Abstract
The NF-κB family of transcription factors plays a critical role in numerous cellular processes, particularly the immune response. Our understanding of how the different NF-κB subunits act coordinately to regulate gene expression is based on a limited set of genes. We used genome-scale location analysis to identify targets of all five NF-κB proteins before and after stimulation of monocytic cells with bacterial lipopolysaccharide (LPS). In unstimulated cells, p50 and p52 bound to a large number of gene promoters that were also occupied by RNA polymerase II. After LPS stimulation, additional NF-κB subunits bound to these genes and to other genes. Genes that became bound by multiple NF-κB subunits were the most likely to show increases in RNA polymerase II occupancy and gene expression. This study identifies NF-κB target genes, reveals how the different NF-κB proteins coordinate their activity, and provides an initial map of the transcriptional regulatory network that underlies the host response to infection.
Keywords: chromatin immunoprecipitation, immune response, transcription factor, macrophage, gene expression
Many transcriptional regulators exist as families sharing similar structures, DNA sequence preferences, and biological functions. The human NF-κB family contains five members: p65 (RelA), p50, c-Rel, RelB, and p52. These subunits form homo- and heterodimers that control a broad spectrum of biological processes, including development, apoptosis, and the immune response (1, 2). The NF-κB regulators have also been implicated in a variety of disease states, such as chronic inflammation, diabetes, and cancer (3–6).
NF-κB is essential for initiation of the innate immune response. Mice deficient in NF-κB family members exhibit defects in macrophage activation and a compromised immune response to pathogens (7). NF-κB is activated in macrophages downstream of pathogen-sensing Toll-like receptors (TLRs) through an evolutionary conserved signaling pathway (8, 9). Lipopolysaccharide (LPS), a component of the Gram-negative bacterial cell wall, binds to TLR-4 and induces a gene expression program almost identical to that induced by whole bacteria (10, 11).
In many cells, NF-κB is sequestered in the cytoplasm through interaction with inhibitory IκB proteins (1). However, the constitutive presence of p50 has been detected in the nucleus of unstimulated monocytes and other cell types where it has been shown to bind to a limited number of promoters (12–18). More recently, NF-κB dimers were revealed to constantly shuttle between the cytoplasm and the nucleus in unstimulated cells (17). p50 lacks a transcriptional activation domain and therefore acts predominantly to repress gene expression, although it can also activate transcription in association with other regulators (16–22). After cellular stimulation, IκB proteins are ubiquitinated and degraded, leading to the increased nuclear presence of all NF-κB subunits and transcriptional activation of their target genes (1).
Chromatin immunoprecipitation (ChIP) has been used successfully to identify direct targets of NF-κB in living cells (16, 18, 23–25). However, these studies have been restricted to either a small set of genes or selected NF-κB subunits. This lack of genomic binding data for each NF-κB subunit limits our understanding of how the different NF-κB family members act coordinately in the cell. To improve our understanding, we have performed a genome-scale survey of NF-κB binding before and after stimulation of human monocytic cells with bacterial LPS. These data identify previously unknown NF-κB target genes and reveal the coordinated action of the five different NF-κB proteins.
Results
NF-κB Binding in Cells Before and After LPS Stimulation.
We used genome-scale location analysis to profile DNA binding of NF-κB proteins in U937 cells before and 1 h after LPS stimulation. Genome-scale location analysis couples ChIP with microarrays to identify DNA sequences bound by transcription factors in living cells (25–30). DNA sequences that are bound by the transcription factor of interest are enriched in the ChIP fraction relative to whole genomic DNA. For this study, we used antibodies that have previously been shown to be specific for each of the five NF-κB proteins (Table 1, which is published as supporting information on the PNAS web site) and microarrays containing probes verified to represent 9,496 proximal promoters (30). All NF-κB binding experiments were performed in triplicate and P values were calculated to assess the significance of each binding event (see Materials and Methods) (26, 28, 30). We selected a stringent P value threshold of 0.002 to identify genes bound by NF-κB (Fig. 6, which is published as supporting information on the PNAS web site).
