Abstract
Development of in vitro resistance to GW640385, a new human immunodeficiency virus type 1 protease inhibitor, was studied. Variants characterized included one with <4-fold resistance and amino acid substitutions Q58E/A71V (protease) and P452K (Gag) and one with >50-fold resistance and amino acid substitutions L10F/G16E/E21K/A28S/M46I/F53L/A71V (protease) and L449F/P453T (Gag). The A28S substitution substantially reduced replication capacity.
The development of resistance during failure of highly active antiretroviral therapy represents a major therapeutic challenge to the long-term suppression of human immunodeficiency virus (HIV) (2). Often substantial levels of cross-resistance render subsequent treatment options less effective than initial treatment regimens, and transmitted resistance can compromise the effectiveness of first-line therapy (6, 13). Consequently, therapies aiming at new antiviral targets (e.g., integrase and CCR5) and new therapies within existing classes with superior potency against resistant viruses are urgently needed (11, 15).
GW640385 is an HIV-1 protease inhibitor (PI) with improved potency against clinical isolates resistant to many of the currently licensed HIV-1 PIs (17). To determine potential resistance mutations that may be observed in the clinical setting, HIV-1HXB2 was passaged in the presence of increasing concentrations of GW640385 (4). Historically, in vitro passage of HIV-1 with PIs has provided some correlation with mutations subsequently observed in the clinic (12, 18), but sometimes important mutations observed in vivo have not been selected in vitro (1). Differences between in vitro and in vivo data may be attributed to stochastic effects, founder effects, or codon bias in viral stocks. In addition, mutations conferring low-level resistance have not always been observed in vitro, possibly because the selection pressure rapidly transitioned above concentrations that would select low-level resistance (9). Virus was therefore passaged using either increasingly high concentrations of GW640385 (high-pressure passage; 0.5 to 120 nM) or using lower incremental increases (low-pressure passage; 0.5 to 5 nM). Clonal sequence analysis was employed to identify minority species and linkage between mutations. Site-directed mutant (SDM) viruses were constructed, and the sensitivity of virus to GW640385 was determined.
For the initial passage, final concentrations of 0.02-, 0.1-, 1-, 2-, 10-, 50-, and 200-fold the 50% inhibitory concentration value for GW640385 were used. MT4 cells were infected with HIV-1HXB2 (100 50% tissue culture infective doses [TCID50]/2 × 106 cells). Samples of cell supernatant were collected at 2- to 4-day intervals, monitored for reverse transcriptase activity, and harvested when activity exceeded >125,000 cpm/30 μl (16). Viral RNA was extracted from harvested virus, reverse transcribed, and amplified by PCR to produce a DNA fragment containing the HIV-1 protease gene and the Gag cleavage sites (CS) p7/p1 and p1/p6 (7, 8). The nucleotide sequence was determined using ABI 3700 technology. SDMs were introduced into HIV-1HXB2 using the QuickChange kit (Stratagene), and recombinant virus was generated (14). Susceptibility assays for all the SDMs were carried out at least twice.
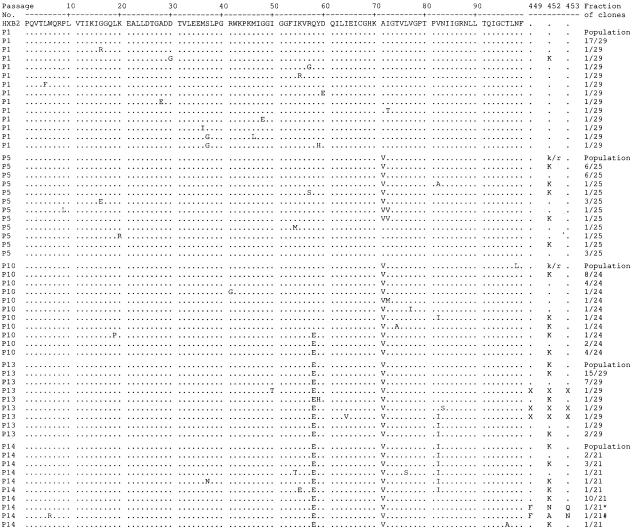
For the low-selection passage, virus was grown in GW640385 over 15 passages for a total of 105 days, to a maximum concentration of 5 nM. The protease amino acid substitutions Q58E and A71V and CS amino acid substitution R452K were selected in the majority of clones (Fig. 1). SDM virus containing these single, double, and triple protease and CS amino acid substitutions had slightly reduced susceptibility to GW640385: Q58E (fold resistance [FR], 2.42 ± 0.07), A71V (FR, 2.03 ± 0.11), Q58E/A71V (FR, 2.37 ± 0.12), A71V/R452K (FR, 2.43 ± 0.23), and Q58E/A71V/R452K (FR, 2.82 ± 0.11). The triple variant when analyzed in further experiments was not cross-resistant to other PIs tested (Table 1). The amino acid substitutions A71V and R452K were also identified during clonal analysis of the control passage in the absence of drug. Analysis using a GlaxoSmithKline database showed that the Q58E and A71V substitutions were present at increased incidence in PI-experienced populations but were not associated with high-level PI resistance. Analysis of the sensitivity to GW640385 of clinical isolates with multiple substitutions (10-11) including Q58E and A71V showed only limited shifts in susceptibility (FR, 2.8- to 4.8-fold). SDM virus with the observed natural variant V82I alone (FR, 1.03 ± 0.1) and with the double variant V82I/A71V (FR, 2.55 ± 0.68) showed thatthis substitution had little or no effect on resistance to GW640385.
