Abstract
The vancomycin resistance vanB2 gene cluster is disseminated worldwide and has been found in phylogenetically remote bacterial genera. The vanB2 operon is part of conjugative transposons Tn1549/Tn5382, but conjugative transposition of these elements has not been demonstrated. We have obtained transfer of a Tn1549-like element (referred to herein as “Tn1549-like”) from Clostridium symbiosum MLG101 to Enterococcus faecium 64/3 and Enterococcus faecalis JH2-2 in the digestive tract of gnotobiotic mice and to E. faecium 64/3 in vitro. Retransfer of Tn1549-like from an E. faecium transconjugant also containing Tn916 to E. faecium BM77 was obtained in vitro, albeit at a very low frequency. Transfer efficiency was found to be both donor and recipient dependent. Pulsed-field gel electrophoresis analysis of total SmaI-digested DNA of 48 transconjugants indicated in 27 instances the acquisition of ca. 34 kb of DNA. Two transconjugants harbored two copies of the transposon. Sequencing of the flanking regions of Tn1549-like in 48 transconjugants revealed 29 integration events in 26 loci in the E. faecium genome, and two hot spots for insertion were identified. Integration of the transposon was associated with the acquisition of 5 (n = 18) or 6 (n = 7) bp of donor DNA or with 5-bp duplications of target DNA in the remaining transconjugants. These data demonstrate functionality of the Tn1549-like element and attest that the transfer of the vanB operon between enterococci and human commensal anaerobes occurs in the intestinal environment.
Emergence of vancomycin resistance in enterococci was reported in 1986, approximately 30 years after the introduction of this antibiotic into clinical practice (25). More recently, vancomycin resistance was detected in strains of Staphylococcus aureus, Oerskovia turbata, Arcanobacterium haemolyticum, Streptococcus bovis, Streptococcus gallolyticus, Streptococcus lutetiensis, Bacillus circulans, Paenibacillus, and Rhodococcus, as well as in anaerobic bacteria belonging to the Clostridium genus and Eggerthella lenta (6, 15, 26, 29, 30, 32, 35, 36, 42). As a consequence, glycopeptide resistance is considered as a global threat to public health, and control of its dissemination constitutes a crucial challenge. Acquired resistance to glycopeptides in enterococci is due to production of modified peptidoglycan precursors ending in d-alanine-d-lactate (d-Ala-d-Lac) (VanA, -B, and -D) or d-alanine-d-serine (d-Ala-d-Ser) (VanC, -E, and -G), to which glycopeptides exhibit low binding affinities, combined with the elimination of high-affinity d-Ala-d-Ala-ending precursors synthesized by the host Ddl ligase (4). Expression of the resistance gene clusters is controlled by two-component regulatory systems that are composed of VanR-type response regulators acting as transcriptional activators and VanS-type histidine kinases that are associated with the membrane (2). The regulatory and resistance genes are transcribed from distinct promoters that are coordinately regulated (1).
Among the glycopeptide resistance determinants, VanA and VanB are the two most commonly encountered in clinical settings (12). VanA-type resistance is mediated in enterococci (3) and, more recently, in Staphylococcus aureus by transposon Tn1546 or closely related elements (13). Two major subtypes of vanB operons, vanB1 and vanB2, have been described so far (9, 16, 33). Clusters related to vanB1 are generally carried by large (90- to 250-kb) elements that are transferable by conjugation from chromosome to chromosome (37). The more common vanB2 operon is generally associated with Tn1549- and Tn5382-like transposons that are closely related (here referred to, for the sake of simplicity, as “Tn1549-like”) (9, 21). Tn1549 is entirely sequenced (accession number AF192329), whereas Tn5382 is partly sequenced (accession numbers AF063010 and AF063900). Minor base differences of the vanB operon were detected in the related elements found in anaerobes, which are structurally similar to Tn1549 (7). These genetic elements possess features of conjugative transposons of the Tn916 family and are capable of excision to form a circular intermediate. Tn1549 and Tn5382 have not been shown to promote conjugative transposition but can be transferred passively as an integral part of variable-size chromosomal fragments or of plasmids (15, 17). The vanB2 gene has been detected in anaerobic bacteria in Australia (42) and, more recently, in Canada (18). The linkage of vanB2 with Tn1549-like elements in members of various genera including Enterococcus, Streptococcus, Clostridium, Eggerthella, and Ruminococcus (6, 16, 17) suggests that the spread of this resistance determinant is due to transposition. The chromosomal sequence of Enterococcus faecalis V583 indicated that more than a quarter of the genome probably consists of mobile foreign DNA and that the vanB operon is part of a mobile element that contains 53 genes, including Tn1549 (34). The aim of this work was to test intergeneric transfer of vancomycin resistance between Clostridium and Enterococcus species and to characterize the mechanism involved in this process.