The classical plant hormones (auxins, ethylene, gibberellins, cytokinins, and abscisic acid [ABA]) were discovered between 40 and 100 years ago, but it is only within the last ∼10 years that their receptors have been identified. The first two classes of receptors to be identified, those for ethylene and cytokinins, were found to encode members of the two-component receptor superfamily (Chang and Stadler, 2001; Inoue et al., 2001). This suggested that, consistent with the fact that these plant hormones resemble metabolites more than classical animal hormones, their signaling mechanisms might be more similar to those of fungi or bacteria than previously suspected. In the last year, however, receptors for the three remaining classical hormone classes have been discovered, and each was found to represent a completely novel mechanism of perception involving binding to nuclear soluble receptors, leading to altered levels of negative regulators. Auxin and gibberellins induce targeted degradation of transcriptional repressor proteins (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Ueguchi-Tanaka et al., 2005), whereas ABA promotes accumulation of a transcriptional repressor via direct effects on RNA processing (Razem et al., 2006).
ABA REGULATES DIVERSE RESPONSES
ABA, misnamed for its presumed role in abscission, functions primarily in plant responses to dehydrating stresses by inducing stomatal closure and production of desiccation protectants and by limiting cell division and expansion (reviewed in Finkelstein et al., 2002; Himmelbach et al., 2003). However, roots and shoots have opposing responses to mild stress and the resulting moderate increases in endogenous ABA: primary root elongation is stimulated, while shoot growth decreases (Sharp, 2002). At the whole-plant level, this apparent contradiction leads to an increased surface area for water absorption without any increased demand from the shoots, poising the plant for rapid growth when water is no longer limiting. Its developmental roles have long appeared more limited than those of other hormones; effects on seed maturation, dormancy and inhibition of germination, and senescence have received the most attention. However, ABA has also been implicated in inhibiting some aspects of photoregulation (Weatherwax et al., 1996; Rohde et al., 2000), inhibiting lateral root initiation (De Smet et al., 2003), and promoting developmental phase changes such as juvenile-to-adult or vegetative-to-reproductive transitions (Rogler and Hackett, 1975), though it may also inhibit flowering (reviewed in Levy and Dean, 1998). Collectively, these responses are sufficiently diverse to require many different signaling mechanisms that are stage, organ, or even cell specific. An obvious question is whether this diversity begins at the receptor or downstream signaling steps.
STRUCTURE/FUNCTION STUDIES OF ABA SIGNALING
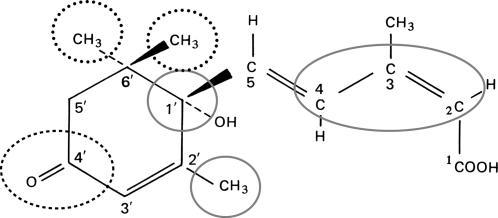
The structural requirements of ABA perception have been studied by testing the activity of the naturally occurring (+)-S form of ABA, its (−)-R enantiomer, and a variety of ABA metabolites and analogs in diverse ABA responses, including stomatal regulation, germination, and gene expression (Walton, 1983; Walker-Simmons et al., 1994, 1997; Nyangulu et al., 2005). Several major conclusions have been derived from these studies: (1) most responses are specifically induced by the (+) form or some of its derivatives, but even (−) ABA is effective for some responses; (2) ABA metabolites, such as 8-hydroxy-ABA and phaseic acid that were initially considered inactivated derivatives of the hormone, still activate some responses; (3) different responses display different specificities; and (4) most changes to ABA decrease, but don't necessarily eliminate, its biological activity. Although disparate metabolism of the analogs and effects on endogenous ABA metabolism complicates interpretation of these studies (Lin et al., 2005), this range of specificities suggests the existence of multiple receptor types or modifications. Based on these studies, the 2-cis, 4-trans side chain configuration, the stereochemistry at C-1′, the presence of the 7′ carbon, and the C4′ ketone have been identified as important for many ABA-like activities, suggesting that they may be involved in an interaction(s) with a receptor(s) (Figure 1). However, one report described the C-8′ and C-9′ methyl groups and the C4′ ketone as slightly less critical and suggested they be used as possible attachment sites for affinity probes (Walker-Simmons et al., 1994).