FIG. 1.
Amino acid PRO (1 to 99) and CS (449, 452, and 453) sequences of clones derived from HIV-1HXB2 passaged under low-pressure selection of GW640385. Two clones (* and #) had a two-nucleotide insertion and a one-nucleotide deletion, respectively, resulting in a frame shift. X, undetermined amino acid. HIV-1HXB2 was passaged in the presence of GW640385 at the following concentrations: passage 1 (P1), 0.5 nM; P5, 1 nM; P10, 2 nM; P13, 3 nM; and P14, 5 nM.
TABLE 1.
Cross-resistance of variants from the high- and low-selection analysisa
| PI | Resistance of variant
|
|||
|---|---|---|---|---|
| L10F/G16E/K20T/A28S/M46I/A71V/L449F/P453T
|
Q58E/A71V/R452K
|
|||
| IC50 (nM) | FR | IC50 (nM) | FR | |
| GW640385 | 45 ± 15 | >52.5 | 0.9 ± 0.2 | 2.4 |
| APV | 283 ± 110 | 8.7 | 20.9 ± 0.3 | 0.4 |
| LPV | 173 ± 98 | 15 | 24.6 ± 4.1 | 1.0 |
| SQV | 22 ± 9 | 2 | 15.7 ± 0.8 | 1.1 |
| NFV | 136 ± 17 | 5 | 26 ± 22.8 | 1.0 |
| IDV | 597 ± 293 | 55 | 56.8 ± 0.6 | 0.9 |
| ATZ | 16 ± 9 | 1 | 20.6 ± 1.1 | 1.4 |
FR, fold resistance, ratio of IC50 of test virus to that of HIV-1 strain HXB2; means and standard deviations for IC50s were calculated from two assays run in parallel for all drugs. APV, amprenavir; LPV, lopinavir; SQV, saquinavir; IDV, indinavir; NFV, nelfinavir; ATZ, atazanavir.
For the high-selection passage, virus was grown in GW640385 over 14 passages for a total of 217 days to a maximum concentration of 120 nM. The protease amino acid substitutions that predominated (Fig. 2) were constructed as single and double SDMs. Analysis for sensitivity to GW640385 showed only small shifts in susceptibility: for L10F, FR of 1.1; for G16E, FR of 1.59 ± 0.03; for A28S, not done; for M46I, FR of 0.83 ± 0.35; for A71V, FR of 2.03 ± 0.11; for M46I/A71V, FR of 1.47 ± 0.35; and with CS substitutions, for L449F, FR of 0.81 ± 0.28, and for P453T, FR of 1.42 ± 0.37. SDM viruses containing A28S failed to replicate sufficiently for evaluation. The A28 residue is located in the protease enzyme active site close to the critical aspartate triad. The amino acid substitution A28S has previously been selected with TMC-126 in an HIV-1NL4-3 background and dramatically reduced viral replication (20). The A28S substitution resulted in a greater than 1,500-fold decrease in kcat/km for peptide substrates (5). Database analysis showed that a substitution at A28 rarely (0.9% after three PIs were received) occurs following treatment with multiple PIs (19).
FIG. 2.
Amino acid PRO (1 to 99) and CS (449, 452, and 453) sequences of clones derived from HIV-1HXB2 passaged under high-pressure selection of GW640385. HIV-1HXB2 was passaged in the presence of GW640385 at the following concentrations: passage 2 (P2), 0.5 nM; P4, 2 nM; P5, 4 nM; P6, 5 nM; P9, 40 nM; P10, 40 nM; P11, 60 nM; P12, 80 nM; P14, 100 nM; and P15, 120 nM.
