a collection of pSKI015 T-DNA–mutagenized Arabidopsis thaliana (ecotype Columbia) lines from the Arabidopsis Biological Resource Center based on the high chlorophyll fluorescence phenotype in the dark under long-wavelength UV radiation (Meurer et al., 1996). Arabidopsis was grown in soil under short-day conditions (10-h-light/14-h-dark cycle) with a photon flux density of 120 μmol m−2 s−1 at a constant temperature of 22°C. To ensure synchronized germination, the seeds were sown in darkness for 48 h at 4°C. For growth on agar plates, seeds were surface-sterilized and sown on Murashige and Skoog (1962) medium containing 3% sucrose and 0.8% agar. To confirm the T-DNA–induced Basta resistance, phosphinotricin was added to the medium to a final concentration of 10 mg/L. Measurement of leaf area was performed with the standard protocols by an LI-3000 A (LI-Cor).
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence was measured using a PAM-2000 portable chlorophyll fluorometer (Walz) connected with a leaf-clip holder (model 2030-B; Walz). Before measurement, leaves were dark-adapted for 30 min. The minimum fluorescence yield (Fo) was measured under measuring light (650 nm) with very low intensity (0.8 μmol m−2 s−1). To estimate the maximum fluorescence yield (Fm), a saturating pulse of white light (3000 μmol m−2 s−1 for 1 s) was applied. The maximal photochemical efficiency of PSII was calculated from the ratio of variable (Fv) to maximum (Fm) fluorescence [Fv/Fm = (Fm − Fo)/Fm]. All the above measurements were performed in a dark room with stable ambient conditions. For the measurement of light-induced P700 absorbance changes at 820 nm, the PAM chlorophyll fluorometer was equipped with an emitter–detector unit ED 800T (Walz), and the measurements were performed according to Meurer et al. (1996). Absorbance changes induced by saturating far-red light represent the relative amounts of photooxidizable P700.
Transmission Electron Microscopy
For transmission electron microscopy processing, the wild-type and mutant rosette leaves from 5-week-old plants were collected. The leaf tissue was cut into small pieces and fixed in 2.5% glutaraldehyde in phosphate buffer for 4 h at 4°C. After fixation, the tissue was rinsed and postfixed in 1% OsO4 overnight at 4°C. After rinsing in phosphate buffer, the samples were dehydrated in an ethanol series, infiltrated with a graded series of epoxy resin in epoxy propane, and embedded in Epon 812 resin. Thin sections were obtained using a diamond knife on a Reichert OM2 ultramicrotome, stained in 2% uranyl acetate, pH 5.0, followed by 10 mM lead citrate, pH 12, and viewed with a transmission electron microscope (JEM-1230; JEOL).
Thylakoid Membrane Preparation
Thylakoid membranes were prepared according to standard methods (Zhang et al., 1999). The leaves were homogenized in an ice-cold isolation buffer containing 400 mM sucrose, 50 mM HEPES-KOH, pH 7.8, 10 mM NaCl, and 2 mM MgCl2 with a chilled mortar and pestle and filtrated through two layers of cheesecloth. The filtrate was centrifuged at 5000g for 10 min. The thylakoid pellets were washed with isolation buffer, recentrifuged, and suspended in isolation buffer. The resulting thylakoid membrane pellets were either used freshly or frozen in liquid N2 and stored at –70°C before use. The chlorophyll content was determined spectrophotometrically according to Porra et al. (1989).
