Abstract
The enzyme strictosidine synthase (STR1) from the Indian medicinal plant Rauvolfia serpentina is of primary importance for the biosynthetic pathway of the indole alkaloid ajmaline. Moreover, STR1 initiates all biosynthetic pathways leading to the entire monoterpenoid indole alkaloid family representing an enormous structural variety of ∼2000 compounds in higher plants. The crystal structures of STR1 in complex with its natural substrates tryptamine and secologanin provide structural understanding of the observed substrate preference and identify residues lining the active site surface that contact the substrates. STR1 catalyzes a Pictet-Spengler–type reaction and represents a novel six-bladed β-propeller fold in plant proteins. Structure-based sequence alignment revealed a common repetitive sequence motif (three hydrophobic residues are followed by a small residue and a hydrophilic residue), indicating a possible evolutionary relationship between STR1 and several sequence-unrelated six-bladed β-propeller structures. Structural analysis and site-directed mutagenesis experiments demonstrate the essential role of Glu-309 in catalysis. The data will aid in deciphering the details of the reaction mechanism of STR1 as well as other members of this enzyme family.
INTRODUCTION
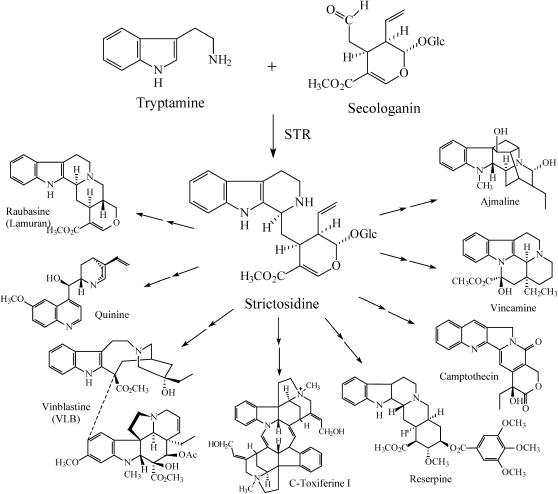
The monoterpenoid-derived indole alkaloids compose one of the structurally largest and pharmacologically most diverse alkaloid families in higher plants. Of the ∼2000 members, which are divided into various structural classes, several have long-standing medical applications. Prominent examples of developed therapeutics include the treatment of cancer (vinblastine or the camptothecin derivative topotecan), malaria (quinine), hypertension (raubasine and reserpine), schizophrenia (reserpine in high dosage), disturbed cerebral blood flow (vincamine), and antiarrhythmic heart disorders (ajmaline, from the Indian medicinal plant Rauvolfia serpentina). Strictosidine synthase (STR1; EC 4.3.3.2) is involved in the biosynthesis of all these alkaloids by catalyzing the condensation of the two initial building blocks, tryptamine and the monoterpenoid secologanin, leading to the glucoalkaloid strictosidine. As the first committed step, STR1 seems to play the principal role in nature's strategy to generate the entire monoterpenoid indole alkaloid family (Figure 1; see reviews in Kutchan, 1993; Stöckigt and Ruppert, 1999).
Figure 1.
STR1 Catalyzes the Stereo-Specific Condensation of Tryptamine and Secologanin Leading to 3α(S)-Strictosidine: A Central Reaction in the Biosynthesis of the Entire Family of Monoterpenoid Indole Alkaloids in Plants.
The reaction type catalyzed by STR1 is so far an exceptional example in the biosynthesis of natural products. It was hitherto known only from synthetic chemistry (Pictet-Spengler–type reaction), where it is applied in alkaloid synthesis, especially of tetrahydroisoquinolines by condensation of an amine and an aldehyde under acidic conditions. The only enzymes known to date that are functionally related to STR1 are deacetylisoipecoside synthase, deacetylipecoside synthase, and norcoclaurine synthase. The first two enzymes catalyze the condensation of dopamine instead of tryptamine with secologanin leading to the family of monoterpenoid tetrahydroisoquinoline alkaloids (De-Eknamkul et al., 2000). Norcoclaurine synthase catalyzes the condensation of dopamine and 4-hydroxyphenylacetaldehyde, resulting in the biosynthesis of ∼6000 benzylisoquinoline alkaloids (such as morphine, sanguinarine, or berberine; Samanani et al., 2004). Although STR1 and its functional related enzymes are key enzymes in the biosynthesis of ∼50% of all alkaloids, very little is known either about their reaction mechanisms or about the amino acids essential for enzymatic activity.
STR1 was first isolated from plant cell suspensions of Catharanthus roseus and R. serpentina and was later detected and purified from many other species of the plant families Apocynaceae and Rubiaceae, such as Vinca, Cinchona, and Ophiorrhiza (Treimer and Zenk, 1979; Hampp and Zenk, 1988; Bracher and Kutchan, 1992; Stevens et al., 1993; DeWaal et al., 1995; Yamazaki et al., 2003). STR1 from R. serpentina is a monomeric precursor protein with 344 amino acids (Kutchan, 1993) that exhibits 100, 79, and 58% identity to STR1 from Rauvolfia mannii, C. roseus, and Ophiorrhiza pumila, respectively. STR1 from these medicinal plants together with >100 STR1-related genes derived from UniProt protein database constitute a sequence-related strictosidine synthase family, which is named after the first enzyme found for this family. Except for STR1, the biological functions of most of the other family members from both plants and animals have not yet been identified. Since no significant sequence homology could be detected between STR1 and known protein structures, the three-dimensional structure of STR1 might help to understand the structural relationship between STR1 and the other family members and would provide a solid base for modeling of other family members.
We recently described an expression system for the production of STR1 from R. serpentina in the milligram range and the protocol for crystallization of this enzyme (Ma et al., 2004; Koepke et al., 2005). Here, we report the crystal structure of R. serpentina STR1 and also the structure of STR1 in complex with each of the natural substrates tryptamine and secologanin. The structure represents a novel six-bladed β-propeller fold in plant proteins. The common features of β-propeller fold proteins are their sequence and functional diversity, which led people to believe that sequence-unrelated propeller structures have evolved from different precursors. However, based on the structure of STR1 and structure-based sequence alignments, we find a possible evolutionary relationship of several sequence-unrelated six-bladed β-propeller structures. The complex structures also allow us to examine issues relating to catalysis, including the architecture of the active site and the nature of the substrate binding pocket, which provide significant hints and the structural basis to unravel the unexplored reaction of this unique plant enzyme. These structures might also serve as a starting point for the rational construction of new STR1 variants with various substrate acceptances, which could lead to novel biologically valuable indole alkaloids.
RESULTS AND DISCUSSION
Overall Architecture of STR1
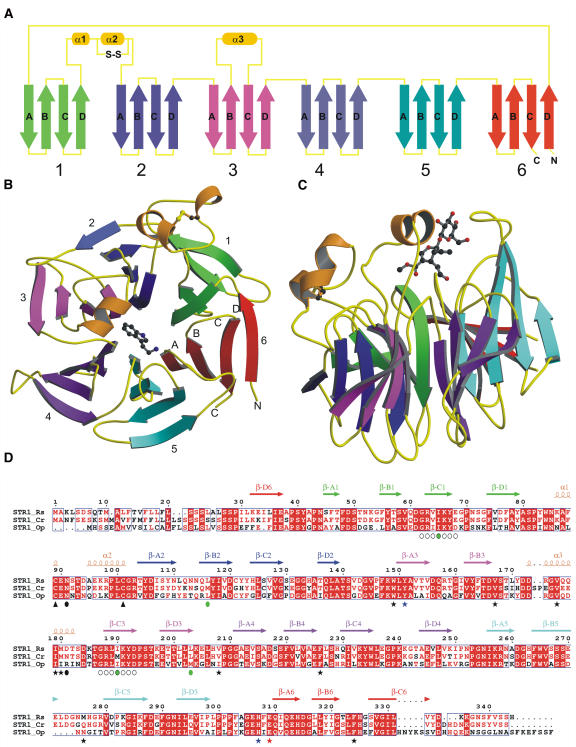
The overall structure of STR1 from R. serpentina resembles a six-bladed four-stranded β-propeller fold (the six blades are indicated from 1 to 6 in Figures 2A and 2B). All six blades are radially arranged around a pseudo six-fold symmetry axis. Each blade contains a twisted four-stranded antiparallel β-sheet. The β-strands in each blade are labeled A to D from the inside to outside of the molecule. The innermost β-strand A (β-A) is closest to the pseudo six-fold axis. The last blade is formed by three strands β-A, β-B, and β-C from the C terminus and one (the outermost strand, β-D) from the N terminus. This type of Velcro closure has already been observed for several other six-bladed β-propeller structures exhibiting different catalytic functions (Scharff et al., 2001; Harel et al., 2004). There are two STR1 molecules in an asymmetric unit. The root mean square deviation (RMSD) of all protein atoms, after superposition of both molecules in the native and ligand complexes, is between 0.19 and 0.24 Å. The contacts between the two molecules are dominated by main chain hydrogen bonds between the β-D strands of the last blade from both monomers. Interfacial area between the two molecules is 1937 Å2, indicating that STR1 might be a dimer. However, STR1 is active as a monomer in solution as determined by size exclusion chromatography (Hampp and Zenk, 1988).