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The origins and properties of bacterial strains and plasmids are listed in Table 1. Eggerthella lenta and Clostridium sp. were grown under anaerobic conditions at 37°C on prereduced brain heart infusion broth (Difco Laboratories, Detroit, MI) and agar supplemented with 5% horse blood (Bio-Rad, Marnes-la-Coquette, France). The MICs of vancomycin were determined by the Etest procedure (AB Biodisk, Solna, Sweden) or by twofold serial dilution in agar.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. lenta sp. | ||
| MLG043 | Vm (vanB2) | 42 |
| Clostridium sp. | ||
| MLG055 | Vm (vanB2) | 42 |
| C. symbiosum spp. | ||
| MLG101 | Vm (vanB2) | 42 |
| MLG101-1 | Vm (vanB2) spontaneous derivative harboring a second copy of Tn1549-like | This study |
| E. faecalis spp. | ||
| BM4110 | Str, JH2 spontaneous mutant | 10 |
| JH2-2 | Fus, Rif, JH2 spontaneous mutant | 24 |
| JH2-2::Tn916 | Fus, Rif, Tet | 20 |
| T1 | JH2-2::Tn1549-like, Fus, Rif, Vm | This study |
| T1::Tn916 | Fus, Rif, Vm, Tet | This study |
| T1(pCM100) | Fus, Rif, Vm, Sp, Cm, xis-intTn1549-like | This study |
| T1::Tn916(pCM100) | Fus, Rif, Vm, Sp, Cm, Tet, xis-intTn1549-like | This study |
| E. faecium spp. | ||
| BM77 | Str spontaneous mutant | Wild-type strain |
| BM4105 | Fus, Rif | 10 |
| BM4105S | Str | 10 |
| 64/3 | Fus, Rif, | 44 |
| T2-28 | 64/3::Tn1549-like, Fus, Rif, Vm | This study |
| T2::Tn916 | Fus, Rif, Vm, Tet | This study |
| T2(pCM100) | Fus, Rif, Vm, Sp, Cm, xis-intTn1549-like | This study |
| T2::Tn916(pCM100) | Fus, Rif, Vm, Tet, xis-intTn1549-like | This study |
| T29 | BM77::Tn1549-like, Fus, Rif, Vm | This study |
| Plasmids | ||
| pAT79 | P2, Cm, Sp | 2 |
| pCM100 | pAT79Ωxis-intTn1549-like | This study |
Abbreviations: Cm, chloramphenicol resistance; Fus, fusidic acid resistance; Rif, rifampin resistance; Sp, spectinomycin resistance; Str, streptomycin resistance; Tet, tetracycline resistance; Vm, vancomycin resistance; xisTn1549-like, excisionase of Tn1549-like; intTn1549-like, integrase of Tn1549-like; P2, promoter of the aphA-3 gene from enterococcal plasmid pJH1 (GenBank accession number V01547).
Conjugation experiments.
Eggerthella lenta MLG043, Clostridium sp. strain MLG055, and Clostridium symbiosum MLG101-1 carrying a vanB2 Tn1549-like transposon were used as donors. Enterococcus faecalis JH2-2 and BM4110 and Enterococcus faecium strains 64/3, BM77, and BM4105 were used as recipients. Transconjugants were used as donors in retransfer experiments. Filter matings were carried out on sterile filter membranes as described previously (14). Antibiotics were used alone or in combination at the following concentrations to counterselect donor strains: vancomycin, 8 μg/ml; streptomycin, 1,000 μg/ml; spectinomycin, 80 μg/ml; and rifampin, 100 μg/ml. Mating experiments were carried out by filter mating in the absence or presence of a subinhibitory level (0.2 μg/ml) of vancomycin.
In vivo transfer.
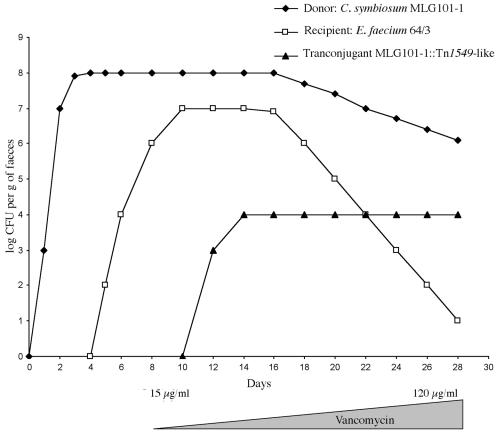
Groups of five germfree consanguineous C3H mice supplied by INRA (Jouy-en-Josas, France) were inoculated intragastrically with a challenge of 108 CFU of donors. After massive and prolonged colonization of the gut by the bacteria, the animals were inoculated with 108 CFU of the recipient enterococci. Mice were maintained in a positive-pressure incubator to prevent any bacterial contamination. After control of the persistence of the association of Enterococcus spp. with the putative anaerobic donor, vancomycin was added to the drinking water at concentrations increasing from 15 to 120 μg/ml throughout the experiment (Fig. 1). Fecal samples were plated on brain heart infusion agar containing 8 μg/ml of vancomycin and incubated at 37°C under aerobic conditions. Attempts to increase retransfer efficiency included introduction in two transconjugants of either Tn916 by conjugative transposition or of a shuttle plasmid by electrotransformation to overexpress IntTn1549-like and XisTn1549-like, as well as both Tn916 and pCM100. Transconjugants were tested for resistance to vancomycin (MIC > 8 μg/ml), and the presence of vanB2 was screened for by PCR with primers VBa and VBb.
FIG. 1.
Bacterial counts in fecal samples from gnotoxenic mice receiving vancomycin in drinking water. Vancomycin was added step-wise in incrementally increasing amounts (15 μg/ml on day 8 to 120 μg/ml on day 24).
PCR amplification.