BN-PAGE, SDS-PAGE, and Protein Identification
BN-PAGE was performed as described (Schägger et al., 1994) with the modification shown by Cline and Mori (2001). The thylakiod membranes were washed in 330 mM sorbitol and 50 mM BisTris-HCl, pH 7.0, and suspended in resuspension buffer (20% glycerol and 25 mM BisTris-HCl, pH 7.0) at 1.0 mg chlorophyll/mL. An equal volume of resuspension buffer containing 2% (w/v) DM was added to the thylakiod suspension in a dropwise manner. After incubation at 4°C for 5 min, insoluble material was removed by centrifugation at 12,000g for 10 min. The supernatant was combined with one-tenth volume of 5% Serva blue G in 100 mM BisTris-HCl, pH 7.0, 0.5 M 6-amino-n-caproic acid, and 30% (w/v) glycerol and applied to 0.75-mm-thick 6 to 12% acrylamide gradient gels in a Hoefer Mighty Small vertical electrophoresis unit connected to a cooling circulator. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer and 5% β-mercaptoethanol for 30 min and layered onto 1-mm-thick 15% SDS polyacrylamide gels containing 6 M urea (Laemmli, 1970). After electrophoresis, the proteins were transferred to nitrocellulose membranes, probed with specific antibodies, and visualized by the enhanced chemiluminescence method. X-ray films were scanned and analyzed using an AlphaImager 2200 documentation and analysis system (Alpha Innotech).
To identify the proteins by MALDI-TOF, in-gel digestion and sample preparations were performed according to the method of Jensen et al. (1999). Samples were loaded onto the MALDI target plates using α-cyano-4-hydroxycinnamic acid as a matrix. MALDI-TOF analysis was performed in reflector mode on the AXIMA-CFR plus mass spectrometer (Kratos Analytical). Proteins were identified as the highest ranking result by the Mascot Wizard freeware (Matrix Science). The following parameters were used for database searches: monoisotopic mass accuracy <50 ppm, one missed cleavage, complete carbamidomethylation of Cys residues, and partial oxidation of Met residues. For positive identification, the score of the result [−10 × Log (P)] had to be over the significance threshold level (P < 0.05).
In Vivo Labeling of Chloroplast Proteins
In vivo protein labeling was performed essentially according to Meurer et al. (1998). Primary leaves of 12-d-old young seedlings were preincubated in 20 μg/mL cycloheximide for 30 min and radiolabeled with 1 μCi/μL [35S]Met (specific activity >1000 Ci/mmol; Amersham Pharmacia Biotech) in the presence of 20 μg/mL cycloheximide for 20 min at 25°C. Four-week-old mature Arabidopsis leaves were also selected for in vivo labeling for 30 min as described above. After washing twice with homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM EDTA), the leaves were ground with a conical stainless steel rod in an Eppendorf tube with 300 μL of the same buffer. Membranes were pelleted by centrifugation at 15,000g for 10 min and resuspended in 100 μL of homogenization buffer. SDS was added to a final concentration of 2% (w/v), and the proteins were solubilized for 10 min at 25°C.
To examine the effects of lincomycin on chloroplast protein synthesis, the leaves were preincubated in the presence of 20 μg/mL cycloheximide and 100 μg/mL lincomycin for 30 min before radiolabeling.
Stability of PSII Subunits
The Arabidopsis leaves were incubated either with 100 μg/mL lincomycin to block the chloroplast-encoded protein synthesis or with 20 μg/mL cycloheximide to block the nuclear-encoded protein synthesis for 30 min and then the leaves were subjected to illumination under growth conditions for various times. After treatments, the thylakoid membranes were isolated, and the contents of PSII proteins were determined through immunoblot analysis.
Antiserum Production
The nucleotide sequences encoding the soluble part of LPA1 (amino acids 307 to 417 corresponding to nucleotide positions 919 to 1251 of the LPA1 gene) were amplified by PCR using the primers 5′-CCGGCTAGCAAAGAAACTGTTACACTTG-3′and 5′-GGCAAGCTTATGATGTACACATCATCG-3′. The resulting DNA fragment was cleaved with NheI and HindIII and fused in frame with the N-terminal His affinity tag of pET28a. The BL21 cells were harvested after the application of 0.4 mM isopropylthio-β-d-galactoside for 3 h and resuspended in 500 mM NaCl and 20 mM NaH2PO4, pH 8.0. After incubation for 30 min at 4°C in the presence of lysozyme at a final concentration of 1 mg/mL, the bacterial lysate was sonicated 12 times for 10 s. The overexpressed proteins in inclusion bodies were centrifuged at 3000g for 30 min, and the pellet was solubilized in 500 mM NaCl, 8 M urea, and 20 mM NaH2PO4, pH 8.0. The fusion protein was purified on a nickel-nitrilotriacetic acid agarose resign matrix, and a polyclonal antibody was raised in rabbit with purified antigen.