Figure 1.
ABA Structure.
Potentially interacting portions are circled. Critical regions indicated with solid lines and less critical regions with dotted lines.
PERCEPTION SITE(S)
Another approach to understanding the perception mechanism is to determine the hormone's site of action and then identify candidate interacting molecules. The structure of ABA is consistent with either intracellular or extracellular action: as a small weak acid, it is likely to be protonated in the relatively acidic apoplast such that it can enter cells as an uncharged molecule but may also remain in the apoplast. Consequently, the structural features of ABA do not dictate its perception site. Studies making use of impermeable ABA derivatives or microinjection and subsequent photoactivation of caged ABA derivatives have demonstrated both intracellular and extracellular sites of perception for distinct responses (reviewed in Finkelstein et al., 2002). Extracellular perception was shown to be sufficient for ABA-induced gene expression and for preventing stomatal opening, whereas intracellular perception was effective for ABA regulation of both stomatal closure and opening. In another approach, biotinylated ABA was shown to induce stomatal closure and, when bound by fluorescently labeled avidin, used to visualize ABA binding sites that appeared to be present in patches on guard cell plasma membranes (Yamazaki et al., 2003). Once again, these results point to the existence of multiple receptor types at distinct locations.
BIOCHEMICAL APPROACHES
Attempts to identify an ABA receptor directly have made use of affinity purification to various ABA derivatives or anti-idiotypic antibodies. Each of these studies depends on the information described in the two preceding sections to design affinity probes that are active and to choose source material likely to express or contain receptors.
More than 20 years ago, ABA binding proteins were first identified, but not purified, by stereospecific photoactivated cross-linking of radiolabeled ABA via the C-4′ ketone to guard cell membrane extracts (Hornberg and Weiler, 1984). Within the last 8 years, several groups have used ABA derivatives linked to carrier molecules via either the C1 carboxyl or the C-4′ ketone moieties for affinity purification of ABA binding proteins. Protein conjugates were used to identify ABA binding proteins preferentially localized in the plasma membrane of Arabidopsis thaliana (Pedron et al., 1998). Affinity chromatography with ABA linked to activated Sepharose via the C1 carboxyl group identified a 42-kD protein from broad bean epidermis that shows saturable, reversible, high affinity, stereospecific binding to (+)-ABA (Zhang et al., 2002). C-4′-linked biotinylated ABA was used for visualizing ABA binding sites (Yamazaki et al., 2003), but no specific proteins were purified in this study. Although each of these studies demonstrated the bioactivity of the relevant ABA derivative, all of these derivatives were coupled through portions of the hormone previously identified as critical for activity and therefore might be compromised in interaction with (some) receptors. An alternate approach was recently described by Nyangulu et al. (2005), who have developed a bioactive ABA derivative that displays all essential features of the hormone and is tethered to biotin via an additional ring structure. Although this affinity probe has been shown to bind anti-ABA antibodies and an ABA-metabolizing enzyme, it has not yet identified any candidate receptors.
Yet another approach has made use of anti-idiotypic antibodies. In this strategy, monoclonal anti-(+)-ABA antibodies were used as antigens to generate new polyclonal antibodies that might recognize an ABA binding site that could be structurally similar in the anti-ABA antibodies and a putative receptor. The anti-idiotypic antibodies (AB2) were used to screen barley aleurone cDNA expression libraries (Liu et al., 1999; Razem et al., 2004), leading to the identification of ABAP1, a barley protein displaying saturable, reversible, high-affinity, stereospecific binding of ABA and apparently localized to the plasma membrane.
While such studies have identified ABA binding proteins, functional demonstration of a role in ABA signaling (e.g., by loss- or gain-of-function studies) is required before they can be designated as true receptors.