Primers used for amplification or sequencing are listed in Table 2. PCR was performed with a Ready-To-Go kit (Amersham Biosciences, Orsay, France) in a GeneAmp 2400 PCR system (Perkin-Elmer Cetus, Norwalk, Conn.). PCR elongation times and temperatures were adjusted according to the expected size of the amplicon and melting temperature of the primers, as recommended by the manufacturer. Circular intermediates of Tn1549-like were screened by amplification and sequencing of the 250-bp PCR product overlapping the joint region using the VB2 and VBR2 primers. In the absence of an amplification product, nested PCR was carried out using internal primers VB1 and VBR3.
TABLE 2.
Oligodeoxynucleotide primers
| Primera | Sequence (5′-3′) | Position in Tn1549 (start, end)b | Source or reference |
|---|---|---|---|
| AF (+) | GCTATGGCAGTTTTCCGTGTG | 483, 504 | 15 |
| AR (−) | AACGCTTCTTCATGGCTCTTG | −33441, −33421 | 15 |
| VB0 (+) | TGCCGGAAAAGCCCGGAAACACG | 33720, 33742 | This study |
| VB1 (+) | GAGGGGGAAATGGTGAGAGGT | 33662, 33682 | This study |
| VB2 (+) | AGAAATGGAACGGCTGGCAGC | 33601, 33621 | This study |
| VB3 (+) | TTCCAATATCACCATGACGCTG | 33541, 33562 | This study |
| VBa (−) | GCTGCGGAGCTTTGAATATC | −29217, −29198 | This study |
| VBb (+) | CGTGTGCTGCAGGATACTAC | 28768, 28789 | This study |
| VBR1 (−) | TGCATCAGCCGTTCAAACGCC | −298, −278 | This study |
| VBR2 (−) | CGCGTTTACGGTGTCGTATTC | −178, −158 | This study |
| VBR3 (−) | TGCGGCTCAATCCGAAAGTAG | −100, −80 | This study |
| VBR4 (−) | TGCGAAATGCCCGTATTTCCGG | −48, −27 | This study |
| xisF (+) | TGAGAGCTCAGGAGGCTGCATTATGAACCATGc | 32139, 32157 | This study |
| intR (−) | ATGTCTAGAGTCAGGCTGCCAGCCGTTCCd | −33626, −33607 | This study |
+, sense primer; −, antisense primer.
Nucleotide numbering according to numbering of GenBank accession no. AF192329.
SacI restriction site underlined.
XbaI restriction site underlined.
Characterization of Tn1549-like targets.
Thermal asymmetric interlaced PCR (TAIL-PCR) and inverse PCR (IPCR) were used to determine the 5′- and 3′-flanking regions of Tn1549-like in the three anaerobic donors and in 48 randomly selected transconjugants (27). TAIL-PCR was performed using an Expand Long Template PCR system kit (Roche, Mannheim, Germany) for strains MLG043, MLG055, and MLG101-1 and an E. faecium 64/3 transconjugant obtained in vivo. Primers AD1, AD2, AD3, and AD4 were used with primers VBR1, VBR2, VBR3, and VBR4, respectively, to examine flanking sequences at the right extremity of Tn1549-like, and VB0, VB1, VB2, and VB3 were used for the left end. IPCR was performed as follows. Total DNA of transconjugants was digested by (i) DraI or HinfI and (ii) AluI, DdeI, or Sau3AI, self-ligated, and used as the template for IPCR with AL/VBR2 and AR/VB2, respectively. PCR products were purified with a High Pure PCR product purification kit (Roche) before sequencing. Total DNA from the strains was prepared with a blood genomics kit (Amersham).
Nucleotide sequencing.
Both DNA strands were sequenced with synthetic oligonucleotides by use of an ABI PRISM 310 automated sequencing apparatus (Perkin-Elmer Applied Biosystems). Determination was carried out by direct sequencing of PCR products or, when necessary, after cloning with TopoTA or TopoXL kits (Invitrogen, Groningen, The Netherlands). Plasmid DNA was purified with a QIAprep Spin Miniprep kit (QIAGEN, Inc., Chatsworth, Calif.).
Analysis of transconjugants by pulsed-field gel electrophoresis (PFGE) and Southern hybridization.
Total DNA from recipients and transconjugants embedded in agarose plugs was digested overnight at 27°C with 25 U of SmaI, and fragments were separated on a 0.8% agarose gel by using a CHEF-DRIII apparatus (Bio-Rad) under the following conditions: total migration, 24 h; initial pulse, 60 s; final pulse, 120 s; voltage, 6 V/cm; included angle, 120°; and temperature, 14°C. Resulting fragments were transferred to a Hybond N+ nylon membrane (Amersham) and hybridized to a probe specific for the xisTn1549 and intTn1549 genes corresponding to a 1,500-bp PCR product obtained from MLG101-1 using the xisF and intR primers. Total DNA from anaerobes and from three transconjugants (T1, T10, and T11) was digested with NdeI. Restricted DNA was electrophoresed on a 0.8% TAE (40 mM Tris-acetate, 1 mM EDTA)-agarose gel, transferred to a nylon membrane, and hybridized under stringent conditions with the xis-int probe (40). Hybridization signals were detected by chemiluminescence using CDP-star reagent (Amersham Biosciences) according to the manufacturer's recommendations.
Cloning and overexpression of the xis and int genes.