Figure 2.
Overview of the R. serpentina STR1 Structure.
(A) The topology of the STR1 structure. Each blade (consisting of four β-strands) of the propeller is shown in various colors (green, blue, magenta, purple, cyan, and red), and the connecting loop is shown in yellow and helices in orange.
(B) Front view of the six-bladed β-propeller in complex with tryptamine. The top of the propeller is, by convention, the face carrying the loops connecting the β-B strand and β-C strand in each blade.
(C) Side view of the propeller in complex with secologanin.
(D) Sequence alignment of STR1 from different plant species. STR1_Rs, STR1 from R. serpentina and from R. mannii; STR1_Cr, STR1 from C. roseus (sequence identity 79%); STR1_Op, STR1 from O. pumila (sequence identity 58%). Four residues that were mutated to Met are marked with green circles; internal repetitive sequences that were used to design the fourth Met mutation I65M are indicted with open circles below the sequence. Residues that form the hydrophobic active site are highlighted with black asterisks; two polar residues within the active site are marked with blue asterisks; the catalytic residue Glu-309 is marked with a red asterisk. The conserved disulfide bridge is indicated by black arrowheads below the Cys residues. Putative N-glycosylation residues are indicated by black circles.
There are three helices in the STR1 β-propeller structure. The first two helices are located between the loops connecting the outermost strand β-D of blade 1 and the innermost strand β-A of blade 2 (Figures 2A to 2C). The two small helices are pulled closer by a disulfide bridge between Cys-89 and Cys-101. This disulfide bridge is conserved throughout the STR1 family, which seems to be a distinct feature of the family (Figure 2D; see Supplemental Figure 1 online). The covalently bound Cys residues play an important role in the integrity of the substrate binding pocket and ultimately to the overall structure. This is confirmed by mutation of Cys-89 to Ser, which resulted in poor expression in Escherichia coli and complete loss of enzyme activity. A striking feature of the STR1 structure is addition of a helix (α3) between strands β-B and β-C in blade 3. The helix together with a loop connecting strands β-B and β-C in blade 5 forms a cap over the active site, thus shaping the substrate binding pocket (Figures 2A to 2C).
STR1 is expressed in the plant as a precursor protein and is glycosylated (Hampp and Zenk, 1988). Signal peptide and glycosylation is not essential for the activity of STR1 but may be important in subcellular compartmentalization of the enzyme to the vacuole (DeWaal et al., 1995; Ma et al., 2004). There is only one potential N-glycosylation site Asn-91 on STR1 from R. serpentina [Nx(S/T) sites]. Interestingly, Asn-91 is located on the surface of the first specific insertion of the STR1 propeller structure between two helices (α1 and α2). This N-glycosylation site is conserved in all three STR1 from different plant species (Figure 2D). In contrast with STR1 in R. serpentina, different isoforms of STR1 have been purified and characterized in C. roseus; these isoforms have been suggested to result from different glycosylation stages since STR1 from both C. roseus and R. serpentina is encoded by a single-copy gene (DeWaal et al., 1995). Analysis of the sequence of STR1 from C. roseus results in an extra potential N-glycosylation site Asn-187, the corresponding position in R. serpentina is Asp-181, that is located on helix α3 (Figure 2D). Therefore, different glycosylation stages at the N-glycosylation site Asn-187 might contribute the different STR1 isoforms from C. roseus.
STR1 Active Site and the Binding of Tryptamine and Secologanin
The substrate binding pocket of STR1 is located near the pseudo six-fold symmetry axis (Figures 2B and 2C). The binding pocket links the active center to the surface of the molecule along a helix (α3) and a connecting loop between strands β-B and β-C of the fifth blade (Figure 2C). The main residues involved in forming the pocket are Tyr-105, Trp-149, Tyr-151, Val-176, Met-180, Val-208, Phe-226, Ser-269, Met-276, Phe-308, His-307, Glu-309, Leu-323, and Phe-324 (Figures 3A to 3C and 4A to 4C). The overall nature of the pocket is primarily hydrophobic (Figures 4A to 4C). Positively charged residues (His-307 and His-277) and hydrophobic residues (Met-276 and Phe-324) are located at the entrance of the pocket.
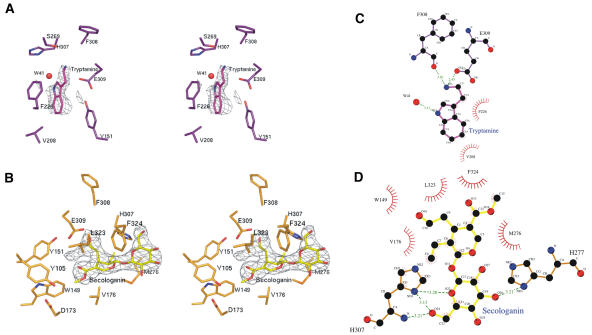
Figure 3.
Overview of Active Site.
(A) Stereoview of tryptamine binding site region. The (Fo-Fc) SIGMAA-weighted electron density of tryptamine contoured at 4 σ is shown in gray. Residues within 4 Å distance from tryptamine are shown in purple and tryptamine in magenta. The water molecule is drawn as a red sphere.
(B) Stereoview of secologanin binding site region. The (Fo-Fc) SIGMAA-weighted electron density of secologanin contoured at 4 σ is shown in gray. Residues within 4 Å distance from secologanin are shown in orange and secologanin in yellow.
(C) Active site interactions of the tryptamine complex.
(D) Active site interactions of the secologanin complex.
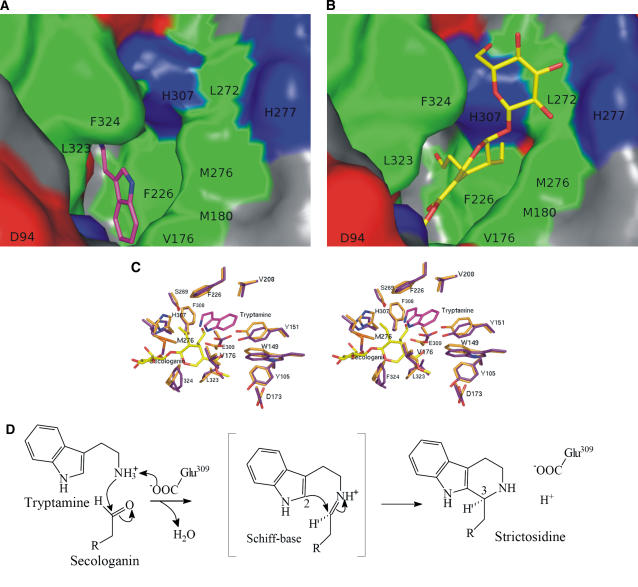
Figure 4.
Surface Representation of STR1-Ligand Complexes.
Hydrophobic residues (Tyr, Trp, Phe, Leu, Met/Mse, Cys, Ile, and Val), positively charged residues (Arg, Lys, and His), negatively charged residues (Asp and Glu), and hydrophilic residues (Ala, Gly, Ser, Thr, Pro, Gln, and Asn) are shown in green, blue, red, and gray, respectively. The surrounding residues are labeled with single-letter codes.