Immunolocalization Studies
The intracellular localization of LPA1 was determined essentially according to Lennartz et al. (2001). The Arabidopsis membranes were suspended to a final concentration of 50 μg chlorophyll/mL in 10 mM HEPES-KOH, pH 8.0, 10 mM MgCl2, 330 mM sorbitol, 1 mM phenylmethylsulfonyl fluoride supplemented with 250 mM NaCl, 200 mM Na2CO3, 1 M CaCl2, or 6 M Urea, respectively. The membrane fractions without supplements were used as a control. These suspensions were sonicated three times for 15 s each while kept on ice. After treatment, the membranes were pelleted at 100,000g for 2 h at 4°C, quickly washed with isolation buffer, and used for SDS-PAGE and immunoblot analysis.
Inverse PCR and Sequence Analysis of lpa1 cDNA
Genomic sequences flanking the T-DNA right border were amplified using inverse PCR (Li et al., 2001). Ten micrograms of lpa1 genomic DNA was digested with KpnI in a 100-μL volume. After digestion, the enzyme was heat-inactivated, and the reaction was passed through a DNA fragment purification kit (TaKaRa). A ligation was set up in a 100-μL volume, and 4 μL of the ligated solution was used as the template for inverse PCR. The ligated DNA yielded a 2.8-kb inverse PCR product with two inverse PCR primers (T3 long primer, 5′-AATTAACCCTCACTAAAGGGAACAAAAG-3′; ACTRB primer, 5′-GTTTCTAGATCCGAAACTATCAGTG-3′). Based on the genomic sequence information, the complete open reading frame of LPA1 was amplified by RT-PCR using total RNA isolated from wild-type plants as a template. The specific primers used were as follows: sense primer (5′-GGCCTCTAGAGTAGAAGAAAATTGAATGGC-3′) and antisense primer (5′-ACGGGATCCTCATCTTTCTAACTTGCTGAGAACGTC-3′).
Complementation of the lpa1 Mutant
For complementation of the lpa1 mutant, the cDNA containing the coding region was amplified by PCR and subcloned into the plant expression vector pBI121 under the control of the cauliflower mosaic virus 35S promoter. The construct was transformed into Agrobacterium tumefaciens EHA105 strain and introduced into Arabidopsis mutant plants by the floral dip method (Clough and Bent, 1998). Transformant plants were selected on Murashige and Skoog medium containing 50 μg/mL kanamycin monosulfate. Ten resistant plants were transformed to soil and grown in a greenhouse to produce seeds. The successful complementation was confirmed by measurements of chlorophyll fluorescence.
Nucleic Acid Preparation and Analysis
Isolation of Arabidopsis DNA was performed as described (Sambrook and Russell, 2001). Total plant RNA was extracted from 100 mg of fresh tissues using Trizol reagent. For the determination of the three genes' expression located around the T-DNA insertion site, RT-PCR was performed using the following primers: At1g02900, 5′-TTCTTAACTCTTACGATTCTCGTCG-3′ and 5′-TAGGTCTTTACTATTGTGCAGAGAC-3′; At1g02910, 5′-GGCCTCTAGAGTAGAAGAAAATTGAATGGC-3′ and 5′-ACGGGATCCTCATCTTTCTAACTTGCTGAGAACGTC-3′; At1g02920, 5′-GAATCAAAGTTTTCGGTCACCCAGC-3′ and 5′-CTAGAAGTGATGTCAGCAACCCAAG-3′. To ensure equal amount of RNA in each sample, RT-PCR analysis of actin cDNA was performed using the following primers: sense (5′-AACTGGGATGATATGGAGAA-3′), antisense (5′-CCTCCAATCCAGACACTGTA-3′).
RNA gel blot analysis was performed essentially as described before (Sambrook and Russell, 2001). After gel electrophoresis with a formaldehyde denaturing 1.2% agarose gel, total RNA was transferred onto nylon membranes (Amersham Pharmacia Biotech). The membranes were probed with 32P-labeled cDNA probes. Following high-stringency hybridization and washing, all the blots were exposed to x-ray film for 1 to 3 d.