A 1,487-bp SacI-XbaI PCR fragment obtained with primers XisF and IntR and containing the xisTn1549 and intTn1549 genes was cloned into shuttle vector pAT79 and resequenced. In this plasmid, designated pCM100, the xis and int genes were placed under the control of transcription and translation signals of gram-positive bacteria. pCM100 was then introduced into E. faecium 64/3::Tn1549-like and E. faecalis JH2-2::Tn1549-like by electrotransformation with selection on 80 μg/ml of spectinomycin.
Nucleotide sequence accession numbers.
The target sequences for Tn1549-like in E. faecalis JH2-2, E. faecium 64/3, and E. faecium BM77 have been deposited in the GenBank data library under accession no. DQ119765 to DQ119820.
RESULTS
In vivo transfer of vancomycin resistance.
Experiments were carried out in the digestive tract of gnotobiotic mice to mimic the natural conditions that prevail in vivo for vancomycin resistance transfer to take place. Irrespective of the donor (C. symbiosum MLG101-1, Clostridium sp. strain MLG055, or E. lenta MLG043) and of the recipient (E. faecium 64/3 or BM4105 or E. faecalis JH2-2) used, feces from challenged mice contained high numbers (109 to 1011 CFU/g of feces) of both donor and recipient (Fig. 1). A high level of colonization by the donor and recipient strains combined with the presence of vancomycin in drinking water created conditions helpful for the selection of transconjugants. Two transconjugants, T2 and T3, were detected using MLG101-1 as a donor and E. faecium 64/3 as a recipient in two independent experiments. A third transconjugant, T1, was obtained following transfer from MLG101-1 to E. faecalis JH2-2 in one out of two experiments (Table 3). Transfer of vancomycin resistance from MLG101-1 to E. faecium BM4105 or from MLG043 or MLG055 to any of the three recipients used was not obtained. All three transconjugants had MICs of vancomycin higher than 8 μg/ml and were shown to contain the vanB gene by PCR. Transfer of vancomycin resistance from T2 to BM77, BM4105, and BM4110 was not obtained in three independent experiments. The level of circular intermediate could be a limiting step in transfer of Tn1549-like. In order to circumvent this possibility, the xis and int genes of Tn1549 were cloned into a shuttle vector generating pCM100 to overproduce the circular intermediate. Attempts to retransfer vancomycin resistance from T2 after the introduction of Tn916 and/or plasmid pCM100 were unsuccessful.
TABLE 3.
Experiments for transfer of vancomycin resistance from anaerobes to Enterococcus spp. in gnotobiotic mice
| Recipient | Resulta for donor
|
||
|---|---|---|---|
| E. lenta MLG043 | C. innocuum MLG055 | C. symbiosum MLG101-1 | |
| E. faecalis JH2-2 | 0/2 | 0/2 | 1/2 |
| E. faecium 64/3 | 0/2 | 0/2 | 2/2 |
| E. faecium BM4105 | 0/2 | 0/2 | 0/2 |
Expressed as number of experiments leading to transconjugants/total number of experiments.
In vitro transfer of vancomycin resistance.
Based on six independent experiments, transfer of vancomycin resistance from MLG101-1 to E. faecium 64/3 was estimated to occur at frequencies of between 10−8 and 10−9 transconjugants per recipient (Table 4). Forty-eight transconjugants resistant to vancomycin and containing vanB were selected for further analysis. Transfer was not detected from MLG101-1 to E. faecium BM4105 or E. faecalis JH2-2 or from MLG043 or MLG055 to any of the recipients. When using T2 as a donor (containing Tn916 and pCM100 or not), a single transconjugant (T29), transferred from T2::Tn916 to E. faecium BM77, could be detected. Subinhibitory concentrations of vancomycin did not significantly influence transfer frequencies.
TABLE 4.
Conjugative transfer of Tn1549-like by filter mating
| Donora | Recipient | Transfer frequencyb |
|---|---|---|
| E. lenta MLG043 | E. faecium 64/3 | <10−9 |
| Clostridium sp. strain MLG055 | E. faecium BM4105 | |
| E. faecalis JH2-2 | ||
| C. symbiosum MLG101-1 | E. faecium 64/3 | 10−9 to <10−7 |
| C. symbiosum MLG101-1 | E. faecium BM4105 | <10−9 |
| E. faecalis JH2-2 | ||
| T1, T1(pCM100), T1::Tn916, T1::Tn916(pCM100) | E. faecium BM77 | <10−9 |
| T2, T2(pCM100), T2::Tn916(pCM100) | E. faecalis BM4110 | |
| T2::Tn916 | E. faecium BM77 | ca. 10−9 |
| T2::Tn916 | E. faecalis BM4110 | <10−9 |
Abbreviations: T1, E. faecalis JH2-2::Tn1549-like; T2, E. faecium 64/3::Tn1549- like.
Transconjugants per recipient.
Characterization of the junction fragments of Tn1549-like in transconjugants.