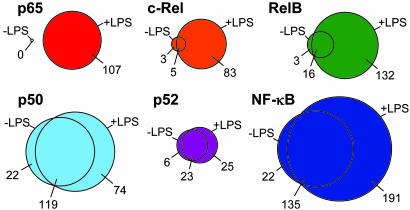
NF-κB subunits together were found to occupy a total of 348 genes at P values below 0.002 (Fig. 1). NF-κB bound 157 genes in unstimulated cells and 326 genes after LPS induction. Our list of NF-κB target genes was enriched for genes with roles in the immune response and transcriptional regulation (Table 2, which is published as supporting information on the PNAS web site) and included 38 previously known NF-κB targets, for example CCL3 (MIP-1α), CCL4 (MIP-1β), CSF2 (GM-CSF), IL8, PTGS2 (COX-2), RAC2, SRF, STAT5A, TNF, and TRAF1. Eleven other previously known target genes were found to be bound at P values between 0.002 and 0.01, including CXCL1, CXCL3, IL10, and IRF1. The identities of the NF-κB target genes identified here (P < 0.002) can be found in Table 3, which is published as supporting information on the PNAS web site. In addition to rediscovering well characterized NF-κB targets, we found many other target genes such as CD58 (LFA3), CREB1, HSPCB (HSP90-β), IL6ST (GP130), ITGA5, JUN, NFKBIB (IκBβ), NRAS, RB1, and VAV1. Gene-specific PCRs of ChIP material compared with whole-cell extract corroborated binding interactions detected by the array (Figs. 6 and 7, which are published as supporting information on the PNAS web site).
Fig. 1.
The overlap between genes bound before and after LPS stimulation by each NF-κB subunit and all NF-κB subunits together, displayed as Venn diagrams. The number of genes occupied by each subunit exclusively in unstimulated cells, exclusively in LPS-stimulated cells, or occupied in both are indicated. All binding events are significant at a P value threshold of 0.002.
p50 and p52 Bind to Significant Numbers of Genes Before Cellular Stimulation.
We found that each of the NF-κB subunits occupied a greater number of genes after LPS stimulation (Fig. 1). However, there were striking differences in the extent to which the various subunits were excluded from binding their target genes in unstimulated cells. Consistent with previous data, p65 binding was not detected in unstimulated cells, but we identified 107 target genes after LPS stimulation. All of the other NF-κB subunits occupied some gene promoters in unstimulated cells and a larger but overlapping set of genes after LPS induction. RelB and c-Rel bound only very small fractions of their target genes in the absence of LPS stimulation (13% and 10%, respectively). In contrast, p50 and p52 occupied a large fraction of their target genes in unstimulated cells (66% and 54%, respectively), the majority of which remained bound after LPS treatment. Genes bound by p50 and p52 in unstimulated U937 cells included the known NF-κB targets CCL3, CSF2, GADD45B, HLA-B, and NFKB2 (p100/p52) itself. The presence of both p50 and p52 in the nucleus of U937 cells was confirmed by cellular fractionation followed by immunoblotting (Fig. 8, which is published as supporting information on the PNAS web site). We conclude that, in unstimulated U937 cells, p50 and p52 not only enter the nucleus but also occupy a significant fraction of their target genes.
NF-κB Co-Occupies Genes with RNA Polymerase II in Unstimulated Cells.
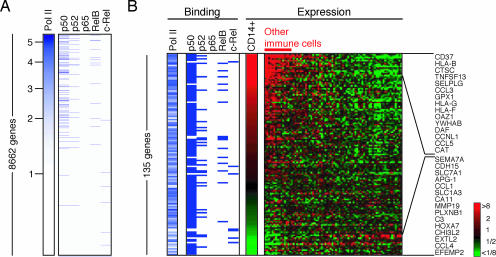
p50 has been associated with both gene activation and gene repression. We therefore examined the transcriptional status of genes bound by NF-κB in unstimulated cells. We first used genome-scale location analysis to identify promoters bound by RNA polymerase II (Fig. 2A). We found that RNA polymerase II was bound to the majority (83%) of p50 targets. Genes bound by p52 and RelB were also generally co-occupied by RNA polymerase II. The absence of any detected p65 binding in our unstimulated cell population indicated that these cells had not been inadvertently activated.
Fig. 2.
Co-occupation of promoters by NF-κB and RNA polymerase II in unstimulated cells. (A) DNA binding of RNA polymerase II and NF-κB subunits in unstimulated cells. The genes are sorted by RNA polymerase II binding ratio (marked on the left). Genes occupied by each NF-κB subunit are marked by a blue dash. (B) Binding and expression data for 135 genes that are bound by at least one NF-κB subunit in unstimulated cells and annotated in the expression data. Expression levels are relative to the average expression of genes across 79 different human tissue or cell types. Red, higher than average expression; green, lower than average, according to the scale shown to the right. Selected overexpressed and underexpressed genes are named on the right.