(A) Close-up view of the substrate binding pocket with tryptamine in stick representation.
(B) Close-up view of the substrate binding pocket with secologanin in stick representation.
(C) Stereoview of tryptamine and secologanin together and superposition of the surrounding residues. The color code is as described in Figure 3.
(D) Schematic presentation of the reaction pathway for the Pictet-Spengler–type reaction with Glu-309 involved in the amine deprotonation.
In the tryptamine complex (STR1-TAM), substrate tryptamine is situated at the bottom of the substrate binding pocket, and its location is fully occupied in the A-molecule (Figure 3A) but partially in the B-molecule, which is observed to have a higher B-factor. The amine group of the tryptamine is clearly coordinated with the side chain of the Glu-309 residue, and water (w41) is bound to the nitrogen atom of the tryptamine indole ring. Residues Phe-226, Val-208, His-307, Glu-309, and Tyr-151 are located in close proximity (4.0 Å) of the tryptamine molecule (Figure 3A). The aromatic ring of the tryptamine ring is stacked between Phe-226 and Tyr-151.
In the secologanin complex (STR1-SEL), the second substrate secologanin is also bound in the same pocket (Figures 2C, 3B, and 4B) but does not occupy the tryptamine position. The location of secologanin is well defined in both molecules. This is perhaps due to soaking of the secologanin complex crystals in 6 mM secologanin solution (see Methods). The secologanin molecule is bound in the substrate binding pocket in extended form. Its ester group is facing toward the bottom of the pocket, with the terpenoid ring in the middle and the aldehyde group pointing toward Glu-309 (Figures 3B, 4B, and 4C). The hydrophilic glucose ring of secologanin is pointing away from the pocket (Figure 4C). It is accessible to the solvent, and two of the oxygen atoms are coordinated with the nitrogen atoms of His-307 (Figure 3D). The aldehyde group of the secologanin is in close proximity to the amine group of tryptamine (Figure 4C).
In the native structure, the substrate binding pocket is occupied by a tartrate molecule that is present in the crystallization conditions. In each molecule, tartrate is coordinated with the hydroxyl group of Tyr-151 and the carbonyl oxygen atom of Phe-308. The tartrate molecule occupies an identical position to the tryptamine ring. Similar to the tryptamine complex, the tartrate molecule is fully occupied in the A-molecule and partially in the B-molecule.
The tryptamine or the tartrate for the B-molecule is partially occupied as suggested by the high B-factor, and this is either due to inadequate ligand concentration during cocrystallization or to the crystal packing environment of the second molecule. The helix (α3) and the loop between strands β-B and β-C of blade 5 contribute to form an entrance for the substrates or the ligand. The loop of A-molecule is involved in crystal contacts and therefore gives rise to a more ordered environment for the pocket. The same loop in the B-molecule is not involved in the crystal contacts, and, consequently, its binding pocked is more flexible.
STR1-SEL complex crystals were soaked in 6 mM secologanin solution together with precipitant and glycerol solution before flash freezing. In the resulting structure, secologanin was found to be well ordered in both molecules, indicating adequate substrate concentration needed for full occupation.
Analysis of Active-Site Amino Acid Residues and Implications for Catalysis
The reaction of STR1 resembles a Pictet-Spengler condensation involving the intramolecular addition of carbon 2 of tryptamine, representing the CH-acidic center, to the Schiff base formed between the aldehyde group of secologanin and the primary amine group of tryptamine. Since the primary alkylamines, such as serotonin and tryptamine, have pKa values close to 10, it is clear that amine deprotonation must occur before Schiff base formation. The substrate binding pocket of STR1 is rather hydrophobic, and there are only three polar residues, Glu-309, Tyr-151, and His-307, around the active site. In the STR1-TAM complex structure (Figure 3C), the amine group of tryptamine is hydrogen bonded with Glu-309. Mutation of Glu-309 to Ala significantly reduces the turnover rate (kcat) (879-fold) but surprisingly has almost no influence on the Km for tryptamine (Table 1), indicating a decisive role for Glu in the amine deprotonation (Figure 4D). After proton abstraction, the free amine function could easily react with the aldehyde group of secologanin to form the enzymatic product, strictosidine, through the step of a Schiff base by loss of a water molecule. The Schiff base and the ring formation steps occur very quickly, since stable intermediates cannot be isolated and are not observed in HPLC analyses. The superposition of both complexes shows that both substrates are clearly arranged in close vicinity but do not overlap. In the superposition, the amine group of tryptamine is only 1.23 Å away from the aldehyde group of secologanin. Moreover, in the superposition, the distance between carbon 2 of tryptamine and the carbon of the aldehyde function of secologanin, which would form the C2-C3 bond in strictosidine, is only 1.03 Å (Figures 4C and 4D). However, the structures of tryptamine and secologanin complex are of rather low resolution (2.8 and 3.00 Å, respectively), and the mean positional error of the atoms as estimated from the Luzatti plot (Luzatti, 1952) is 0.37 and 0.46 Å, respectively. Additionally, the RMSD of all protein atoms, after superposition of STR1-TAM complex and STR1-SEL complex, is 0.4 Å. Therefore, the close contacts between tryptamine and secologanin do not necessarily imply substantial structural rearrangement in the transition state. Relative to the tryptamine complex, there are small side chain movements of the surrounding residues upon secologanin binding in the secologanin complex. It is expected that the binding pocket of strictosidine should be similar as shown in Figure 4C, but there could be a minor positional shift of the tryptamine and/or secologanin during the catalysis to form strictosidine.
Table 1.
Kinetic Parameters for Wild-Type STR1 and Its Mutants
|
Km (μM)
|
|||
|---|---|---|---|
| STR1 | kcat (min−1) | Tryptamine | Secologanin |
| Wild type | 78.200 | 6.2 | 39 |
| Y151F | 57.700 | 17.2 | 44 |
| H307A | 1.800 | 7.9 | 5070 |
| E309A | 0.089 | 5.4 | 95 |
The other polar residue close to tryptamine is Tyr-151. However, a minor role for this residue in the catalysis is suggested because a mutation of Tyr-151 to Phe (Table 1) causes an increase in the Km for tryptamine (2.8-fold) without significantly altering the kcat. The 25% decrease in kcat might reflect a subtle influence on the orientation of the catalytic residue Glu-309 since the structure the OH group of Tyr-151 is hydrogen bonded with the carboxyl group of Glu-309. The closest imidazole nitrogen of His-307 is still 4.5 Å away from the amine group of bound tryptamine in the complex structures, too far for His-307 to be directly involved in the catalysis. Site-directed mutagenesis studies also support the conclusion that His-307 is not involved in the binding of tryptamine since mutation of this residue to Ala has no substantial effect on the Km value for tryptamine. Instead, the mutant H307A significantly increases the Km for secologanin (130-fold), suggesting an important role of His-307 in the binding of this substrate or in maintaining the geometry of the binding pocket for secologanin. In the secologanin complex structure, His-307 is hydrogen bonded with two glucose oxygens of secologanin (Figure 3D). The significant decrease in kcat caused by the H307A mutation might reflect a substantial conformational change in the active site that could influence the orientation of the proposed catalytic residue Glu-309.
Therefore, Glu-309 is the only essential catalytic residue of STR1 we have identified to date, although apart from amine deprotonation, other specific functions of Glu-309 remain unclear. This result is also in agreement with our preliminary biochemical results from selective modification of reactive amino acid residues of STR1 by adding various group-selective labeling reagents. Among the reagents used to chemically modify STR1, only a few reduced the enzyme activity (Table 2). STR1 was not inactivated by Tyr, Lys/Arg, and Ser/Cys-selective reagents and was only partially inactivated by Cys- and His-selective reagents. By contrast, STR1 was totally inactivated by the Asp/Glu-selective reagent N,N′-dicyclohexylcarbodiimide (DCC). Interestingly, STR1 activity was affected differently by the carboxylate-selective reagents DCC and N-ethyl-5-phenylisoxazolium-3′-sulfonate (Woodward's reagent K) and was more effectively inactivated by the hydrophobic reagent DCC than by the hydrophilic Woodward's reagent K. Inactivation by DCC could be avoided by addition of the substrate tryptamine or secologanin (Table 3). These results suggest that at least one essential Asp or Glu is located in the active site of STR1, and it is likely that the active site is hydrophobic, which is consistent with our crystal structure analysis.