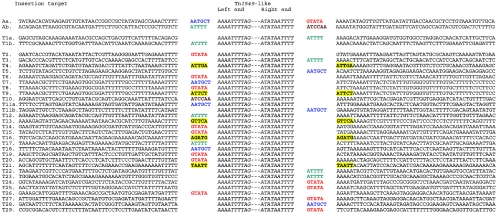
The junction fragments of Tn1549-like in MLG101-1, MLG043, and MLG055, in the three transconjugants obtained in vivo, and in the randomly selected 45 clones obtained in vitro were characterized by using TAIL-PCR or IPCR. For all transconjugants obtained from in vivo, in vitro, and retransfer experiments, a total of 29 unique integration events were observed from a total of 48 transconjugants studied (Fig. 2). In order to rule out mismatches during amplification, the nucleotide sequences of the target sites were confirmed by sequencing the conventional PCR products obtained with new primers from E. faecium 64/3 DNA. Quasi-identity with sequences available from a partial genome sequencing of E. faecium (GenBank accession no. AAAK00000000) was detected for 20 transconjugants, and homology with sequences in GenBank is summarized in Table 5. The flanking junctions of Tn1549-like in the 29 distinct transconjugants showed acquisition of five (18 transconjugants) or six (7 transconjugants) base pairs originally flanking the transposon in MLG101-1. In the remaining transconjugants, surprisingly, 5-bp duplications of the target site were detected. Except for two hot spots for insertion common to three (T7 to T9) and two (T6 and T12) independent events and easily recognized at the coupling sequence level, 24 different integration sites were recovered for E. faecium, whereas two integration sites were detected in the E. faecalis JH2-2 transconjugant T1 (Fig. 2). These data showed that Tn1549 can transpose at multiple sites in the E. faecium genome, although only two hot spots for insertion were identified.
FIG. 2.
Target DNA sequences at the integration sites of Tn1549-like in C. symbiosum MLG101-1, E. faecalis JH2-2, E. faecium 64/3, and E. faecium BM77. Insertion target rows: A, C. symbiosum MLG101-1; T1, E. faecalis JH2-2; T2 to T28, E. faecium 64/3; T29, E. faecium BM77. Coupling sequences are colored, each color corresponds to a specific sequence, and target duplication is indicated by yellow boxes.
TABLE 5.
Insertion targets of Tn1549-like in E. faecium strains 64/3 and BM77
| Transconjugant | E. faecium strain DOa | Target |
|---|---|---|
| E. faecium 64/3 | ||
| T2 | Contig 636 (19731) | Hypothetical protein |
| T3 | Contig 646 (32512) | G2160: l-arabinose isomerase |
| T5 | Contig 655 (25838) | Noncoding sequence |
| T6 and T12 | Contig 605 (10224) | Noncoding sequence |
| T7, T8, and T9 | Contig 626 (20593) | Noncoding sequence |
| T10 | Contig 651 (27393) | COG0642: signal transduction histidine kinase |
| T11 | Contig 596 (8946) | COG4698: uncharacterized protein conserved in bacteria |
| T13 | Contig 578 (8952) | Noncoding sequence |
| T14 | Contig 635 (19630) | COG3964: predicted amidohydrolase |
| T15 | Contig 632 (3516) | Noncoding sequence |
| T16 | Contig 618 (10216) | COG0751: glycyl-tRNA synthetase, beta subunit |
| T17 | Contig 627 (18298) | COG3279: response regulator of the LytR/AlgR family |
| T19 | Contig 633 (17428) | COG1593: TRAP-type C4-dicarboxylate transport system, large permease component |
| T20 | Contig 621 (11540) | COG1597: sphingosine kinase and enzymes related to eukaryotic diacylglycerol kinase |
| T24 | Contig 619 (13913) | Noncoding sequence |
| T25 | Contig 611 (3757) | Noncoding sequence |
| T26 and T27b | Contig 559 (T26, 6622; T27, 6628) | COG0556: helicase subunit of the DNA excision repair complex |
| T28 | Contig 655 (59076) | Hypothetical protein |
| E. faecium BM77 | ||
| T29 | Contig 603 (17495) | Hypothetical protein |
Identity with E. faecium strain DO (GenBank accession no. AAAK00000000) was 98% to 100% in all sequences from E. faecium 64/3 and 88% with contig 603 from E. faecium BM77. The position of the insertion in E. faecium DO is shown in parentheses.
Insertions differ by a 6-bp shift.
DNA analysis of transconjugants by PFGE.
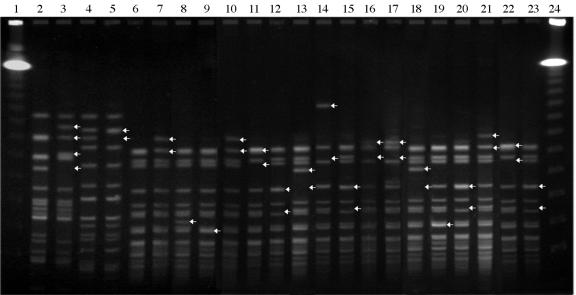
Total DNA from E. faecium 64/3 and BM77, E. faecalis JH2-2, and transconjugants T1 to T29 was analyzed by PFGE after digestion by SmaI. The acquisition of a ca. 34-kb fragment corresponding to Tn1549 was detected in 27 out of the 29 transconjugants (Fig. 3). The restriction profile of T1, compared to that of E. faecalis JH2-2, suggested the integration of two copies of Tn1549-like. By contrast, acquisition of a fragment larger than 68 kb was detected in T11, suggesting cotransfer of additional genetic material to the two copies of the transposon. Acquisition of additional material was also detected in T16 (data not shown).
FIG. 3.