RNA polymerase II occupancy suggested that these NF-κB target genes may be transcribed. To investigate, we used gene expression microarrays to assess whether RNA transcripts for these genes were present in unstimulated U937 cells. Of the 79 genes bound by NF-κB and RNA polymerase II and represented on the microarray, transcripts for 62 (78%) were called present by the microarray software, compared with 50% for all genes. Therefore, the majority of genes bound by NF-κB and RNA polymerase II in unstimulated cells are transcribed. To assess the level at which these genes were expressed, we aligned our binding data with expression data depicting the relative expression level of genes in 79 different human cell and tissue types (Fig. 2B) (31). About one-third of NF-κB-bound genes were expressed in monocytes at levels over twice that of other cell types, including CCL3, CD58, DAF, HLA-B, HLA-F, and ITGA5. Conversely, 10% of genes, including CCL4 and C3, were occupied by NF-κB and expressed in monocytes at levels less than half that of other cell types. Therefore, in unstimulated cells, NF-κB is associated with a set of RNA polymerase II-bound genes that exhibit a range of expression levels.
p50 Targets in Unstimulated Cells Are Bound by Other NF-κB Subunits After LPS Induction.
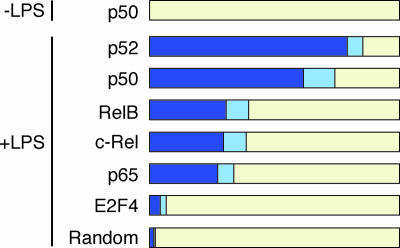
The genes IL6 and IFNB1 (IFN-β) are known to be occupied by p50 in unstimulated cells and then by additional NF-κB family members after cellular stimulation (16, 18). Therefore, the initial binding of p50 to genes in unstimulated cells may be a common event at NF-κB target genes. To test this hypothesis, we analyzed the fraction of genes bound by NF-κB in LPS-stimulated cells that were prebound by p50 in unstimulated cells (Fig. 3). We found that 67 of the 141 p50 targets in unstimulated cells were bound by other NF-κB subunits after LPS treatment (P = 5 × 10−20). p50 remained bound to 64 of these 67 genes. Different subunits showed varying preferences for p50 prebound genes. p52 had the strongest preference for p50 prebound genes, with 79% of p52 targets in LPS-induced cells being prebound by p50, due to the fact that p52 was often already bound with p50 in unstimulated cells. p65, RelB, and c-Rel showed less preference for p50 prebound genes; ≈30% of the targets of each of these subunits were prebound by p50. As a negative control, we also compared the p50 data with the DNA binding pattern of E2F4 (Table 4, which is published as supporting information on the PNAS web site). There was only a slightly higher correlation between the set of p50 prebound genes and E2F4 targets than expected by chance (randomized data; Fig. 3). We conclude that many NF-κB target genes are bound by p50 before cellular stimulation.
Fig. 3.
After LPS stimulation, NF-κB subunits bind to promoters that were previously bound by p50. Fraction of bound genes in activated cells that were prebound by p50 before stimulation at two significance levels, 0.002 (dark blue) and 0.01 (light blue). All NF-κB subunits bound to significant numbers of p50 target genes. In contrast, the binding pattern of E2F4 was not related to p50 binding and followed the pattern expected by chance (randomized data).
Analyses of single and multiple NF-κB subunit knockout mice indicate that individual subunits fulfill both unique and shared functions with other family members (7). To assess the role that coordinated DNA binding might play in these phenomena, we compared the degree with which the targets of the different NF-κB subunits overlapped after LPS stimulation. We found that, in stimulated cells, the majority of binding events occurred at gene promoters that were occupied by more than one NF-κB family member. Whereas 191 binding events occurred at genes bound only by a single subunit, 289 binding events occurred at genes bound by two or more subunits (135 genes). Strikingly, we identified 22 genes that were bound by four different NF-κB subunits and 15 genes that were bound by all five NF-κB family members. These genes included CCL3 (MIP-1α), ICAM1, NFKB2 (p52), and TRAF1 and are listed in Table 5, which is published as supporting information on the PNAS web site. Therefore, genes are often occupied by multiple NF-κB family members in stimulated cells.
Genes Bound by Multiple NF-κB Subunits Exhibit Increased RNA Polymerase II Occupancy and Gene Expression.