Table 2.
Effects of Group-Selective Reagents on STR1 Activity
| [Reagent] (mM)
|
[Reagent]/[STR1] (mol/mol)
|
Group Selectivitya
|
Inactivation (%)
|
|||
|---|---|---|---|---|---|---|
| Reagent | 30 min | 2 h | Buffer | |||
| DCC | 0.05 | 50 | Asp/Glu | 100 | A | |
| WK | 1.00 | 1,000 | Asp/Glu | 48 | 53 | A |
| 5.00 | 5,000 | 80 | 100 | |||
| DEPC | 1.00 | 1,000 | His | 49 | 63 | B |
| PCMB | 0.20 | 200 | Cys | 34 | 35 | B |
| PMSF | 1.00 | 1,000 | Ser/Cys | 0 | 0 | C |
| TPCK | 0.12 | 120 | Ser/Cys | 0 | 0 | C |
| TLCK | 0.25 | 250 | Ser/Cys | 0 | 0 | C |
| AEBSF | 4.00 | 4,000 | Ser/Cys | 4 | 12 | C |
| NAI | 10.00 | 10,000 | Tyr | 5 | 12 | D |
| DNFB | 5.00 | 5,000 | Lys/Arg | 0 | 15 | D |
Selective modification of reactive residues of STR1 was performed by adding various amounts (0.05 to 10 mM) of reagents to 1 μM enzyme solution in appropriate buffers. Aliquots of incubation mixture were taken at time intervals, and STR1 activity was measured. The pH of the reaction as well as the molar excess of reagent over the enzyme was designed to optimize the group selectivity of each reagent. DCC, N,N′-dicyclohexylcarbodiimide; WK, N-ethyl-5-phenylisoxazolium-3′-sulfonate (Woodward's reagent K); DEPC, diethylpyrocarbonate; PCMB, p-chloromercuribenzoate; PMSF, phenylmethanesulfonyl fluoride; TPCK, l-chloro-3-(4-tosyl-amido)-4-phenyl-2-butanone; TLCK, l-chloro-3-(4-tosyl-amido)-7-amino-2-heptanone; AEBSF, 4-(2-aminoethyl)-benzenesulfonyl-fluoride; NAI, N-acetylimidazole; DNFB, dinitrofluorobenzene. Buffer A, 50 mM MES, pH 6.0; buffer B, 100 mM potassium phosphate buffer, pH 6.0; buffer C, 100 mM potassium phosphate buffer, pH 7.0; buffer D, 10 mM Tris-HCl, pH 8.0.
The most likely side chains modified under the conditions of the experiment.
Table 3.
Protection of STR1 against Chemical Modification of DCC
| Residual Activity (%) after 15 and 30 min Reaction
|
||
|---|---|---|
| Experiment | 15 min | 30 min |
| No protection | 0 | 0 |
| +Tryptamine (5 mM) | 81 | 52 |
| +Secologanin (5 mM) | 91 | 81 |
The chemical inactivation of STR1 was started after 20-min preincubation of the enzyme with either 5 mM tryptamine or 5 mM secologanin. Molar excess of DCC (100 μM) over enzyme was 100-fold in 50 mM MES, pH 6.0.
It is also noteworthy that STR1 exhibits high substrate specificity, accepting only a few hydroxylated, fluorinated, or methylated tryptamine derivatives (Table 4), but it does not accept dopamine. Dopamine is, however, the substrate for similar reactions catalyzed by the STR1-related enzymes deacetylisoipecoside synthase, deacetylipecoside synthase, and norcoclaurine synthase. Deacetylipecoside synthase, which catalyzes the condensation of dopamine and secologanin to form deacetylipecoside, has recently been purified from Alangium lamarckii (Alangiaceae) (De-Eknamkul et al., 2000). Both STR1 and deacetylipecoside synthase are similar with respect to molecular size, temperature, and pH optimum, exhibiting high substrate specificity and sharing one common substrate secologanin, but they exist in different plant species and have different substrate specificity. Whether both enzymes, leading in each species to different alkaloid types, are evolved from the same ancestor is not known. This can only be elucidated when the amino acid sequence of deacetylipecoside synthase and the structures of both enzymes are available. Norcoclaurine synthase catalyzes the Pictet-Spengler–type condensation of dopamine and 4-hydroxyphenylacetaldehyde to form (S)-norcoclaurine. Molecular cloning and characterization of norcoclaurine synthase from Thalictrum flavum cell culture have been reported recently (Samanani et al., 2004). The open reading frame encoded a protein of 210 amino acids. However, the amino acid sequence of norcoclaurine synthase showed no sequence identity with STR1; instead, it belongs to the pathogenesis-related 10 and Bet v1 protein family. These results strongly suggest that the same reaction type catalyzed by STR1 and norcoclaurine synthase could also be convergently evolved from different ancestors. But the question whether norcoclaurine synthase also uses the same reaction mechanism as that of STR1 can only be answered when the structure of norcoclaurine synthase becomes available.
Table 4.
Substrate Specificity of STR1
| Substrate | Relative Activity (%) |
|---|---|
| Tryptamine | 100.0 |
| 5-Hydroxytryptamine (serotonin) | 9.9 |
| 5-Fluorotryptamine | 9.5 |
| 6-Fluorotryptamine | 6.6 |
| 7-Methyltryptamine | 8.9 |
| 5,6-Dihydroxytryptamine | 7.0 |
| 6-Methoxytryptamine | 2.1 |
| 5-Methyltryptamine | 0.0 |
| 5,7-Dihydroxytryptamine | 0.0 |
| N-Acetyl-5-hydroxytryptamine | 0.0 |
| Phenylalanine | 0.0 |
| Histamine | 0.0 |
| Dopamine | 0.0 |
| 5-Methoxytryptamine | 0.0 |
| N-Methyltryptamine | 0.0 |
| N-ω-Methyltryptamine | 0.0 |
| 2-Methyl-5-hydroxytryptamine | 0.0 |
Recently, it has been proven by isotope-labeled precursors that not only plants but also human cells are capable of synthesizing morphine alkaloids, and the biosynthetic pathway involves a Pictet-Spengler–type reaction, similar to the catalytic mechanism of STR1 (Boettcher et al., 2005). It seems that nature is utilizing this reaction type more universally in both the plant and animal kingdoms.
Gene-Related STR1 Family Members
Enzymes with STR1 activity have been found in several species of plant families Apocynaceae and Rubiaceae (Kutchan, 1993). To date, only the cDNA sequences of STR1 from C. roseus (McKnight et al., 1990), R. serpentina (Kutchan et al., 1988), R. mannii (Bracher and Kutchan, 1992), and O. pumila (Yamazaki et al., 2003) have been reported. The residues maintaining the hydrophobic core of the β-propeller, the hydrophobic active site, and two prominent helix insertions are highly conserved throughout these family members. The characteristic disulfide bond and the catalytic residue Glu-309 are completely conserved (Figure 2D). Therefore, it is likely that STR1s from different plant species diverged while maintaining their overall active site architecture and using a similar catalytic mechanism.
It seems that nature has used the STR1 scaffold for a great variety of functions since many new members of STR1-related genes have been found both in plants and animals that are known to be unrelated to alkaloid biosynthesis (appropriate sequence alignments with STR1 are available in Supplemental Figure 1 online). For example, in Arabidopsis thaliana, STR1-like genes (26 to 39% identity to STR1) form a multigene family, which may be divided into different groups, and may perform different functions and are involved in more than one biochemical pathway (Fabbri et al., 2000). In Drosophila melanogaster, a novel cell surface molecule hemomucin was isolated from a hemocyte-like cell line (Theopold et al., 1996). Hemomucin, which may be involved in the induction of immune responses mediated by lectin in insects, is composed of two domains, one with a mucin-like sequence and the other domain similar to STR1 (31% identity to STR1). An adipocyte plasma membrane–associated protein BSCv (see Supplemental Figure 1 online), which may play a role in adipocyte differentiation, has also been found in human (Morita et al., 2000). These STR1-related proteins maintain the key structural elements of STR1, mostly, the C89-C101 disulfide bridge and the hydrophobic core residues. However, the catalytic residue Glu-309 of STR1 is no longer present in the family members of STR1-related genes (see Supplemental Figure 1 online).