SmaI PFGE profiles of Enterococcus recipient strains and of the transconjugants. Lanes: 1, 48.5- to 970-kb lambda ladder (Bio-Rad); 2, E. faecalis JH2-2; 3, T1 (JH2-2::Tn1549-like); 4, E. faecium BM77; 5, T29 (BM77::Tn1549-like); 6, E. faecium 64/3; 7, T2 (64/3::Tn1549-like); 8, T10; 9, T7; 10, T4; 11, T5; 12, T6; 13, T25; 14, T11; 15, T12; 16, T13; 17, T14; 18, T21; 19, T17; 20, T19; 21, T20; 22, T24; 23, T15; 24, lambda ladder. Arrows indicate the fragments that varied in size by acquisition of the ca. 34-kb Tn1549-like element. In some cases, comigrating fragments were observed. For T1 and T11, two copies of Tn1549-like were detected.
Copy number of Tn1549-like in donors and transconjugants.
Total DNA of MLG101-1, MLG043, MLG055, T1, T10, and T11 was analyzed after digestion by NdeI and hybridized with a probe specific for the intTn1549 and xisTn1549 genes. Since NdeI does not cut in these genes, the presence of two hybridizing fragments in MLG101-1, T1 (data not shown), and T11 indicated that these strains harbored two copies of Tn1549-like; by contrast, a single copy of the transposon was detected in T10 (data not shown). MLG101 was previously reported as harboring a single copy of the transposon (7), indicating that duplication in MLG101-1 had spontaneously occurred during subculturing.
Detection of circular intermediates.
The presence of circular intermediates of Tn1549 was detected in MLG101-1, MLG043, and MLG055 and in all 29 transconjugants by PCR or nested PCR using primers designed to direct polymerization outward from the ends of the transposon. The circularized end products from MLG101-1, MLG043, MLG055, T1, T2, T3, and T11 and from T1(pCM100) and T2(pCM100) overproducing IntTn1549 and XisTn1549, which led as expected to an increased amount of the circular intermediate (data not shown), were sequenced. Analysis of these products showed that the coupling sequences which joined the ends of the circularized transposon consisted of 5 or 6 bp. These data are in agreement with that for the base pairs acquired at the target level during insertion (Fig. 2).
DISCUSSION
The presence of Tn1549-like elements in many distinct strains of various genera, including enterococci and anaerobic bacteria, suggests that this element is a functional conjugative transposon. Until now, however, transfer of Tn1549 was associated with the movement of plasmids or of large elements of various sizes (9, 17, 21, 37, 39). To study the intergeneric transfer of vancomycin resistance, we took into account the failure of previous in vitro experiments to demonstrate the active movement of Tn1549-like elements (9, 15, 21) and the fact that the intestinal ecosystem is the most probable habitat for meetings between gram-positive anaerobic bacteria and Enterococcus species. The digestive tract of humans and animals, in which very dense and diverse microbial populations live in intimate contact, sometimes as part of biofilms, constitutes an extremely favorable ecosystem for gene transfer (5, 19, 31). We have performed experiments in the digestive tract of gnotoxenic mice to mimic the natural conditions that prevail in vivo for vancomycin resistance transfer to take place. Intergeneric transfer was achieved both in vivo and in vitro into E. faecium 64/3. A single E. faecalis transconjugant was obtained in vivo, whereas in vitro experiments led to higher numbers of events in E. faecium 64/3. It is unclear whether the Tn1549-like elements included in this study differ from Tn1549 and Tn5382 with respect to transfer ability. Our results indicate that the nature of the strains involved in conjugation was the main factor influencing the occurrence of transfer, with MLG101-1 being the most efficient donor and E. faecium 64/3 being the best recipient. This suggests that host factors may be involved in the transfer process.
Interestingly, MLG101-1 harbored two copies of Tn1549-like elements. It has been proposed for Tn916 that multiple copies of the transposon can increase the transfer frequency (38). Although two copies of Tn1549 were also detected in T1 and T11, all attempts to transfer resistance in vivo and in vitro from these transconjugants were unsuccessful. We recently performed conjugation between MLG101 and E. faecium 64/3 in an in vitro experiment and showed that transfer also occurred. This indicated that a second copy of Tn1549-like was not essential for conjugative transposition. To account for transfer of Tn1549-like elements from C. symbiosum to Enterococcus spp. we hypothesize that conjugative transposition could be more easily detected after intergeneric transfer, since plasmid replication or homologous recombination via chromosomal fragments should be strongly reduced, two features which could impair the detection of the conjugative transposition of Tn1549 in enterococci. Furthermore, the detection of transconjugants is also made difficult because vanB can confer a low level of resistance to vancomycin and requires induction for phenotypic expression (23).
In order to explain the weak transfer efficiency of Tn1549-like elements, we have carried out in vitro experiments and showed that vancomycin did not increase transfer frequency. In contrast, tetracycline induces transfer of Tn916 by the formation of a large transcript through the circular intermediate (11). In addition, overproduction of Int and Xis in T1 and T2 did not influence transfer or the rate thereof. A similar feature has been reported for Tn916 excision, which is necessary but not sufficient for the occurrence of conjugal transfer (28). Because of the failure to retransfer vancomycin resistance from the transconjugants, we introduced Tn916, which is capable of mobilizing plasmids or other transposons (20), into T1 and T2 and into their derivatives overexpressing Int and Xis. Despite numerous in vivo and in vitro attempts, we obtained only a single transconjugant from T2::Tn916, which indicates that retransfer from E. faecium 64/3 was possible but did not give evidence for a role of Tn916 in mobilization of Tn1549-like.