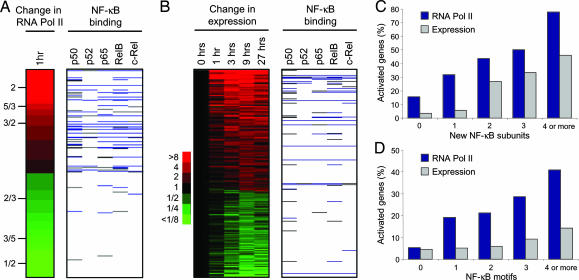
Upon LPS stimulation, additional NF-κB molecules enter the nucleus and activate transcription (1, 17). We therefore questioned whether the binding of NF-κB subunits was associated with increases in the transcription of their target genes after LPS treatment. Using genome-scale location analysis, we measured the difference in RNA polymerase II occupancy between activated cells and resting cells (Fig. 4A). We found that NF-κB was more often associated with genes to which RNA polymerase II was recruited than with those where it was lost. When ordered by change in polymerase II occupancy, the top 20% of genes comprised 50% of all NF-κB binding events, whereas the bottom 20% of genes accounted for only 6%. This increase in RNA polymerase II was correlated with the recruitment of p65, c-Rel, and RelB subunits, NF-κB proteins that contain transcriptional activation domains (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 4.
Recruitment of multiple NF-κB family members leads to increased RNA polymerase II occupancy and gene expression. (A) NF-κB binding at genes to which RNA polymerase II is recruited and to genes where it is lost (top 5% of genes for both categories). The change in RNA polymerase binding is represented as a ratio, with red indicating an increase and green indicating a decrease, according to the scale on the left. Only genes bound by RNA polymerase II in either unstimulated or activated cells were used. Gray, binding of NF-κB subunits at P values < 0.01; blue, binding at P values < 0.002. (B) NF-κB binding at genes that are up-regulated or down-regulated over time in response to LPS in U937 cells. Genes were either up-regulated by 2-fold or down-regulated by 2-fold at two consecutive time points. Red, increase in expression; green, decrease in expression, according to the scale on the left. (C) The percentage of genes to which 1, 2, 3, or 4 or more different NF-κB family members are recruited that show an increase in RNA polymerase II occupancy (blue bars) or an increase in RNA abundance (gray bars) after LPS stimulation. (D) The percentage of genes whose promoters contain 1, 2, 3, or 4 or more NF-κB binding motifs that show an increase in RNA polymerase II occupancy (blue bars) or an increase in RNA abundance (gray bars) after LPS stimulation. The binding motif was defined as GGGRNNYYCC and was counted if present within or 200 bp either side of the promoter element represented on the array.
We next examined whether NF-κB binding was associated with changes in gene expression. We harvested U937 cells before and 1, 3, 9, and 27 h after exposure to LPS and measured changes in RNA abundance by using DNA microarrays (Fig. 4B). As also shown by the RNA polymerase II data, NF-κB was more often associated with genes that became activated than with genes that were repressed. Furthermore, p65, c-Rel, or RelB were recruited to 92% of the NF-κB target genes that were up-regulated after LPS stimulation (P = 1 × 10−6). We conclude that transcriptional activation of NF-κB target genes is associated with the recruitment of NF-κB subunits that contain a transcriptional activation domain.
We noticed that genes to which RNA polymerase II was recruited or which increased in expression tended to be bound by multiple NF-κB family members. To examine this relationship in more detail, we calculated the fraction of genes that exhibited increases in gene activity upon the recruitment of different numbers of subunits after LPS stimulation (Fig. 4C). Gene activity was assessed by two measures, an increase in RNA polymerase II occupancy and an increase in mRNA abundance. We found that the greater the number of different family members that were recruited, the greater the fraction of genes that became activated. Increases in RNA polymerase II occupancy and mRNA abundance were also found to correlate with the number of NF-κB motifs present in the gene promoter, indicating that multiple NF-κB subunits could be binding within single cells (Fig. 4D). Taken together, these data suggest that the presence of multiple NF-κB subunits enhances the recruitment of RNA polymerase II and gene transcription.
NF-κB Target Genes Form Part of the Transcriptional Program That Underlies the Host Response to Pathogens.