GENETIC APPROACHES (FORWARD AND REVERSE)
Loss of function for hormone receptors or downstream signaling components was initially expected to result in decreased hormone sensitivity, so numerous screens were performed to identify mutants with impaired hormonal responses. Although this approach led to identification of ethylene and cytokinin receptors, even highly pleiotropic mutations affecting signaling by ABA were not due to receptor defects. Instead, the ABA response loci identified genetically included transcription factors, protein phosphatases, a farnesyl transferase subunit, and several RNA processing enzymes affecting transcript splicing and stability (reviewed in Finkelstein and Rock, 2002; Kuhn and Schroeder, 2003). A variation on this strategy tried to exploit differential changes in sensitivity to (+)- and (−)-ABA as a means of focusing on receptor mutants (Nambara et al., 2002). Although this screen identified two new loci whose products have not yet been identified, it also identified many new alleles of the ABI transcription factor loci; the explanation for the distinct stereospecificities of different alleles is unknown.
A surprising early observation was that the lines displaying the greatest hormone resistance carried dominant mutations and were therefore unlikely to represent simple loss of function. For example, the abi1-1 mutant, which produces a dysfunctional PP2C, is impaired in control of >90% of ABA-regulated genes (Hoth et al., 2002), but a loss of function for this gene results in only mild hypersensitivity to ABA (Gosti et al., 1999). This phenomenon was later explained by genetic redundancy: ABI1 is a member of a clade of nine PP2Cs, five of which have already been reported as negative regulators of ABA response (Schweighofer et al., 2004).
An alternate approach made use of reverse genetics to test the roles of candidate early signaling molecules identified biochemically or pharmacologically. These included G-protein–coupled receptor-like (GPCR) molecules (Pandey and Assmann, 2004) and G-proteins (both subunits of heterotrimers and monomeric) (Wang et al., 2001; Ullah et al., 2002; Zheng et al., 2002) and other ABA-regulated receptor-like kinases (Osakabe et al., 2005). Many of these were shown to play some role in ABA response, but the effects were often mild and limited to a few responses. Furthermore, the G-protein and GPCR appeared to have opposing functions in regulating ABA response of guard cells, suggesting that the GPCR (GCR1) might negatively regulate G-protein–mediated ABA signaling (Pandey and Assmann, 2004). Although any of these limitations might still be consistent with effects on function of only one of multiple possible receptors, none were reported to have ABA binding activity.
BIOCHEMISTRY AND GENETICS CONVERGE
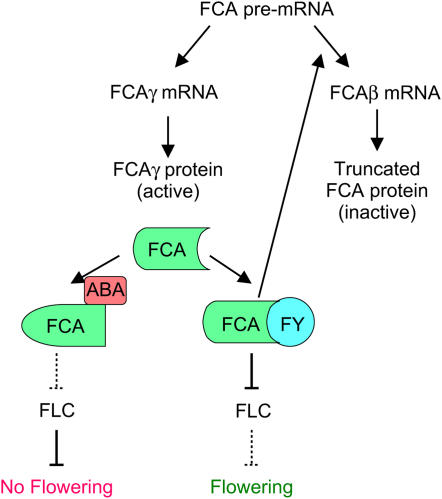
Most recently, an Arabidopsis RNA binding protein initially identified for its role in promoting flowering, FCA (for flowering time control protein A), was identified as a candidate ABA receptor based on homology to ABAP1, the barley aleurone protein identified by ABA-mimicking anti-idiotypic antibodies (Razem et al., 2006). This was a bit surprising because the barley ABA binding protein was described as plasma membrane associated and lacking a consensus RNA binding motif (Razem et al., 2004), whereas Arabidopsis FCA is a nuclear protein with two conserved RNA recognition motifs (RRMs) (Macknight et al., 1997). FCA was already well characterized in terms of the target and effects of its RNA binding activity in the context of flowering (Putterill et al., 2004), so Razem et al. tested whether ABA affects these known activities. Their studies showed that FCA, like its barley homolog, stereospecifically binds (+)-ABA with high affinity (Kd = 19 nM for FCA and ∼28 nM for ABAP1) and that this binding inhibits association of FCA with FY (for flowering locus Y), a 3′ RNA polyadenylation factor (Razem et al., 2006). In the absence of ABA, the FCA-FY complex directly inhibits production of active FCA by promoting alternative processing of the FCA pre-mRNA. In addition, the complex inhibits accumulation of the transcript for the floral repressor FLC (for flowering locus C), possibly indirectly. Consistent with FCA function as an ABA receptor, inhibition of FCA-FY complex formation by ABA enhances accumulation of the FLC transcripts and consequently delays bolting, but these effects are lost in an fca mutant. Production of full-length FCA is also enhanced by ABA and can act as a negative feedback loop to reduce the effect of ABA. However, counterintuitively, loss of FCA-based perception and decreased ABA levels have opposite effects on flowering time because ABA binding inhibits FCA function (Figure 2). In this respect, ABA perception is similar to that of ethylene, whose binding also inactivates its receptor. It is not known whether all ABA-regulated FCA functions depend on interaction with FY, but recent studies show that null alleles of FY are embryo lethal, demonstrating that this locus is required for far more functions than flowering control (Henderson et al., 2005).
