Abstract
Morganella morganii produces an inducible, chromosomally encoded AmpC β-lactamase. We describe in this study three new variants of AmpC within this species with apparent pIs of 6.6 (M19 from M. morganii strain PP19), 7.4 (M29 from M. morganii strain PP29), and 7.8 (M37 from M. morganii strain PP37). After gene sequencing, deduced amino acid sequences displayed one to six substitutions when compared to the available Morganella AmpC sequences. An AmpR-encoding gene was also found upstream of ampC, including the LysR regulators' helix-turn-helix DNA-binding domain and the putative T-N11-A-protected region in the ampR-ampC intercistronic sequence. All three AmpC variants were purified from in vitro-generated derepressed mutants and showed overall similar kinetic parameters. None of the observed amino acid changes, occurring at the surface of the protein, appear to have a major influence in their catalytic properties. Morganella AmpCs exhibit the highest catalytic efficiencies (kcat/Km) on classical penicillins, cefoxitin, narrow-spectrum cephalosporins, and cefotaxime. Cefotaxime was more effectively hydrolyzed than other oxyimino-cephalosporins, whereas cefepime was 3 log-fold less efficiently hydrolyzed than other cephalosporins such as cephalothin. Several differences with other AmpC β-lactamases were found. Ampicillin was more efficiently hydrolyzed than benzylpenicillin. High kcat/Km values were observed for oxacillin and piperacillin, which are usually poor substrates for AmpC. A fairly efficient hydrolysis of imipenem was detected as well. Aztreonam, carbenicillin, and tazobactam were effective transient inactivators of these variants.
In gram-negative microorganisms that are naturally resistant to aminopenicillins and narrow-spectrum cephalosporins (sometimes resistant even to cephamycins and other expanded-spectrum cephalosporins), production of chromosome-encoded β-lactamases (AmpC) belonging to Bush-Jacoby-Medeiros group 1 or the Ambler class C is typically responsible for this resistance (7, 16).
In general, AmpC β-lactamases are poorly inhibited by mechanism-based β-lactamase inhibitors such as clavulanate and hydrolyze cephalosporins and penicillins with similar catalytic efficiencies (7, 21). Morganella morganii displays an unusual inhibition by tazobactam combinations, with lower MICs than other AmpC producers (1, 20).
With some exceptions, chromosome-encoded class C β-lactamases are inducible, but a majority of those clinical isolates that are highly resistant exhibit (in the absence of other acquired β-lactamases) a derepressed constitutive phenotype which results in a large production of enzyme in the periplasm (18, 20).
In Morganella morganii, the inducible phenotype is the most frequent for the production of chromosomally encoded AmpC (20). Nucleotide sequences corresponding to several enzymes from Morganella, both chromosomally and plasmid encoded, have been reported and deposited in databases (3, 4, 11, 31). Reported pIs for these enzymes usually range between 7.2 and 7.6, although other pIs were also reported (3, 31, 36, 38). The plasmid-encoded DHA-1 and DHA-2 β-lactamases, apparently derived from the Morganella AmpC enzyme and reported in several countries around the world, are always inducible, like their chromosome-encoded counterparts (2, 12, 19, 37, 39).
In the present study, we describe the phenotypic characteristics of three new variants of chromosomally encoded AmpC β-lactamases from clinical isolates of Morganella morganii, including their kinetic properties toward selected β-lactam compounds.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Morganella morganii strains PP19, PP29, and PP37 were clinical isolates from diarrheal, urinary tract, and skin infections, respectively. These strains were identified using API system 20E (BioMérieux). Morganella morganii strains PP19C, PP29II, and PP37B were in vitro-generated derepressed mutants derived from corresponding wild-type strains (PP19, PP29, and PP37, respectively). Selection of isogenic mutants of M. morganii strains with the derepressed hyperproduction of AmpC was performed by growing wild-type microorganisms with increasing concentrations of cefoperazone or ceftazidime, reaching up to 100 times the corresponding parental strain MIC. Escherichia coli Top10 F′ (Invitrogen) was used as the recipient for transformation experiments. Escherichia coli TCR192 and TC192-2 were Top10 derivatives harboring recombinant plasmids pMCR19-2 and pMC192-2, respectively. Plasmid vectors pGEM-T-Easy (Promega) and chloramphenicol-resistant vector pMCL210 (28) were used for cloning ampC genes from M. morganii strains PP29 and PP37 and ampC/ampR genes from M. morganii strain PP19, respectively.
Antimicrobial susceptibility.
MICs were determined by the agar dilution method, following CLSI's guidelines, using a Steers multipoint inoculator (29). Detection of inducible β-lactamases was performed by a double-disk test, placing 30 μg cefoxitin and 60 μg 6-amino-penicillanic acid (6-APA) disks 20 mm away from either 30-μg cefotaxime or ceftazidime disks as previously described (9). All disks were from Britania, Argentina.
Production and induction of β-lactamases.