The hybridization probes were labeled by random priming and prepared from the PCR fragments of the chloroplast genome (GenBank accession number AP000423). The sequences of the PCR primers used for amplification of chloroplast genes are given in Supplemental Table 1 online.
Sucrose Gradient Fractionation of Thylakoid Membranes
Thylakoids (0.5 mg chlorophyll/mL) were solubilized for 5 min on ice in 5 mM MgCl2, 10 mM NaCl, and 25 mM MES-NaOH, pH 6.0, containing 1% DM. After centrifugation at 14,000g for 10 min at 4°C, the supernatant of 500 μL was loaded onto a linear 0.1 to 1 M sucrose gradient in 5 mM MgCl2, 10 mM NaCl, 0.06% DM, and 25 mM MES-NaOH, pH 5.7. The gradient was centrifuged for 22 h at 180,000g at 4°C on an SW40 Ti rotor (Beckman). After centrifugation, 20 fractions were collected from the top to the bottom of the gradient. For further analysis, the proteins in each fraction were separated by SDS-PAGE and characterized by immunoblot analysis.
For molecular weight estimation of the localized protein, a mixture of standard proteins (Amersham Pharmacia Biotech) containing protein molecular mass standards of 669 kD (thyroglobulin), 440 kD (ferritin), 232 kD (catalase), 140 kD (lactate dehydrogenase), and 66 kD (BSA) was separated in a parallel gradient.
Split Ubiquitin Assays
The yeast two-hybrid assay was performed using the yeast strain NMY32 supplied by Dualsystems Biotech (Stagljar et al., 1998). To construct the bait plasmids, the vector pCCW-SUC encoding the Cub-LexA-VP16 fragment was used. The prey plasmids were constructed from the vector pDSLNx, which encodes the NubG fragment (Dualsystems Biotech). The mature full-length LPA1 gene and the complete gene sequences of D1, D2, cytochrome b6, and Alb3 were obtained by PCR amplification. The LPA1 gene was cloned in Cub vector, and the D1, D2, cytochrome b6, and Alb3 genes were cloned in NubG vectors. Interaction was determined by growth of diploid yeast colonies on SD-His-Leu-Trp plates and also by β-Gal activity through X-Gal filter assay (Stagljar et al., 1998).
Other Methods
The conditions for the growth of Arabidopsis seeding in darkness and greening of etiolated plants under light were as described by Meurer et al. (1998).
Polysomes were isolated from leaf tissue of wild-type and mutant plants under conditions that maintain polysome integrity as described by Barkan (1988). RNA was isolated, fractionated, and transferred onto nylon membranes. The filters were hybridized with 32P-labeled cDNA probes for psbA and psbD.
Thylakoid membrane–associated RNC fractions were isolated essentially according to Zhang et al. (1999).
Accession Numbers
Sequence data used for the alignment from this article can be found in the Genbank/EMBL data libraries under the following accession numbers: NP_171790, Arabidopsis At1g02910; BAD52962, Oryza sativa (japonica cultivar group); C_130158, Chlamydomonas reinhardtii (from http://www.chlamy.org database); XP_473105, O. sativa (japonica cultivar group); and NP_567817 Arabidopsis At4g28740.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Oligonucleotides Used for the Preparation of the Hybridization Probes.
Supplementary Material
Acknowledgments
We thank Tingyun Kuang for constant support and encouragement, Eva-Mari Aro for providing the antibodies as well as for valuable advice and discussion, and Peter Westhoff for the in vivo protein labeling protocol. We also thank Jixing Liu, Feng Sun, and Wenhe Cai for making the antibodies and the Nottingham Arabidopsis Stock Centre for the Arabidopsis seeds. This research was supported by the National Natural Science Foundation of China (30500037), by the Frontier Project of the Knowledge Innovation Engineering of the Chinese Academy of Sciences, and by grants from the Institute of Botany.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lixin Zhang (zhanglixin@ibcas.ac.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037689.