The fact that in our experiments MLG101-1 was the only efficient donor for conjugative transposition could also be due to a polar effect of the genomic environment. The location and orientation of transfer genes in Tn1549 are in part similar to those of Tn916, and transcription of these genes may have resulted following transposon insertion near an active promoter. Examination of the sequence in MLG101-1 for a putative promoter sequence upstream from the genes involved in transfer of Tn1549 did not show consensus promoter motifs. Thus, the reason why MLG101-1 could act as a donor whereas MLG055 and MLG043 did not remains unexplained.
Conjugative transposition of Tn916 has been studied extensively (38). Transposition of Tn916 proceeds by excision and formation of a circular intermediate in which the ends of the transposon are separated by six nucleotides, resulting from staggered cleavages by Int (43). The excised transposon contains a heteroduplex consisting of the 6-bp sequences termed coupling sequences. Insertion is the reverse of excision, since staggered cuts occur at the junction of the circular molecule and the new target site. This produces an integrated transposon flanked by heteroduplex coupling sequences which are resolved by DNA replication. Analysis of the junction fragments flanking the end of Tn1549-like in 29 different transconjugants indicated that 24 coupling sequences were inherited from the donor. Both GTATA and AATGCT sequences originated from flanking sequences of the first copy of the transposon in MLG101, whereas ATTTT and ATCCAA were attributed to the second copy (Fig. 2). This resulted in asymmetric acquisition of either five or six nucleotides from the donor, depending on the level of DNA cutting. In addition, five events corresponded to 5-bp nucleotide duplications of the target site, similar to what was seen for duplications produced by other kinds of transposons, although the mechanism is completely different. Such a feature has been reported for Tn916 and was attributed to mismatch repair following integration of the transposon (41). This could have also been the result of two successive transposition events, in which the transposon brings a coupling sequence from the intermediate integration site, which is identical to that of the final target. However, the fact that five independent transposition events led to exact 5-bp duplication does not favor this hypothesis. Moreover, we might have predicted that random cutting upon insertion would have generated distinct 5-bp flanking regions, which were not observed. Interestingly, three different events, either with acquisition of five or six nucleotides or with duplication of five base pairs, were observed in T7, T8, and T9 at the same locus. Two other events with acquisition of five nucleotides in T6 and T12 were detected at another locus. These two loci are likely to be two hot spots for Tn1549-like insertion (Fig. 2).
The acquisition of fragments larger than 68 kb by T11 and T16 is not clearly understood and is probably due to chromosomal rearrangement during DNA replication, since analysis of the flanking regions of Tn1549 gave evidence of a transposition process.
Surprisingly, T1 and T11 contained two copies of Tn1549-like elements, as did MLG101-1. Since conjugative transposons do not transpose in a replicative manner, the presence of multiple copies could be explained either by intracellular transposition between daughter chromosomes after passage of the replication fork (38), by cotransfer, or by independent acquisition of the two copies by horizontal transfer.
The presence of vanB2 on a functional conjugative transposon accounts for its spread in both anaerobes and enterococci. These elements have a broad host range that includes numerous genera of gram-positive bacteria. It has been recently reported that mobile elements designated as integrative and conjugative elements encode integrative and putative transfer functions related to those of conjugative transposons (8). Such structures have been found in the genomes of various bacteria with low G+C content, including Bacillus subtilis, Butyrivibrio fibrisolvens, Clostridium difficile, E. faecalis, Listeria monocytogenes, S. aureus, Streptococcus mutans, and Streptococcus thermophilus. A similar structure or a conjugative transposon has probably recruited the vanB operon from a glycopeptide producer to form a Tn1549-like element.
Recently, operons closely related to vanA and vanB have been reported for Paenibacillus thiaminolyticus and Paenibacillus apiarius (22). Anaerobic bacteria could have played a role as an intermediate for the transfer of vanB-mediated glycopeptide resistance from glycopeptide producers to enterococci. Anaerobes which constitute the majority of the bacteria in the digestive tract and enterococci largely present are also very common in soil. The high density and the promiscuity of these bacteria in the digestive tract, associated with the presence of glycopeptide, create favorable conditions for the occurrence of transfer. Our data provide further support for the hypothesis that vanB-containing, naturally occurring anaerobes may be associated with the emergence of new strains of vancomycin-resistant enterococci under appropriate clinical selective conditions (6, 42). Due to their broad host range, conjugative transposons are exceedingly important in bacterial evolution, and the fact that Tn1549 is a genuine conjugative transposon highlights the transfer potential of VanB-type glycopeptide resistance in pathogenic species. This fact argues in favor of detecting the carriage of the vanB gene directly from the feces by PCR in order to control the spread of this resistance determinant.
Acknowledgments
This work was partially supported by the European Community's Fifth Framework Programme “Quality of Life and Management of Living Resources” contract QLK2-CT-2002-00843-ARTRADI.
We thank Anne Collignon for the coordination of ARTRADI and P. Courvalin for helpful discussions.