Figure 2.
Model of FCA Interaction with ABA and Effects on Flowering.
ABA binding prevents interaction with FY required to reduce production of FLC and full-length FCA, thereby maintaining FLC-mediated repression of flowering. Arrows indicate positive regulation, solid bars indicate repression, and dotted bars indicate derepressed steps.
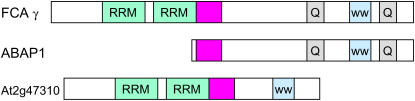
Several other ABA response loci have been implicated in aspects of RNA processing, including Cap binding, intron processing, and miRNA production (Kuhn and Schroeder, 2003), but identification of FCA as an ABA receptor constitutes the most direct link between ABA signaling and RNA processing. Given the high degree of conservation among FCAs from monocots and dicots (Figure 3), it is striking that ABAP1 appears to lack the RRMs. The simplest explanation for this discrepancy, consistent with the fact that the ABAP1 cDNA is the same size as the smaller of two prominent transcripts detected in barley aleurone (Razem et al., 2004), is that this cDNA may correspond to a truncated splice variant. Alternatively, ABAP1 may have an as yet unidentified RNA binding motif, may interact with distinct RNA binding protein(s), or may have an entirely different mode of action. Isolation of a cDNA clone corresponding to the longer ABAP1 homologous transcript, analysis of the genomic sequence adjacent to the ABAP1 encoding region, and identification of proteins that interact with ABAP1 will help distinguish among these possibilities.
Figure 3.
Domain Structures of Full-Length Active FCA, Barley ABA Binding Protein (ABAP1), and Predicted Product of the Closest Arabidopsis Homolog.
Shaded boxes are highly conserved across FCAs from wheat, rice, Lolium, pea, Brassica, and Arabidopsis. WW, protein interaction domain; Q, Gln-rich domain.
A traditional assumption about receptors is that each can initiate multiple responses to a ligand via branching signaling pathways. In recent years, it has become clear that this may be complicated by the existence of multiple classes of receptors with some distinct and some redundant functions. So far, the role of FCA has been tested in only four ABA-regulated responses: flowering, germination, stomatal regulation, and lateral root formation. Although essential for ABA-delayed flowering and ABA-inhibited lateral root formation, FCA does not appear to play a role in the two best-characterized ABA responses: germination inhibition and stomatal regulation. This spectrum of effects is consistent with the expression pattern of the full-length FCA transcript required for production of a functional protein, which is strongest in shoot and root apices and young flower buds but very low or undetectable in seeds and leaf tissue (Macknight et al., 2002). It is not yet clear which of the many ABA response loci act downstream of this receptor. The highly pleiotropic abi1-1 and abi2-1 mutants have been described as early flowering (Martinez-Zapater et al., 1994), like the ABA-deficient mutants, or unaffected in flowering (Razem et al., 2006), so their involvement remains controversial. However, there are dozens of other loci known to function in ABA responses that might be good candidates for mediators of FCA-dependent signaling, including some already implicated in control of flowering or ABA-dependent nitrate inhibition of lateral root production (e.g., ABI3 or ABI5) (Kurup et al., 2000; Signora et al., 2001).
Because FCA regulates RNA processing, a global approach, such as transcriptional profiling of wild-type versus loss- or gain-of-FCA expression lines, would probably have detected the misregulation of FLC and might identify additional FCA-regulated transcripts that could implicate FCA in other processes. In particular, overexpression of a full-length cDNA would avoid complications from autoregulation of alternative splicing and could reveal additional functions of FCA that might be masked by redundantly acting receptors providing these functions in mutant lines.