References
- Adir, N., Schochat, S., and Ohad, I. (1990). Light-dependent D1 protein synthesis and translocation is regulated by reaction centre II. Reaction centre II serves as an acceptor for the D1 precursor. J. Biol. Chem. 265 12563–12568. [PubMed] [Google Scholar]
- Aro, E.-M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Auchincloss, A.H., Zerges, W., Perron, K., Girard-Bascou, J., and Rochaix, J.-D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González, E., and Aro, E.-M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572. [DOI] [PubMed] [Google Scholar]
- Bennoun, P., and Delepelaire, P. (1982). Isolation of photosynthetic mutants in Chlamydomonas. In Methods in Chloroplast Molecular Biology, M. Edelman, ed (Amsterdam: Elsevier Science Publishers), pp. 25–38.
- Boudreau, E., Nichelsen, J., Lemaire, S.D., Ossenbühl, F., and Rochaix, J.-D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., and Rochaix, J.-D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewski, G.J., et al. (2001). Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 159 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., and Vallon, O. (2002). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82 615–634. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529 39–42. [DOI] [PubMed] [Google Scholar]
- Chua, N.H., and Gillham, N.W. (1977). The sites of synthesis of the principal thylakoid membrane polypeptides in Chlamydomonas reinhardtii. J. Cell Biol. 74 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., and Mori, H. (2001). Thylakoid delta pH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry, C., Olive, J., Drapier, D., Recouvreur, M., and Wollman, F.-A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder, S., Meierhoff, K., Sane, A.P., Meurer, J., Driemel, C., Plücken, H., Klaff, P., Stein, B., Bechtold, N., and Westhoff, P. (2001). The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13 2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, K.N., Iverson, T.M., Maghlaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838. [DOI] [PubMed] [Google Scholar]
- Goebl, M., and Yanagida, M. (1991). The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16 173–177. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177 115–180. [DOI] [PubMed] [Google Scholar]
- Gong, H., and Ohad, I. (1991). The PQ/PQH2 ratio and occupancy of photosystem II-QB site by plastoquinone control the degradation of D1 protein during photoinhibition in vivo. J. Biol. Chem. 266 21293–21299. [PubMed] [Google Scholar]
- Greenberg, B.M., Gaba, V., Mattoo, A.K., and Edelman, M. (1987). Identification of a primary in vivo degradation product of the rapidly- turning-over 32 kd protein of photosystem II. EMBO J. 6 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.K., Zhang, Z.Z., Bi, Y.R., Yang, W., Xu, Y.N., and Zhang, L.X. (2005). Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett. 579 3619–3624. [DOI] [PubMed] [Google Scholar]
- Jensen, K.H., Herrin, D.L., Plumley, F.G., and Schmidt, G.W. (1986). Biogenesis of photosystem II complexes: Transcriptional, translational and posttranslational regulation. J. Cell Biol. 103 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, O.N., Wilm, M., Shevchenko, A., and Mann, M. (1999). Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. In Methods in Molecular Biology, Vol. 112: 2-D Proteome Analysis Protocols, J.A. Link, ed (Totowa, NJ: Humana Press), pp. 513–530. [DOI] [PubMed]
- Klinkert, B., Ossenübhl, F., Sikorski, M., Berry, S., Eichacker, L., and Nickelsen, J. (2004). Prat A, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in synechocystis sp. PCC 6803. J. Biol. Chem. 279 44639–44644. [DOI] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. 42 313–349. [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19 47–56. [DOI] [PubMed] [Google Scholar]
- Lennartz, K., Plücken, H., Seidler, A., Westhoff, P., Bechtold, N., and Meierhoff, K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001). BRS1, a serine carboxypeptidase, regulates BRI1 signalling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll, B., Kern, J., Saeger, W., Zouni, A., and Biesiadka, J. (2005). Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Meierhoff, K., and Westhoff, P. (1996). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and northern hybridization. Planta 198 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, D. (1982). The use of mutations to probe photosynthesis in higher plants. In Methods in Chloroplast Molecular Biology, M. Edelman, R. Hallick, and N.-H. Chua, eds (Amsterdam: Elsevier Biomedical Press), pp. 75–107.