Evolutionary Relevance to Other Six-Bladed Four-Stranded β-Propeller Structures
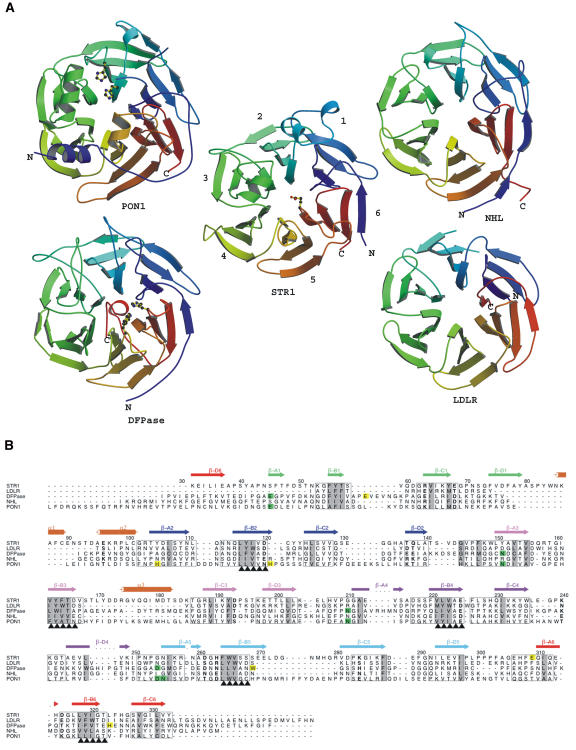
STR1 does not show clear sequence similarities nor does it share functional homologies with other six-bladed β-propeller structures. Among all the six-bladed β-propeller structures, the closest structures that could be aligned to STR1 include diisopropylfluorophosphatase (DFPase) from Loligo vulgaris (Protein Data Bank [PDB] code 1e1a; Scharff et al., 2001), brain tumor NHL domain (PDB code 1q7f; Edwards et al., 2003), serum paroxonase (PON1; PDB code 1v04; Harel et al., 2004), and low-density lipoprotein receptor YWTD domain (LDLR; PDB code 1ijq; Jeon et al., 2001) (Figure 5A). Structures of these proteins could be aligned to STR1 with an RMSD between 1.89 and ∼2.41 Å over 207 to ∼241 amino acids, but structure-based sequence alignments show only 11 to ∼16% sequence identities (Figure 5B). Both DFPase and PON1 are calcium-dependent phosphotriesterases. The residues used to bind catalytic calcium Ca1 are structurally conserved in both enzymes (Figure 5B), although sequence similarity between them is only 25%. Another significant difference between the two enzymes is that the helices H2 and H3 in PON1 are not present in DFPase, leaving DFPase with an open active site and PON1 with a closed one (Figure 5A). In the PON1 structure, the closure of the active site is achieved by the two helices H2 and H3. H2 is located between strands β-B and β-C in blade 3, which corresponds to the helix α3 of the STR1 structure. H3 of PON1 is situated between the strands β-B and β-C in blade 5. Although the corresponding helix is not present in STR1, the loop at the same position contributes to the cap formation by extending to the top of the active site. The comparable closed active site observed in STR1, especially in respect to the unique helix addition in STR1 used to cover the active site, also exists at the same position in the PON1 structure, suggesting that both enzymes might be evolutionarily related. Both enzymes have a similar molecular architecture; however, they differ greatly in their primary sequences and catalytic function. STR1 catalyzes the Pictet-Spengler–type reaction, while PON1 is an esterase. Incubation of STR1 with 10 mM EDTA for even 1 week has no influence on the activity, indicating that metal ions are not required for the catalytic activity or for the structural stability of STR1. This result is also supported by the crystal structure of STR1, where there is no trace of metal ions in the electron density near the binding pocket, while PON1 contains two calcium atoms in its central tunnel. One of them is catalytic calcium and the other is structurally important. Helix H2 in PON1 is thought to be involved in high-density lipoprotein anchoring by pointing its hydrophobic face toward the solvent, while in STR1, the α3 helix is important to create a highly hydrophobic environment around the active site, with its hydrophobic face pointing toward the substrate binding pocket. Interestingly, the ligand binding residues in these structures are located either between the strands β-D and β-A or β-B and β-C (Figure 5A).
Figure 5.
Structural Comparison.
(A) Structural comparison of different six-bladed four-stranded β-propeller fold proteins. Illustrations show the N termini (blue) and C termini (red) of the structures. STR1 (this work) is in the center; PON1, DFPase, NHL, and LDLR are at the top left, bottom left, top right, and bottom right, respectively. The putative catalytic residues of STR1, PON1, and DFPase are shown as ball-and-stick representations.
(B) Structure-based sequence alignment of R. serpentina STR1 with several other six-bladed four-stranded β-propeller structures. Sequence motifs are marked with black arrowheads. Catalytic calcium Ca1 binding residues in PON1 and DFPase are highlighted in green. Putative catalytic residues in PON1, DFPase, and STR1 are in yellow. The sequence numbering is shown according to STR1. Secondary elements of STR1 are shown above the sequence, and their coloring is the same as shown in Figure 2D. Conserved hydrophobic residues are shaded. The boxed residues are within 1.2 Å from the residues of STR1.
Among all the structures, only LDLR contains an easily identifiable internal repetitive sequence XYWTD (X is a hydrophobic residue). STR1 also exhibits a similar XYFTD repeat on β-B3 (Figure 5B). Structure-based sequence alignment showed that this XYFTD repeat is located at the equivalent position of the XYWTD repeat in LDLR, which led us to consider whether there is any evolutionary relationship between the sequences of STR1 and LDLR. Further analysis of sequences in STR1 was performed at those sections where five XYWTD repeats are located in LDLR. In fact, two other similar sequence repeats XYIVD (β-B2) and XYIGT (β-B6) were detected in STR1, which correspond to XYWSD (β-B2) and XFWTD (β-B6) in LDLR (Figure 5B). In addition, the rest of the equivalent positions in STR1 contain the following sequences: XLVAE (β-B4) and XWVSS (β-B5). At first glance, it seems that the last two sequences are unrelated to the XYWTD repeat. However, further inspection shows that there is a common motif for these sequences, XXX#§, in which three hydrophobic residues (XXX) are followed by a small residue (#; Ser/Thr/Ala/Val/Gly) and a hydrophilic residue (§; in most cases Asp or Glu). Surprisingly, this common motif also fits all the relevant sequences in PON1, DFPase, and NHL (Figure 5B). These observations indicate that all these structurally related proteins are in fact evolutionary related; it is likely that they have divergently evolved from a common ancestral β-sheet gene. The internal repeats may suggest that a six-bladed β-propeller might be first formed by duplication of an ancestral β-sheet gene that contains the YWTD repeat. During evolution, the ancestral structure undergoes insertions and deletions to shape the central tunnel and to adopt different functions, for example, the cap region in PON1 and STR1. As suggested before, propeller topology does not impose specific constraints on sequence (Murzin, 1992), so these sequence repeats gradually lose their sequence identity, but they still keep some common sequence homology features that are important to maintain the overall structural stability of the fold, as can be seen by the XXX#§ consensus at the second strand of each blade (Figure 5B).
Extreme sequence diversity accompanied by greatly different functions and phylogenetic origins usually makes people believe that these sequence-unrelated propeller structures are unlikely to have a common precursor and that the evolution may have taken place from distinct ancestral β-sheet genes (Jawad and Paoli, 2002). Now, based on the structure of STR1 and structure-based sequence alignment between STR1 and four other six-bladed β-propeller structures, a possible evolutionary relationship of several sequence-unrelated six-bladed β-propeller structures is proposed.