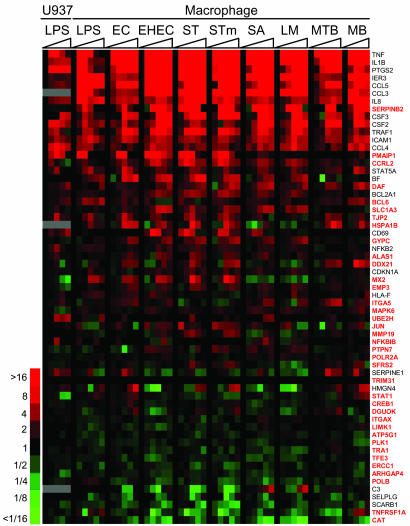
Macrophages and other host cells respond to a range of different pathogens by inducing a core gene expression program (10, 11). To determine whether the NF-κB targets discovered from our analysis formed part of the host transcriptional response to pathogens, we collated gene expression data from primary macrophages exposed to a number of different bacterial species (10) and compared this information with our NF-κB binding data. Fig. 5 depicts NF-κB targets that have been described to change their expression in macrophages in response to bacterial stimuli in refs. 10 and 11. Within the set of regulated genes were many previously known NF-κB targets, for example CCL1 (TCA3), CCL3 (MIP-1α), CCL4 (MIP-1β), CCL5 (RANTES), IL1B, IL8, NFKB2 (p52), PTGS2 (COX-2), STAT5A, and TNF. There were also several genes known to have important roles in the immune response that have not been defined previously as NF-κB targets, including DAF (CD55), MX2, NFKBIB (IκBβ), PMAIP1, SERPINB2 (PAI-2), and STAT1 (gene names are in red in Fig. 5). In addition, there were genes for which a role in the immune response has been suggested only by their expression profile (CCRL2, GYPC, MAPK6, TJP2, and UBE2H). The identification of these genes as direct NF-κB targets supports the hypothesis that they constitute an important part of the immune response.
Fig. 5.
NF-κB target genes in the host response to pathogens. Changes in the expression of 61 NF-κB target genes during the response of U937 cells to LPS and the response of macrophages to LPS and different bacterial species. The genes have previously been identified as regulated in response to pathogens (10, 11). Genes named in red have not previously been identified as NF-κB targets. Expression changes are shown over time (0–24 h) and are colored according to the scale shown to the left. EC, Escherichia coli; EHEC, Enterohemorrhagic Escherichia coli; ST, Salmonella typhi; STm, Salmonella typhimurium; SA, Staphylococcus aureus; LM, Listeria monocytogenes; MTB, Mycobacterium tuberculosis; MB, Mycobacterium bovis.
Discussion
We have performed a genome-scale analysis of DNA binding for all five members of the human NF-κB family in unstimulated and LPS-induced monocytic cells. In unstimulated cells, p50 and p52 are bound to the promoters of many NF-κB target genes. Upon cellular stimulation with LPS, other NF-κB family members enter the nucleus and bind to those genes and to other genes, leading to increases in RNA polymerase II occupancy and gene expression.
Our analysis was focused on the binding of NF-κB to ≈10,000 well defined proximal promoter elements during the initial response to LPS stimulation. We identified binding events by using a stringent statistical threshold to minimize our false positive rate. These restrictions guard against inaccuracies in the data and aid in its interpretation but may also cause us to miss some genuine NF-κB binding events. Even with these restrictions, we have identified 348 genes bound by NF-κB, doubling the number of experimentally derived direct targets and increasing our understanding of the role of NF-κB in the initiation of the immune response.
Binding of p50 and p52 to NF-κB Target Genes Before LPS Stimulation.
We discovered that NF-κB target genes can be divided into two sets; those that are bound before cellular stimulation and those that are only bound after cellular stimulation. p50 accounted for the majority of NF-κB binding activity in unstimulated cells. The constitutive presence of p50 has been documented in a number of cell types, including U937 cells (12). However, previously only two genes, IFN-β and IL-6, had been shown by ChIP to be bound directly by p50 under resting conditions (16, 18). Our results support a model in which p50 not only shuttles between the cytoplasm and nucleus in unstimulated cells (17) but also binds to a substantial number of promoters.
We also detected p52 bound to DNA in unstimulated U937 cells. In most cell types, the processing and nuclear localization of p52 are considered to be under tighter control than for p50 (1), although p52 is constitutively nuclear in Panc1 and U937 cells (12, 32) (Fig. 8). Some lymphoma cell lines contain a mutant form of p100 that is constitutively active, but these proteins are smaller than the 100-kDa form present in U937 cells (33). The vast majority of the genes bound by p52 in unstimulated cells were also bound by p50. We believe that this relationship is unlikely to be because of the p52 antibody cross-reacting with p50 because the peptide sequence to which the p52 antibody was raised bears no similarity to any part of p50 and does not recognize p50 in Western blots (Table 1 and Fig. 8). The shared targets of p50 and p52 may instead reflect an association between these two proteins (34).
p50 and Gene Expression in Unstimulated Cells.