REFERENCES
- 1.Arthur, M., F. Depardieu, G. Gerbaud, M. Galimand, R. Leclercq, and P. Courvalin. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 179:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, M., P. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 5.Bahl, M. I., S. J. Sørensen, L. H. Hansen, and T. R. Licht. 2004. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl. Environ. Microbiol. 70:758-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballard, S. A., E. A. Grabsch, P. D. R. Johnson, and M. L. Grayson. 2005. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 49:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballard, S. A., K. K. Pertile, M. Lim, P. D. R. Johnson, and M. L. Grayson. 2005. Molecular characterization of vanB elements in naturally occurring gut anaerobes. Antimicrob. Agents Chemother. 49:1688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICEStl element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 9.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlier, C., and P. Courvalin. 1990. Emergence of 4′,4′′-aminoglycoside nucleotidyltransferase in enterococci. Antimicrob. Agents Chemother. 34:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 12.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, N. C., L. M. Weigel, J. B. Patel, and F. C. Tenover. 2005. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 49:470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clewell, D. B., F. Y. An, B. A. White, and C. Gawro-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl, K. H., T. P. Røkenes, E. W. Lundblad, and A. Sundsfjord. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl, K. H., G. S. Simonsen, O. Olsvik, and A. Sundsfjord. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl, K. H., and A. Sundsfjord. 2003. Transferable vanB2 Tn5382-containing elements in fecal streptococcal strains from veal calves. Antimicrob. Agents Chemother. 47:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domingo, M. C., A. Huletsky, A. Bernal, R. Giroux, D. K. Boudreau, F. J. Picard, and M. G. Bergeron. 2005. Characterization of a Tn5382-like transposon containing the vanB2 gene cluster in a Clostridium strain isolated from human faeces. J. Antimicrob. Chemother. 55:466-474. [DOI] [PubMed] [Google Scholar]
- 19.Doucet-Populaire, F., P. Trieu-Cuot, I. Dosbaa, A. Andremont, and P. Courvalin. 1991. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 35:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannagan, S. E., and D. B. Clewell. 1991. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J. Bacteriol. 173:7136-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 22.Guardabassi, L., H. Christensen, H. Hasman, and A. Dalsgaard. 2004. Members of the genera Paenibacillus and Rhodococcus harbor genes homologous to enterococcal glycopeptide resistance genes vanA and vanB. Antimicrob. Agents Chemother. 48:4915-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayden, M. K., R. N. Picken, and D. F. Sahm. 1997. Heterogeneous expression of glycopeptide resistance in enterococci associated with transfer of vanB. Antimicrob. Agents Chemother. 41:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 26.Ligozzi, M., G. Lo Cascio, and R. Fontana. 1998. vanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob. Agents Chemother. 42:2055-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 28.Marra, D., B. Pethel, G. G. Churchward, and J. R. Scott. 1999. The frequency of conjugative transposition of Tn916 is not determined by the frequency of excision. J. Bacteriol. 181:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall, C. G., I. A. Lessard, I. Park, G. D. Wright, and P. J. Wright. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mevius, D., L. Devriese, P. Butaye, P. Vandamme, M. Verschure, and K. Veldman. 1998. Isolation of glycopeptide resistant Streptococcus gallolyticus strains with vanA, vanB, and both vanA and vanB genotypes from faecal samples of veal calves in The Netherlands. J. Antimicrob. Chemother. 42:275-276. [DOI] [PubMed] [Google Scholar]
- 31.Moubareck, C., N. Bourgeois, P. Courvalin, and F. Doucet-Populaire. 2003. Multiple antibiotic resistance gene transfer from animal to human enterococci in the digestive tract of gnotobiotic mice. Antimicrob. Agents Chemother. 47:2993-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, R., K. Piper, F. R. Cockerill III, J. M. Steckelberg, and A. A. Yousten. 2000. The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob. Agents Chemother. 44:705-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, J. M. Steckelberg, B. Kline, and F. R. Cockerill III. 1998. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob. Agents Chemother. 42:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 35.Power, E. G., Y. H. Abdulla, H. G. Talsania, W. Spice, S. Aathithan, and G. L. French. 1995. vanA genes in vancomycin-resistant clinical isolates of Oerskovia turbata and Arcanobacterium (Corynebacterium) haemolyticum. J. Antimicrob. Chemother. 36:595-606. [DOI] [PubMed] [Google Scholar]
- 36.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintiliani, R., Jr., and P. Courvalin. 1994. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol. Lett. 119:359-364. [DOI] [PubMed] [Google Scholar]
- 38.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, L. B., L. L. Carias, C. L. Donskey, and S. D. Rudin. 1998. Transferable, plasmid-mediated VanB-type glycopeptide resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 42:963-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 42.Stinear, T. P., D. C. Olden, P. D. R. Johnson, J. K. Davies, and M. L. Grayson. 2001. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357:855-856. [DOI] [PubMed] [Google Scholar]
- 43.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1993. Sequence requirements for target activity in site-specific recombination mediated by the Int protein of transposon Tn1545. Mol. Microbiol. 8:179-185. [DOI] [PubMed] [Google Scholar]
- 44.Werner, G., R. J. L. Willems, B. Hildebrandt, I. Klare, and W. Witte. 2002. Influence of transferable genetic determinants on the outcome of typing methods commonly used for Enterococcus faecium. J. Clin. Microbiol. 41:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]