METHODS
Expression, Purification, Mutagenesis, and Enzyme Activity Assay
Rauvolfia serpentina STR1 excluding the signal peptide (first 28 residues) was subcloned into pQE-2 vector. Only two additional residues (Gly and Ala) are left on the N terminus of STR1 when the N-terminal His tag is cleaved with dipeptidyl aminopeptidase (Qiagen). Expression in Escherichia coli strain M15 and purification of native STR1 have been described previously (Ma et al., 2004). Site-directed mutants were constructed using the Quickchange method (Stratagene), verified by sequencing the complete gene and purified as for the wild-type enzyme.
Selenomethionyl-STR1 (SeMet-STR1) was obtained by the Met pathway inhibition technique (Van Duyne et al., 1993) with the same vector and E. coli strain used for native STR1. The purification procedure for SeMet-STR1 was the same as that used for the native enzyme, except that 1 mM DTT was added to all of the buffers.
Native STR1 contains only two Met residues. Crystals of this SeMet-STR1 were proven to be insufficient to result in the useful anomalous signal in solving the structure. Therefore, two mutants with four Met residues (STR1-L116MI190M) and six Met residues (STR1-L116M I190ML203MI65M) were prepared; these two mutants are designated here as 4Met-STR1 and 6Met-STR1, respectively. Three positions of Leu or Ile for mutations (Leu-116, Ile-190, and Leu-203) were chosen based on the sequence alignment with STR1 from species Catharanthus roseus and Ophiorrhiza pumila (see the sequence alignment in Figure 2D). The fourth one (Ile-65) was chosen based on the high sequence similarity between 62GRVI65KYE68 and 187GRLI190KYD193 inside the R. serpentina STR1 sequence alone, since mutation of Ile-190 to Met in the mutant 4Met-STR1 does not influence the activity or change the crystallization behavior of this mutant. Both 4Met-STR1 and 6Met-STR1 mutants are active with only a slight decrease on the activity (∼10%) compared with native STR1. Selenomethionyl-4Met-STR1 (4SeMet-STR1) and selenomethionyl-6Met-STR1 (6SeMet- STR1) were prepared as mentioned above for the SeMet-STR1.
The activity of STR1 was determined by incubating 1 mM tryptamine, 2 mM secologanin, and 50 mM phosphate buffer, pH 7.0 (total volume 50 μL), for 15 min at 35°C while shaking (400 rpm). The reaction was terminated by addition of 100 μL methanol. After centrifugation (11,000g; 5 min), the supernatant was analyzed by HPLC. For HPLC, a Merck Hitachi instrument and a Lichrospher 60 RP select B column (125 × 4 mm; 5 μm) were used: injection volume 60 μL; flow rate 1 mL min−1; absorption measured at 250 nm; acetonitrile/H2O (pH 2.3 adjusted by H3PO4) as solvent system, gradient 10:90 → 50:50 within 8 min → 80:20 within 3 min → 10:90 within 0.5 min → 10:90 for 3.5 min. Retension time values are 5.47 min (tryptamine), 6.57 min (secologanin), and 8.65 min (strictosidine). To determine Km and kcat values, the enzyme assay was performed using different concentrations of tryptamine and secologanin.
Crystallization
Crystals of STR1 and its complexes were grown by the hanging drop vapor diffusion method (Ma et al., 2004; Koepke et al., 2005). The reservoir solution contained 0.8 M potassium sodium tartrate tetrahydrate and 0.1 M HEPES, pH 7.5. The enzyme concentration was 4 to 5 mg/mL. Crystals of STR1 in complex with substrates tryptamine (STR1-TAM complex) and secologanin (STR1-SEL complex) were obtained by cocrystallization in the presence of 1.0 mM tryptamine and secologanin, respectively, in the enzyme solution.
Data Collection and Processing
Native, SeMet, and tryptamine complex crystals were cryo-protected by addition of 25% glycerol to the precipitant buffer before being flash-cooled in a stream of cold nitrogen at 100K. The secologanin complex crystals were flash-cooled after 8 min of soaking in the solution of 25% glycerol, 6.0 mM secologanin, and the precipitant buffer. Three multiwavelength anomalous diffraction (MAD) experiments were performed near the selenium edge. The MAD data sets from 4SeMet-STR1 were collected at the PX beamline at the Swiss Light Source in Villigen, Switzerland, and another three wavelength MAD data sets from 6SeMet-STR1 were collected at the BW7A beamline of EMBL Hamburg at the DORIS storage ring of the Deutsches Elektronen Synchrotron DESY (Hamburg, Germany). All the x-ray data from cocrystals were collected either on the BW7A, BW7B, X11, or X13 beamline of EMBL Hamburg. The diffraction data were processed and scaled using HKL (Otwinowski and Minor, 1997) in the R3 space group. The data collection statistics are illustrated in Table 2. The preliminary data analysis showed that the crystals contain ∼66% solvent content with a Matthews coefficient of 3.6 and two molecules of STR1 in an asymmetric unit.
Structure Determination, Refinement, and Quality of the Models
The structure was solved by the combination of MAD and molecular replacement methods using SeMet-substituted STR1. Initially, the interpretable electron density was obtained at 3.2-Å resolution for the 6SeMet-STR1 data set using the three wavelengths MAD protocol of AUTO-RICKSHAW: an automated crystal structure determination platform (Panjikar et al., 2005) in the R3 space group. Within this procedure, the structure factors were calculated from the measured intensity by employing the CCP4 programs (Collaborative Computation Project Number 4, 1994), and 8 out of 12 selenium atoms were located using SHELXD (Schneider and Sheldrick, 2002). The positions of these sites were refined using the program MLPHARE (Collaborative Computation Project Number 4, 1994). Density modification and twofold noncrystallographic symmetry (NCS) averaging were performed using DM (Cowtan, 1994) in the correct enantiomorph. Subsequently, it was possible to trace most of the protein backbone using the molecular graphics program XTALVIEW/XFIT (McRee, 1999). However, side chains and some loops could not be placed unambiguously in the electron density. Therefore, MAD phases were calculated with the 4SeMet-STR1 data set in the similar manner as described above and were significantly improved when it was combined with the phase generated from the 6SeMet-STR1 data set using the program SIGMAA (Collaborative Computation Project Number 4, 1994) and continued with the phase extension to 2.95 Å by NCS averaging and density modification. The model was further improved by building missing loops. Some of the side chains could be recognized and were placed in the electron density. At this stage of the model building, it became evident that the structure looked like a six-bladed β-propeller fold. A quick search with Structural Classification of Proteins database (http://scop.berkeley.edu/) showed many structures (i.e., PDB codes 1v04, 1ijq, 1e1a, and 1q7f) containing such a fold. Although molecular replacement against the STR1 data was unsuccessful using any of the structures as a search model, the 1v04 structure could be placed in the experimental electron density successfully using the phased molecular replacement protocol of the program MOLREP (Vagin and Isupov, 2001). The resultant model was used as a guideline for further model building, which helped unambiguous tracing of β-strands in the correct direction. The side chain was fitted in the electron density wherever the map was interpretable. The model was further improved using the graphics program COOT (Emsley and Cowtan, 2004) and its real space fitting and interactive manual building. The model was subjected to multiple rounds of simulated annealing, followed by positional and restrained B-factor refinement as implemented in CNS (Brünger et al., 1998) using the data from 20 to 2.95 Å. The alternate refinement and manual rebuilding of the SeMet-STR1 model resulted in a nearly complete backbone with a majority of side chains positioned; however, the quality of the electron density maps did not permit the rapid convergence of a complete STR1 model. Therefore, further refinement of the model was halted. As 2.3-Å x-ray data (STR1-Highres) were available from a tryptamine complex, the complex was solved by molecular replacement based on this partial SeMet-derived structure using rigid body refinement in CNS. The resultant model was used as a starting model for the automated model building program ARP/wARP (Perrakis et al., 1999; Morris et al., 2004) using the data from 20- to 2.3-Å resolution. Eighty-five percent of the complete model was built automatically, and the model was completed further with manual building using COOT alternating with additional REFMAC cycles, which included bulk solvent correction, anisotropic scaling, NCS restraints, and with each molecule defined as a TLS group in the modeling of anisotropy in the program REFMAC5 (Murshudov et al., 1997; Winn et al., 2001).
Native and all other ligand structures were solved by molecular replacement using the high-resolution structure (2.3-Å resolution) as a search model, as the data were nonisomorphous to each other. The structures were refined using a similar refinement protocol as described above for the high-resolution structure.