Considering that neither p50 nor p52 contain a transcriptional activation domain, these proteins may not be responsible for the basal level of target gene activity in unstimulated cells. Consistent with this model, the expression levels of MHC class I and TNF, whose genes are bound by p50 and RNA polymerase II in U937 cells, are unaltered in cells from p50-null mice (20). p50 is generally considered to repress transcription in unstimulated cells (16–18). Ishikawa and colleagues (21) generated mice that lack the inhibitory p50 precursor p105 but maintain expression of p50 itself. Macrophages from these mice express lower levels of the NF-κB target genes GM-CSF (CSF2), ICAM1, IL6, and TNF than wild-type mice. However, although these data indicate that p50 is repressive, we note that expression of these genes was detected by the authors in wild-type macrophages (21). Consistent with these data, we detect both p50 and RNA polymerase II at the CSF2, ICAM1, and TNF gene promoters in U937 cells. Therefore, although p50 may have a repressive effect on gene expression, it does not completely prevent RNA polymerase II binding in wild-type cells.
NF-κB Binding After LPS Stimulation.
LPS stimulation leads to an increase in NF-κB binding, with p65, c-Rel, and RelB exhibiting the greatest increases in gene occupancy (Fig. 1). These subunits contain transcriptional activation domains, and their recruitment leads to an increase in RNA polymerase II binding and gene expression (Fig. 4). Therefore, genes bound by p50 are not fully activated in unstimulated cells, and this activation may require the replacement of the p50 homodimer with more activating heterodimers (1, 17).
Interestingly, the greater the number of NF-κB subunits that are recruited, the greater the fraction of genes exhibiting increases in RNA polymerase II binding and gene expression. We have profiled protein-DNA interactions in a cell population, so it is possible that genes bound by multiple NF-κB subunits are actually bound by different subunits in different cells. However, we found that RNA polymerase II binding and gene expression were also correlated with the number of NF-κB binding sites present in the proximal promoter. Therefore, the simplest explanation for the observed correlation between the number of different subunits bound and RNA polymerase II binding is that these different subunits co-occupy genes within single cells. One model that fits these data is that the different NF-κB family members can activate gene expression cumulatively, possibly through their interaction with different chromatin modifiers (35).
A Transcriptional Regulatory Network Involving NF-κB Family Members.
The experimental approaches we have undertaken allow us to begin reconstructing the transcriptional regulatory network that underlies the host response to infection (Fig. 10, which is published as supporting information on the PNAS web site). NF-κB is known to induce the expression of genes encoding other transcription factors involved in the immune response, for example IRFs, SRF, and STAT5A (36–38). We discovered additional NF-κB target genes that encode transcription factors known to be involved in the regulation of the immune response, among them CREB1, JUN, STAT1, STAT6, and TFAP2A (AP-2). Indeed, “transcription factor activity” was one of the most significantly enriched gene ontology terms associated with the set of genes bound by NF-κB (Table 2). Thus, the LPS-stimulated gene expression program in U937 cells is the product of both direct regulation of a set of genes by NF-κB and indirect regulation of an additional set through the action of transcription factors downstream of NF-κB. Some genes are targets of both NF-κB and of NF-κB-targeted regulators and are thereby controlled by a transcriptional feed-forward loop. For example, CDKNA1 and IL10 are regulated by both NF-κB and STAT1 (39, 40). The feed-forward loop is used to generate outputs only after prolonged activation (30) and may be necessary for IL-10 because early activation of this antiinflammatory cytokine could compromise the initial phase of the immune response.
NF-κB activation is autoregulatory because of the induction of genes encoding the inhibitor IκBα, its own subunits, and many other proteins modulating NF-κB activity (41, 42). Our list of NF-κB targets contains additional genes affecting the activity of NF-κB, including NFKBIB (IκBβ), S100A12, and TANK. It was suggested that NFKBIA, but not NFKBIB, is regulated by NF-κB, based on studies with LPS-stimulated pre-B-cells (41). However, gene-specific ChIP confirmed that p65 and p50 occupied the NFKBIB promoter in LPS-stimulated U937 cells (Fig. 7). Furthermore, this result is consistent with the observation that the levels of both IκBα and IκBβ are reduced in p65/p50 double-knockout embryonic stem cells (43).
The majority of human transcription factors can be separated into families in which members share similar structures, DNA binding specificities, and biological functions. Comparative genomics indicates that many families evolved through gene duplication events, but it is often unclear how the functions of individual family members have diverged and how they work together to regulate gene expression. Our results reveal the manner in which NF-κB family members act coordinately to regulate gene expression. Extending this work to other transcription factor families will reveal whether they adopt similar or alternative strategies for controlling gene expression.