It is feasible that the A28S substitution requires an additional substitution in the CS coding region and other substitutions within protease to increase viral fitness and replicative capacity, as observed previously with other PIs (8). However, additional amino acid substitutions in the protease/CS only marginally increased growth (1.8 log10 TCID50) of the A28S-containing SDM virus (G16E/K20T/A28S/M46I/A71V/L449F/P453T). Addition of the L10F amino acid substitution, which was observed at late passages, to a clone extracted from passage 9 caused a further small increase in virus growth (2.5 log10 TCID50), which enabled phenotypic analysis to be carried out. Sensitivity assays showed high-level resistance to GW640385 (>52.5-fold) and cross-resistance to some PIs tested (Table 1). The poor growth of the A28S-containing virus extracted from in vitro passage may indicate that upstream mutations facilitate viral replication, as has been reported (3, 10). A repeat of the high-selection passage with GW640385 and HIV-1HXB2 again selected the A28S amino acid substitution, despite the effect of this mutation on the replicative capacity of the virus.
In conclusion, using two in vitro selection strategies, two alternative resistance pathways were identified. Low-pressure selection gave rise to three protease amino acid substitutions (Q58E, A71V, and V82I) and one CS substitution (R452K) that conferred low-level resistance. In contrast, during high-pressure passage, seven protease amino acid substitutions (L10F, G16E, E21K, A28S, M46I, F53L, and A71V) and two CS substitutions (L449F and P453T) were selected. Virus containing the A28S substitution replicated extremely poorly, but in the presence of other protease substitutions, high-level resistance to GW640385 was detected. These observations are consistent with high-level drug resistance developing at a replicative cost to the virus and highlight the complex balance between resistance, drug pressure, and replicative capacity.
REFERENCES
- 1.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In Vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks, S. G. 2000. Determinants of virological response to antiretroviral therapy: implications for long-term strategies. Clin. Infect. Dis. 30(Suppl. 2): S177-S184. [DOI] [PubMed] [Google Scholar]
- 3.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 4.Hazen, R., M. St Clair, M. Hanlon, S. Danehower, I. Kaldor, V. Samano, J. Miller, J. Ray, A. Spaltenstein, D. Todd, and M. Hale. 2003. GW0385, a broad spectrum, ultrapotent inhibitor of wild-type and protease inhibitor-resistant HIV-1. Second International AIDS Society Conference on HIV Pathogenesis and Treatment, Paris, France.
- 5.Hong, L., J. A. Hartsuck, S. Foundling, J. Ermolieff, and J. Tang. 1998. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci. 7:300-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 8.Maguire, M. F., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. T. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J. P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maguire, M. F., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myint, L., M. Matsuda, Z. Matsuda, Y. Yokomaku, T. Chiba, A. Okano, K. Yamada, and W. Sugiura. 2004. Gag non-cleavage site mutations contribute to full recovery of viral fitness in protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrill, A. L. 2003. HIV-1 integrase inhibition: binding sites, structure activity relationships and future perspectives. Curr. Med. Chem. 10:1811-1824. [DOI] [PubMed] [Google Scholar]
- 12.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. D. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. J. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richman, D. D., S. C. Morton, T. Wrin, N. Hellmann, S. Berry, M. F. Shapiro, and S. A. Bozzette. 2004. The prevalence of antiretroviral drug resistance in the United States. AIDS 18:1393-1401. [DOI] [PubMed] [Google Scholar]
- 14.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retrovir. 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 15.Rusconi, S., E. Bulgheroni, and P. Citterio. 2004. Entry and fusion inhibitors of HIV. Expert Opin. Ther. Patents 14:733-748. [Google Scholar]
- 16.Schwartz, O., Y. Henin, V. Marechal, and L. Montagnier. 1988. A rapid and simple colorimetric test for the study of anti-HIV agents. AIDS Res. Hum. Retrovir. 4:441-448. [DOI] [PubMed] [Google Scholar]
- 17.Spaltenstein, A., W. Andrews, E. Furfine, R. Hazen, M. Hale, M. Hanlon, I. Kaldor, J. Miller, V. Samano, J. Ray, and R. Xu. 2003. Discovery and evaluation of GW0385, a third-generation HIV protease inhibitor: broad spectrum inhibition of wild-type and multi-PI-resistant viruses by femtomolar HIV protease inhibitors. Second IAS Conference on HIV Pathogenesis and Treatment, Paris, France.
- 18.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retrovir. 15:355-363. [DOI] [PubMed] [Google Scholar]
- 19.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimura, K., R. Kato, M. F. Kavlick, A. Nguyen, V. Maroun, K. Maeda, K. A. Hussain, A. K. Ghosh, S. V. Gulnik, J. W. Erickson, and H. Mitsuya. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 76:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]