The final native STR1 structure was refined to Rcryst 18.9% and Rfree 23.6% using the data from 20 to 2.96 Å. The structure comprises residues 29 to 333 per molecule and includes 99 water molecules, and the electron density unambiguously showed the residual density of tartrate molecules in each molecule.
STR1-Highres comprises residues 32 to 333 and includes 468 water molecules. The structure was refined with Rcryst 18.8% and Rfree 21.7% in the resolution range 20 to 2.3 Å. STR1-Highres data were collected from a STR1-tryptamine complex crystal, and the residual density was observed to 8σ. However, the density could not be explained only with tryptamine, as the density was certainly bigger than the expected tryptamine density (stereoview of the residual density is shown in Supplemental Figure 2 online). All our interpretations of the residual density were not convincing, even with multiple conformations of tryptamine. The density was not modeled with a single tryptamine, as the residual density was not fully interpreted. The data played a main role in resolving the structure. Therefore, another data set of STR1-TAM was collected and refined (see below). The located tryptamine in the STR1-TAM complex fits the residual density with room of some unknown molecule.
The STR1-TAM complex comprises residues 35 to 333 as well as one tryptamine per STR1 molecule. The model includes 173 water molecules. The model is refined with Rcryst 16.4% and Rfree 21.7% in the resolution range 20 to 2.8 Å.
The structure of the STR1-SEL complex comprises residues 32 to 333 and one secologanin per STR1 molecule and includes 115 water molecules. The model is refined with Rcryst 18.6% and Rfree 24.0% in the resolution range 20 to 3.00 Å.
Comparisons of the backbone trace of the partial SeMet-STR1 and the reported structures displayed nearly identical architecture and no evidence for conformational differences between the native and complex forms of STR1. Therefore, refinement of the original SeMet-STR1 structure was not continued. The overall geometric quality of resultant models was assessed using PROCHECK (Laskowski et al., 1993). Table 5 summarizes the data collection and refinement statistics on each of the final models.
Table 5.
Data Collection and Refinement Statistics of STR1
| STR1-TAM Complex
|
STR1-SEL Complex
|
4SeMet-STR1
|
6SeMet-STR1
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| STR1-Native | STR1-Highres | Peak | Inflection | Remote | Peak | Inflection | Remote | |||
| Wavelength (Å) | 0.9330 | 0.9714 | 0.9714 | 0.8057 | 0.9791 | 0.9793 | 0.9537 | 0.9813 | 0.9822 | 0.9714 |
| Unit cell (Å) | a = 150.3 | a = 147.3 | a = 148.9 | a = 148.60 | a = 149.93 | a = 150.24 | a = 152.00 | a = 149.46 | a = 149.41 | a = 151.10 |
| (Space group R3) | c = 122.4 | c = 122.3 | c = 121.3 | c = 121.73 | c = 121.39 | c = 121.49 | c = 121.88 | c = 121.45 | c = 121.38 | c = 122.75 |
| Total reflections | 43,518 | 110,461 | 107,648 | 235,230 | 50,199 | 56,136 | 44,692 | 98,520 | 98,445 | 118,859 |
| Unique reflections | 21,253 | 45,003 | 30,858 | 19,913 | 25,356 | 26,779 | 23,394 | 16,656 | 16,656 | 20,193 |
| Mosaicity | 0.91 | 0.21 | 1.10 | 0.375 | 0.97 | 0.98 | 1.1 | 0.71 | 0.73 | 0.76 |
| Resolution (Å) | 20–2.96 | 20–2.30 | 20–2.80 | 20–3.00 | 50–2.95 | 50–2.80 | 20–3.2 | 20–3.2 | 30–3.24 | 20–3.1 |
| Completeness (%) | 98.4 (99.3) | 99.9 (100) | 99.0 (100) | 100 (100) | 98.4 (96.9) | 97.2 (95.3) | 98.2 (97.3) | 99.8 (100) | 99.8 (100) | 99.7 (99.2) |
| I/σ (I) | 14.3 (1.9) | 15.8 (3.2) | 10.8 (3.0) | 18.6 (3.2) | 11.8 (1.7) | 14.7 (1.9) | 10.6 (2.3) | 13.3 (3.6) | 15.1 (4.0) | 15.8 (3.0) |
| Rmerge (%)a | 7.4 (38.0) | 5.2 (37.7) | 9.9 (43.6) | 12.3 (70.0) | 8.9 (18.8) | 6.6 (27.6) | 7.4 (28.9) | 14.1 (54) | 11.7 (48.1) | 10.8(45.4) |
| Refinement | ||||||||||
| Resolution (Å) | 20–2.96 | 20–2.30 | 20–2.80 | 20–3.00 | ||||||
| Rcryst (Rfree) (%)b | 18.9/23.6 | 18.8/21.7 | 16.4/21.7 | 18.6/24.0 | ||||||
| Number of nonhydrogen atoms | 4,811 | 4,922 | 4,959 | 4,926 | ||||||
| Number of water molecules | 99 | 468 | 173 | 115 | ||||||
| B+ (Å2) for protein | 20.7 | 22.2 | 22.0 | 26.5 | ||||||
| B+ (Å2) for ligand | 72.0 | – | 67.7 | 60.4 | ||||||
| B+ (Å2) for water | 52.4 | 50.8 | 45.9 | 56.3 | ||||||
| RMSD of bond (Å) | 0.021 | 0.021 | 0.021 | 0.021 | ||||||
| RMSD of angles (°) | 1.99 | 1.95 | 1.95 | 1.96 | ||||||
| ++Most favorable region (%) | 84.4 | 87.6 | 85.5 | 81.9 | ||||||
| ++Additionally allowed region (%) | 14.8 | 11.8 | 13.5 | 17.3 | ||||||
| ++Generously allowed region (%) | 0.8 | 0.6 | 1.0 | 0.8 | ||||||
The values in parentheses correspond to the last resolution shell. +Average B-factor. ++Residues in Ramachandran plot.
Rmerge = ΣhklΣi|Ii (hkl) – < I(hkl) >|/ΣhklΣi< I(hkl) >, where < I(hkl) > is the average intensity over symmetry equivalent reflections.
Rcryst (Rfree) = Σhkl||Fo(hkl)| – |Fc(hkl)|| /Σhkl|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
Structurally related proteins were retrieved from secondary structure matching (http://www.ebi.ac.uk/msd-srv/ssm/cgi-bin/ssmserver) servers using the single STR1 monomer model. Structure-based sequence alignment was performed using the program STAMP (Russell and Barton, 1992), and the picture of the sequence alignment was made using the program ALSCRIPT (Barton, 1993). All figures were produced using MOLSCRIPT (Kraulis, 1991), RASTER3D (Merritt and Murphy, 1994), PyMol (DeLano, 2002), and LIGPLOT (Wallace et al., 1995).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers P68175 (STR1 from R. serpentina), P18417 (STR1 from C. roseus), and Q94LW9 (STR1 from O. pumila). The model coordinates and structural factor amplitudes have been deposited in the PDB for structures of the STR1-Native (2FP9), STR1-Highres (2FP8), STR1-TAM complex (2FPB), and STR1-SEL complex (2FPC).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of STR1 from Different Plant Species with Other STR1-Related Gene Family Members.
Supplemental Figure 2. The Unexplained Fo-Fc Electron Density of a Ligand at 4.0 σ and Residues Nearby the Binding Pocket in STR1-Highres.
Supplementary Material
Acknowledgments
We thank T.M. Kutchan (Leibniz Institute of Plant Biochemistry, Halle/Saale, Germany) for kindly sharing the STR1 clone with us. The continuous interest of H. Michel and G. Fritzsch (Max Plank Institute of Biophysics, Frankfurt, Germany) and U. Pindur (Institute of Pharmacy, Mainz, Germany) and linguistic advice as well as helpful discussions by P.A. Tucker (European Molecular Biology Laboratory) are greatly appreciated. We also thank Verena Linhard (Max-Planck-Institute of Biophysics, Frankfurt, Germany) for excellent technical assistance. Staff members of the EMBL BW7A, BW7B, and X13 beamline at the DORIS storage ring (DESY, Hamburg, Germany) and PX beamline at the Swiss Light Source in Villigen (Switzerland) are appreciated for their help. This work was supported by the Deutsche Forschungsgemeinschaft (Bonn, Bad-Godesberg, Germany) and the Fonds der Chemischen Industrie (Frankfurt/Main, Germany) together with the Bundesministerium für Bildung und Forschung (Berlin, Germany).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joachim Stöckigt (stoeckig@mail.uni-mainz.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038018.