THE SEARCH CONTINUES…
Regardless of whether the limited effects of FCA are due to its limited expression, redundancy, or specialized functions of distinct receptors, it is clear that additional receptors must exist. Obvious candidates to start analyzing are close homologs of FCA. The most recognizable conserved domains in FCA are the RRMs, which are shared with 197 other Arabidopsis proteins, and the WW domain implicated in protein–protein interactions. All known FCAs also have Gln-rich domains surrounding the WW domain and additional highly conserved regions adjacent to the RRMs. Although essential to FCA function, the N-terminal RRMs do not confer ABA responsiveness and were not found in ABAP1. The ABA binding domain has been mapped to an unspecified C-terminal portion of FCA, in close proximity to the FY binding WW domain, but the specific residues involved in ABA binding have not yet been identified (Figure 3). BLAST analyses identify at least half a dozen Arabidopsis proteins with homology to the C-terminal part of FCA, with homology concentrated in the FY binding region, but only one of these includes the RRMs and adjacent conserved regions. Presumably, tighter mapping and structural analysis of the ABA binding domain would aid efforts to identify more candidate receptors.
Even if this approach identifies additional ABA receptors related to FCA, the diversity of stereospecific requirements among ABA responses suggests that there may be other completely unrelated proteins functioning as ABA receptors for other responses. Biochemical approaches with carefully designed affinity probes, such as that described by Nyangulu et al. (2005), or follow-up on the ABA binding proteins that have been reported by Zhang et al. (2002) and Yamazaki et al. (2003) are likely paths to success.
Finally, studies reported in the last several years have demonstrated the prevalence of previously underappreciated mechanisms of regulated RNA processing. Alternative splicing affects 30 to 60% of mammalian genes (Lee et al., 2003) and ∼10% of Arabidopsis and rice transcripts (Iida et al., 2004). Hundreds of microRNAs regulate translation or stability of many more transcripts (Bartel, 2004), many of which are involved in stress responses (Sunkar and Zhu, 2004). It has long been assumed that the hundreds of RNA binding proteins encoded in any genome were likely to participate in regulating function of specific transcripts, but nobody had suggested that the RNA binding proteins would be hormone receptors themselves. It is now an intriguing possibility that at least some of these proteins constitute a direct mechanism for modifying expression of specific genes in response to hormones or other small metabolites or signals.
Acknowledgments
I thank T. Lynch, E. Garcia, I. Brocard-Gifford, W. Reeves, and several anonymous reviewers for critical review of the manuscript. Research in the author's laboratory was supported by grants from the U.S. Department of Energy, the National Science Foundation, and the USDA.
References
- Bartel, D. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Chang, C., and Stadler, R. (2001). Ethylene hormone receptor action in Arabidopsis. Bioessays 23 619–627. [DOI] [PubMed] [Google Scholar]
- De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development in Arabidopsis. Plant J. 33 543–555. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R., Gampala, S., and Rock, C. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Rock, C.D. (2002). Abscisic acid biosynthesis and signaling. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0058, http://www.aspb.org/publications/arabidopsis/.
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I., Liu, F., Drea, S., Simpson, G., and Dean, C. (2005). An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132 3597–3607. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. [DOI] [PubMed] [Google Scholar]
- Hornberg, C., and Weiler, E. (1984). High-affinity binding sites for abscisic acid on plasmalemma of Vicia faba guard cells. Nature 310 321–324. [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.-P., Hanafey, M., Tingey, S., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115 4891–4900. [DOI] [PubMed] [Google Scholar]
- Iida, K., Seki, M., Sakurai, T., Satou, M., Akiyama, K., Toyoda, T., Konagaya, A., and Shinozaki, K. (2004). Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 32 5096–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kuhn, J., and Schroeder, J. (2003). Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 6 463–469. [DOI] [PubMed] [Google Scholar]
- Kurup, S., Jones, H., and Holdsworth, M. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21 143–155. [DOI] [PubMed] [Google Scholar]
- Lee, C., Atanelov, L., Modrek, B., and Xing, Y. (2003). ASAP: The Alternative Splicing Annotation Project. Nucleic Acids Res. 31 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-L., Wang, H.-J., Wang, J.-S., Zaharia, L., and Abrams, S. (2005). Abscisic acid regulation of heterophylly in Marsilea quadrifolia L.: Effects of R-(-) and S-(+) isomers. J. Exp. Bot. 56 2935–2948. [DOI] [PubMed] [Google Scholar]
- Liu, J.-H., Luo, M., Cheng, K.-J., Mohapatra, S.S., and Hill, R.D. (1999). Identification and characterization of a novel barley gene that is ABA-inducible and expressed specifically in embryo and aleurone. J. Exp. Bot. 50 727–728. [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Coupland, G., Dean, C., and Koornneef, M. (1994). The transition to flowering in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 403–433.