- Minai, L., Wosterikoff, K., Wollman, F.A., and Choquet, Y. (2005). Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18 159–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B., and Eichacker, L.A. (1999). Assembly of the D1 precursor in monomeric photosystem II reaction centre precomplexes precedes chlorophyll a-triggered accumulation of reaction centre II in barley etioplasts. Plant Cell 11 2365–2377. [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15 473–497. [Google Scholar]
- Nanba, O., and Satoh, K. (1987). Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc. Natl. Acad. Sci. USA 84 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naver, H., Boudreau, E., and Rochaix, J.-D. (2001). Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13 2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Björkman, O., and Grossman, A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl, F., Göhre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J.-D., and Eichacker, L.A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasch, J.C., Nickelsen, J., and Schünemann, D. (2005). The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl. Microbiol. Biotechnol. 69 440–447. [DOI] [PubMed] [Google Scholar]
- Plücken, H., Müller, B., Grohmann, D., Westhoff, P., and Eichacker, L.A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532 85–90. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Prasil, O., Adir, N., and Ohad, I. (1992). Dynamics of photosystem II: Mechanisms of photoinhibition and recovery process. In The Photosystems: Structure, Function and Molecular Biology, Vol. 11, J. Barber, ed (Amsterdam: Elsevier Science Publishers), pp. 295–348.
- Rochaix, J.D. (2001). Assembly, function, and dynamics of the photosynthetic machinery in Chlamydomonas reinhardtii. Plant Physiol. 127 1394–1398. [PMC free article] [PubMed] [Google Scholar]
- Rokka, A., Suora, M., Saleem, A., Battchikova, N., and Aro, E.-M. (2005). Synthesis and assembly of thylakoid protein complexes: Multiple assembly steps of photosystem II. Biochem. J. 388 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217 220–230. [DOI] [PubMed] [Google Scholar]
- Shi, L.X., and Schröder, W.P. (2004). The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta 1608 75–96. [DOI] [PubMed] [Google Scholar]
- Shikanai, T., Munekage, Y., Shimizu, K., Endo, T., and Hashimoto, T. (1999). Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol. 40 1134–1142. [DOI] [PubMed] [Google Scholar]
- Smith, D.F. (2004). Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi, A., Barnes, D., and Mayfield, S.P. (2005). A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of chloroplast psbA mRNA. Plant J. 42 341–352. [DOI] [PubMed] [Google Scholar]
- Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Boudreau, E., Lemaire, S.D., Goldschmidt-Clermont, M., and Rochaix, J.D. (2000). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk, K.J., Roobol-Boza, M., Kettunen, R., Andersson, B., and Aro, E.-M. (1997). Synthesis and assembly of the D1 protein into photosystem II: Processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36 6178–6186. [DOI] [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Maiwald, D., Kurth, J., Salamini, F., and Leister, D. (2000). Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38 497–504. [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman, F.A., Minai, L., and Nechushtai, R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1141 21–85. [DOI] [PubMed] [Google Scholar]
- Wostrikoff, K., Girad-Bascou, J., Wollman, F.A., and Choquet, Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, A., Wu, E., and Lennarz, W.J. (2005). Studies of yeast oligosaccharyl transferase subunits using the split-ubiquitin system: Topological features and in vivo interactions. Proc. Natl. Acad. Sci. USA 102 7121–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., and Vermaas, W. (1990). Transcript levels and synthesis of photosystem II components in cyanobacterial mutants with inactivated photosystem II genes. Plant Cell 2 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., and Vermaas, W. (1993). Synthesis and turnover of photosystem II reaction centre polypeptides in cyanobacterial D2 mutants. J. Biol. Chem. 268 7407–7413. [PubMed] [Google Scholar]
- Zhang, L.X., and Aro, E.-M. (2002). Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett. 512 13–18. [DOI] [PubMed] [Google Scholar]
- Zhang, L.X., Paakkarinen, V., van Wijk, K.J., and Aro, E.-M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274 16062–16067. [DOI] [PubMed] [Google Scholar]
- Zhang, L.X., Paakkarinen, V., van Wijk, K.J., and Aro, E.-M. (2000). Biogenesis of the chloroplast-encoded D1 protein: Regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12 1769–1781.11006346 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.