Materials and Methods
Cells and Growth Conditions.
U937 cells were obtained from American Type Culture Collection (ATCC) and cultured according to ATCC recommendations. Cells were treated with 2.5 μg/ml LPS (E. coli 05:88; Sigma) for 1 h for genome scale location analysis experiments and for 1, 3, 9, and 27 h for expression analysis experiments.
Antibodies.
Specific antibodies were as follows: p50, sc-1190 (C-19; Santa Cruz Biotechnology); p52, 06–413 (Upstate Biotechnology, Lake Placid, NY); p65, sc-372 (C-20; Santa Cruz Biotechnology); RelB, sc-226 (C-19; Santa Cruz Biotechnology); c-Rel, sc-71 (C terminus; Santa Cruz Biotechnology); E2F4, sc-1082 (Santa Cruz Biotechnology; ref. 28); RNA polymerase II, 8WG16 (30). The affinity-purified NF-κB antibodies were raised against nonconserved N- or C-terminal regions of the proteins to avoid cross-reactivity with other NF-κB subunits. Their specificities have been documented by numerous publications (Table 1).
Human Promoter Array.
We constructed a custom DNA microarray that contains the promoter proximal regions of 9,496 human genes when mapped to the August 2003 assembly of the human genome and with PCR products verified by agarose gel electrophoresis. We targeted the region spanning 700 base pairs upstream and 200 base pairs downstream of the transcription start sites best characterized by National Center for Biotechnology Information annotation. The array production process has been described in detail (30).
Genome-Scale Location Analysis.
The genome-scale location analysis protocol is described in detail as Supporting Text, which is published as supporting information on the PNAS web site. To begin, 5 × 107 cells were cross-linked with 1% formaldehyde for 10 min and the reaction stopped by adding 1/20 vol 2.5 M glycine. The cross-linked material was washed with PBS, lysed, and sonicated 15 times for 20 s at 40W. The sheared chromatin was incubated with specific antibodies coupled to protein G magnetic beads (Invitrogen) for 16 h at 4°C. After washing and eluting with 1% SDS, the DNA was recovered by reversing the cross-links overnight at 65°C. The DNA isolated by ChIP was quantified by using Picogreen (Molecular Probes) and, together with an equal amount of input DNA, was blunted and amplified by ligation-mediated PCR. The material was Cy5 (ChIP) or Cy3 (WCE) dye labeled with the Klenow fragment of DNA polymerase and hybridized to the human promoter array. Genes bound by the protein of interest exhibit high signals in the immunoprecipitation channel relative to the WCE channel.
Analysis of Location Data.
A whole-chip error model was used to combine data for the triplicate experiments and obtain a final average binding ratio and P value for each promoter region (26, 28, 30). The P value is a statistical measure of the significance of binding, based on the magnitude of the enrichment ratio and the strength of the signal from the microarray element. Genes with low P values are more likely to be bound than genes with high P values. Selecting a low P value threshold, identifies a relatively low number of genes with a low false positive rate but a high false negative rate, whereas selecting a higher P value, identifies a larger number of genes but with a correspondingly higher false positive rate and a lower false negative rate.
Expression Analysis.
We harvested total RNA from 5 × 106 untreated cells or cells treated with LPS (2.5 μg/ml) by TRIzol extraction. Ten micrograms of total RNA was labeled according to Affymetrix protocols and hybridized to Affymetrix HG-U95Av2 arrays. The data were analyzed by using Affymetrix Microarray Suite software. Each array was scaled to 150, and ratios were taken to the average of two duplicate time 0 controls. Data for primary macrophages were taken from ref. 10.
Supplementary Material
Acknowledgments
We thank E. Herbolsheimer for help with web site design. This work was supported by the National Institute of Allergy and Infectious Diseases and a Deutsche Forschungsgemeinschaft fellowship (to J.S.).
Abbreviation
- ChIP
chromatin immunoprecipitation.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The microarray data have been deposited in ArrayExpress, www.ebi.ac.uk/arrayexpress (accession no. E-WMIT-6).