- Nambara, E., Suzuki, M., Abrams, S., McCarty, D.R., Kamiya, Y., and McCourt, P. (2002). A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyangulu, J., Galka, M., Jadhav, A., Gai, Y., Graham, C., Nelson, K., Cutler, A., Taylor, D., Banowetz, G., and Abrams, S. (2005). An affinity probe for isolation of abscisic acid-binding proteins. J. Am. Chem. Soc. 127 1662–1664. [DOI] [PubMed] [Google Scholar]
- Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). Leucine-rich repeat Receptor-Like Kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., and Assmann, S. (2004). The Arabidopsis putative G protein–coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedron, J., Brault, M., Nake, C., and Miginiac, E. (1998). Detection of abscisic-acid-binding proteins in the microsomal protein fraction of Arabidopsis thaliana with abscisic-acid-protein conjugates used as affinity probes. Eur. J. Biochem. 252 385–390. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Laurie, R., and Macknight, R. (2004). It's time to flower: The genetic control of flowering time. Bioessays 26 363–373. [DOI] [PubMed] [Google Scholar]
- Razem, F., El-Kereamy, A., Abrams, S., and Hill, R. (2006). The RNA binding protein, FCA, is an abscisic acid receptor. Nature 439 290–294. [DOI] [PubMed] [Google Scholar]
- Razem, F., Luo, M., Liu, J.-H., Abrams, S., and Hill, R. (2004). Purification and characterization of a barley aleurone abscisic acid-binding protein. J. Biol. Chem. 279 9922–9929. [DOI] [PubMed] [Google Scholar]
- Rogler, C., and Hackett, W. (1975). Phase change in Hedera helix: Stabilization of the mature form with abscisic acid and growth retardants. Physiol. Plant. 34 148–152. [Google Scholar]
- Rohde, A., De Rycke, R., Beeckman, T., Engler, G., Van Montagu, M., and Boerjan, W. (2000). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer, A., Hirt, H., and Meskiene, I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9 236–243. [DOI] [PubMed] [Google Scholar]
- Sharp, R.E. (2002). Interaction with ethylene: Changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 25 211–222. [DOI] [PubMed] [Google Scholar]
- Signora, L., Smet, I., Foyer, C., and Zhang, H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28 655–662. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.-y., Hsing, Y., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.-G., Wang, S., and Jones, A. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons, M., Rose, P., Shaw, A., and Abrams, S. (1994). The 7′-methyl group of abscisic acid is critical for biological activity in wheat embryo germination. Plant Physiol. 106 1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons, M.K., Holappa, L.D., Abrams, G.D., and Abrams, S.R. (1997). ABA metabolites induce group 3 LEA mRNA and inhibit germination in wheat. Physiol. Plant. 100 474–480. [Google Scholar]
- Walton, D. (1983). Structure-activity relationships of abscisic acid analogs and metabolites. In Abscisic Acid, F. Addicott, ed (New York: Praeger Publishers), pp. 113–146.
- Wang, X.-Q., Ullah, H., Jones, A., and Assmann, S. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072. [DOI] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, D., Yoshida, S., Asami, T., and Kuchitsu, K. (2003). Visualization of abscisic acid-perception sites on the plasma membrane of stomatal guard cells. Plant J. 35 129–139. [DOI] [PubMed] [Google Scholar]
- Zhang, D.-P., Wu, Z.-Y., Li, X.-Y., and Zhao, Z.-X. (2002). Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol. 128 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.-L., Nafisi, M., Tam, A., Li, H., Crowell, D., Chary, S., Schroeder, J., Shen, J., and Yang, Z. (2002). Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14 2787–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]