References
- Barton, G.J. (1993). ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6 37–40. [DOI] [PubMed] [Google Scholar]
- Boettcher, C., Fellermeier, M., Boettcher, C., Dräger, B., and Zenk, M.H. (2005). How human neuroblastoma cells make morphine. Proc. Natl. Acad. Sci. USA 101 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher, D., and Kutchan, T.M. (1992). Strictosidine synthase from Rauvolfia serpentina: Analysis of a gene involved in indole alkaloid biosynthesis. Arch. Biochem. Biophys. 294 717–723. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computation Project Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. (1994). ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31 34–38.
- De-Eknamkul, W., Suttipanta, N., and Kutchan, T.M. (2000). Purification and characterization of deacetylipecoside synthase from Alangium lamarckii Thw. Phytochemistry 55 177–181. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. (2002). The PyMOL Molecular Graphics System. (San Carlos, CA: DeLano Scientific).
- DeWaal, A., Meijer, A.H., and Verpoorte, R. (1995). Strictosidine synthase from Catharanthus roseus: Purification and characterization of multiple forms. Biochem. J. 306 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, T.A., Wilkinson, B.D., Wharton, R.P., and Aggarwal, A.K. (2003). Model of the brain tumor-pumilio translation repressor complex. Genes Dev. 17 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan, K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D 60 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fabbri, M., Delp, G., Schmidt, O., and Theopold, U. (2000). Animal and plant members of a gene family with similarity to alkaloid-synthesizing enzymes. Biochem. Biophys. Res. Commun. 271 191–196. [DOI] [PubMed] [Google Scholar]
- Hampp, N., and Zenk, M.H. (1988). Homogeneous strictosidine synthase from cell suspension cultures of Rauvolfia serpentina. Phytochemistry 27 3811–3815. [Google Scholar]
- Harel, M., et al. (2004). Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 11 412–419. [DOI] [PubMed] [Google Scholar]
- Jawad, Z., and Paoli, M. (2002). Novel sequences propel familiar folds. Structure 10 447–454. [DOI] [PubMed] [Google Scholar]
- Jeon, H., Meng, W., Takagi, J., Eck, M.J., Springer, T.A., and Blacklow, S.C. (2001). Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat. Struct. Biol. 8 499–504. [DOI] [PubMed] [Google Scholar]
- Koepke, J., Ma, X.Y., Fritzsch, G., Michel, H., and Stöckigt, J. (2005). Crystallization and preliminary x-ray analysis of strictosidine synthase and its complex with the substrate tryptamine. Acta Crystallogr. D 61 690–693. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Kutchan, T.M. (1993). Strictosidine: From alkaloid to enzyme to gene. Phytochemistry 32 493–506. [DOI] [PubMed] [Google Scholar]
- Kutchan, T.M., Hampp, N., Lottspeich, F., Beyreuther, K., and Zenk, M.H. (1988). The cDNA clone for strictosidine synthase from Rauvolfia serpentina. DNA sequence determination and expression in Escherichia coli. FEBS Lett. 237 40–44. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Luzatti, P.V. (1952). Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr. D 5 802–810. [Google Scholar]
- Ma, X.Y., Koepke, J., Fritzsch, G., Diem, R., Kutchan, T.M., Michel, H., and Stöckigt, J. (2004). Crystallization and preliminary x-ray crystallographic analysis of strictosidine synthase from Rauvolfia – The first member of a novel enzyme family. Biochim. Biophys. Acta 1702 121–124. [DOI] [PubMed] [Google Scholar]
- McKnight, T.D., Roessner, C.A., Devagupta, R., Scott, A.I., and Nessler, C.L. (1990). Nucleotide sequence of a cDNA encoding the vacuolar protein strictosidine synthase from Catharanthus roseus. Nucleic Acids Res. 18 4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee, D.E. (1999). XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Murphy, M.E. (1994). Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50 869–873. [DOI] [PubMed] [Google Scholar]
- Morita, M., Hara, Y., Tamai, Y., Arakawa, H., and Nishimura, S. (2000). Genomic construct and mapping of the gene for CMAP (Leukocystatin/Cystatin F, CST7) and identification of a proximal novel gene, BSCv (C20orf3). Genomics 67 87–91. [DOI] [PubMed] [Google Scholar]
- Morris, R.J., Zwart, P.H., Cohen, S., Fernandez, F.J., Kakaris, M., Kirillova, O., Vonrhein, C., Perrakis, A., and Lamzin, V.S. (2004). Breaking good resolutions with ARP/wARP. J. Synchrotron. Radiat. 11 56–59. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53 240–255. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G. (1992). Structural principles for the propeller assembly of β-sheets: The preference for seven-fold symmetry. Proteins 14 191–201. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Panjikar, S., Parthasarathy, V., Lamzin, V.S., Weiss, M.S., and Tucker, P.A. (2005). Auto-Rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D 61 449–457. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R.J., and Lamzin, V.S. (1999). Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6 458–463. [DOI] [PubMed] [Google Scholar]
- Russell, R.B., and Barton, G.J. (1992). Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins Struct. Funct. Genet. 14 309–323. [DOI] [PubMed] [Google Scholar]
- Samanani, N., Liscombe, D.K., and Facchini, P.J. (2004). Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40 302–313. [DOI] [PubMed] [Google Scholar]
- Scharff, E.I., Koepke, J., Fritzsch, G., Lücke, C., and Rüterjans, H. (2001). Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure 9 493–502. [DOI] [PubMed] [Google Scholar]
- Schneider, T.R., and Sheldrick, G.M. (2002). Substructure solution with SHELXD. Acta Crystallogr. D 58 1772–1779. [DOI] [PubMed] [Google Scholar]
- Stevens, L.H., Giroud, C., Pennings, E.J.M., and Verpoorte, R. (1993). Purification and characterization of strictosidine synthase from a suspension culture of Cinchona robusta. Phytochemistry 33 99–106. [Google Scholar]
- Stöckigt, J., and Ruppert, M. (1999). Strictosidine: The biosynthetic key to monoterpenoid indole alkaloids. In Comprehensive Natural Products Chemistry: Amino Acids, Peptides, Porphyrins and Alkaloids, Vol. 4, D.H.R. Barton, K. Nakanishi, O. Meth-Cohn, and J.W. Kelly, eds (Amsterdam: Elsevier), pp 109–138.
- Theopold, U., Samakovlis, C., Erdjument-Bromage, H., Dillon, N., Axelsson, B., Schmidt, O., Tempst, P., and Hultmark, D. (1996). Helix pomatia lectin, an inducer of Drosophila immune response, binds to hemomucin, a novel surface mucin. J. Biol. Chem. 271 12708–12715. [DOI] [PubMed] [Google Scholar]
- Treimer, J.F., and Zenk, M.H. (1979). Purification and properties of strictosidine synthase, the key enzyme in indole alkaloid formation. Eur. J. Biochem. 101 225–233. [DOI] [PubMed] [Google Scholar]
- Vagin, A.A., and Isupov, M.N. (2001). Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D 57 1451–1456. [DOI] [PubMed] [Google Scholar]
- Van Duyne, G.D., Standaert, R.F., Karplus, A., Schreiber, S.L., and Clardy, J. (1993). Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and Rapamycin. J. Mol. Biol. 229 105–124. [DOI] [PubMed] [Google Scholar]
- Wallace, A.C., Laskowski, R.A., and Thornton, J.M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8 127–134. [DOI] [PubMed] [Google Scholar]
- Winn, M.D., Isupov, M.N., and Murshudov, G.N. (2001). Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57 122–133. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Sudo, H., Yamazaki, M., Aimi, N., and Saito, K. (2003). Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: Cloning, characterization and differential expression in tissues and by stress compounds. Plant Cell Physiol. 44 395–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.