References
- 1.Hayden M. S., Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 2.Bonizzi G., Karin M. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Shoelson S. E., Lee J., Yuan M. Int. J. Obes. Relat. Metab. Disord. 2003;3:49–52. doi: 10.1038/sj.ijo.0802501. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal B. B. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H. Cell. 2004;118:671–674. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Karin M., Greten F. R. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 7.Gerondakis S., Grossmann M., Nakamura Y., Pohl T., Grumont R. Oncogene. 1999;18:6888–6895. doi: 10.1038/sj.onc.1203236. [DOI] [PubMed] [Google Scholar]
- 8.Silverman N., Maniatis T. Genes Dev. 2002;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 9.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 10.Nau G. J., Richmond J. F., Schlesinger A., Jennings E. G., Lander E. S., Young R. A. Proc. Natl. Acad. Sci. USA. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenner R. G., Young R. A. Nat. Rev. Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman P. A., Weinberg J. B., Greene W. C. J. Clin. Invest. 1992;90:121–129. doi: 10.1172/JCI115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang S. M., Tran A. C., Grilli M., Lenardo M. J. Science. 1992;256:1452–1456. doi: 10.1126/science.1604322. [DOI] [PubMed] [Google Scholar]
- 14.Ten R. M., Paya C. V., Israel N., Le Bail O., Mattei M. G., Virelizier J. L., Kourilsky P., Israel A. EMBO J. 1992;11:195–203. doi: 10.1002/j.1460-2075.1992.tb05042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaltschmidt C., Kaltschmidt B., Neumann H., Wekerle H., Baeuerle P. A. Mol. Cell Biol. 1994;14:3981–3992. doi: 10.1128/mcb.14.6.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senger K., Merika M., Agalioti T., Yie J., Escalante C. R., Chen G., Aggarwal A. K., Thanos D. Mol. Cell. 2000;6:931–937. doi: 10.1016/s1097-2765(05)00081-x. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh S., Karin M. Cell. 2002;109:81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhong H., May M. J., Jimi E., Ghosh S. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 19.Israel N., Gougerot-Pocidalo M. A., Aillet F., Virelizier J. L. J. Immunol. 1992;149:3386–3393. [PubMed] [Google Scholar]
- 20.Sha W. C., Liou H. C., Tuomanen E. I., Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa H., Claudio E., Dambach D., Raventos-Suarez C., Ryan C., Bravo R. J. Exp. Med. 1998;187:985–996. doi: 10.1084/jem.187.7.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dechend R., Hirano F., Lehmann K., Heissmeyer V., Ansieau S., Wulczyn F. G., Scheidereit C., Leutz A. Oncogene. 1999;18:3316–3323. doi: 10.1038/sj.onc.1202717. [DOI] [PubMed] [Google Scholar]
- 23.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 24.Saccani S., Pantano S., Natoli G. Mol. Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 25.Martone R., Euskirchen G., Bertone P., Hartman S., Royce T. E., Luscombe N. M., Rinn J. L., Nelson F. K., Miller P., Gerstein M., et al. Proc. Natl. Acad. Sci. USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 27.Iyer V. R., Horak C. E., Scafe C. S., Botstein D., Snyder M., Brown P. O. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 28.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinmann A. S., Yan P. S., Oberley M. J., Huang T. H., Farnham P. J. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandler N. M., Canete J. J., Callery M. P. J. Surg. Res. 2004;118:9–14. doi: 10.1016/S0022-4804(03)00354-8. [DOI] [PubMed] [Google Scholar]
- 33.Migliazza A., Lombardi L., Rocchi M., Trecca D., Chang C. C., Antonacci R., Fracchiolla N. S., Ciana P., Maiolo A. T., Neri A. Blood. 1994;84:3850–3860. [PubMed] [Google Scholar]
- 34.Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 35.Leung T. H., Hoffmann A., Baltimore D. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Harada H., Takahashi E., Itoh S., Harada K., Hori T. A., Taniguchi T. Mol. Cell Biol. 1994;14:1500–1509. doi: 10.1128/mcb.14.2.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franzoso G., Carlson L., Brown K., Daucher M. B., Bressler P., Siebenlist U. EMBO J. 1996;15:3403–3412. [PMC free article] [PubMed] [Google Scholar]
- 38.Hinz M., Lemke P., Anagnostopoulos I., Hacker C., Krappmann D., Mathas S., Dorken B., Zenke M., Stein H., Scheidereit C. J. Exp. Med. 2002;196:605–617. doi: 10.1084/jem.20020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin Y. E., Kitagawa M., Su W. C., You Z. H., Iwamoto Y., Fu X. Y. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 40.Ziegler-Heitbrock L., Lotzerich M., Schaefer A., Werner T., Frankenberger M., Benkhart E. J. Immunol. 2003;171:285–290. doi: 10.4049/jimmunol.171.1.285. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J. E., Phillips R. J., Erdjument-Bromage H., Tempst P., Ghosh S. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann A., Levchenko A., Scott M. L., Baltimore D. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann A., Leung T. H., Baltimore D. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.