Overnight cultures at 37°C in Luria-Bertani (LB) broth were diluted 1/50 into fresh LB broth and incubated on an orbital shaker (300 rpm) at 37°C until an optical density at 600 nm of 0.3 to 0.4 was reached. A subinhibitory concentration of inducer (5 μg/ml cefoxitin or 50 μg/ml 6-APA) was added, and incubation was continued for 2 to 4 h. Crude lysates were obtained as described before (32).
Determination of β-lactamase activity.
β-Lactamase activity was routinely determined spectrophotometrically by measuring the hydrolysis of 100 μM cephalothin as a substrate (λ = 273 nm; ΔɛM = −6,300 M−1 · cm−1). One unit of β-lactamase activity was defined as the amount of enzyme which hydrolyzes 1 μmol of substrate per min (in 20 mM phosphate buffer, pH 7.0) at 30°C; the specific activity was defined as the units of AmpC per milligram of protein determined by using the Bio-Rad Protein Assay kit (Bio-Rad).
IEF.
Analytical isoelectric focusing (IEF) was performed in broad-range (pH 3 to 10) polyacrylamide gels (24), using protein standards (Pharmacia) and enzymes of known pIs (TEM-1, SHV-2, P99, CTX-M-2) as markers. β-Lactamase activity was detected by an iodometric overlay system using 500 μg/ml cephalothin and 500 μg/ml benzylpenicillin as substrates (32). In situ inhibition of AmpC β-lactamases was assayed after IEF by first soaking the gel in 1 mM β-lactamase inhibitor solutions (lithium clavulanate, EDTA, and aztreonam) for 30 min and detection of the remaining β-lactamase activity as described above.
Recombinant DNA methodology and DNA sequencing.
Basic recombinant DNA procedures were carried out as described by Sambrook et al. (33). Chromosomal DNA was extracted by a modification of the original method described by Gerhardt et al. (15). For cloning ampC-ampR genes from M. morganii strain PP19, genomic DNA was partially digested with KpnI (Amersham Pharmacia) and resulting fragments (between 1 and 5 kb) were purified from 1% agarose gels and ligated to KpnI-linearized and dephosphorylated pMCL210 vector. For PCR-based cloning, reaction mixtures contained 3 U Pfu polymerase (Promega), 0.2 to 0.5 μg DNA, 2 mM MgSO4, 0.3 mM (each) deoxynucleoside triphosphate, and 0.6 μM (each) primer. PCRs were performed in a Biometra T-Gradient apparatus; cycling parameters were as follows: initial denaturation for 3 min at 95°C; 30 cycles of denaturation at 95°C for 1 min, annealing at 55 to 60°C for 1 to 2 min, and polymerization at 73°C for 3 to 4 min; and final polymerization at 73°C for 20 min. Construction of pMC-1922: the ampC gene was amplified from pMCR19-2 by PCR using primers MOR-2F (5′-TCT GTC TGG TGA ATC TGA CGA-3′; accession no. AF055067) and MOR-E3R (5′-ACA CAG TGA ATT CCG GTT CAG CGG-3′, EcoRI site underlined). The fragment was digested with EcoRI (Boehringer, Germany) and subcloned into the pMCL210 vector digested with SmaI and EcoRI. M. morganii strain PP29 and PP37 ampC genes were amplified using primers MOR-H2F (5′-CTG GTG AAG CTT ACG ATA CTT GCC-3′; accession no. AF055067) and MOR-E3R and ligated in a pGEM-T Easy Vector System (Promega) following manufacturer's guidelines. Ligation mixtures were used to transform competent E. coli Top10 F′ cells (Invitrogen), and recombinant clones were selected on LB agar plates supplemented with 30 μg/ml chloramphenicol and 50 μg/ml ampicillin (for pMCL210-based constructions) or 100 μg/ml ampicillin, 30 μM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma Chemical Co.), and 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Boehringer, Germany). The DNA sequences on both strands were determined by the automated dideoxy-chain termination method of Sanger (34) in an ABI PRISM 3700 DNA Analyzer (Applied Biosystems, Hitachi). Sequence analyses were performed by using NCBI (http://www.ncbi.nlm.nih.gov/) and EBI (http://www.ebi.ac.uk/) analysis tools.
Purification of AmpC β-lactamases.
Clear supernatants containing the AmpC β-lactamases were loaded onto a Sephadex G-100 column (2.0 by 20 cm; Pharmacia-LKB, Sweden) equilibrated with 20 mM Tris-HCl buffer, pH 7.5. Elution was performed with the same buffer, and active fractions were collected. The sample was then loaded onto a CM-Sepharose column (5.0 by 20 cm; Pharmacia-LKB, Sweden) and connected to a Pharmacia LC-250 fast-protein liquid chromatography system equilibrated with the same buffer. The column was extensively washed to remove unbound proteins, and β-lactamases were eluted with a linear gradient of NaCl (0 to 1 M) in the same buffer. Fractions containing β-lactamase activity were pooled, dialyzed overnight at 4°C against 10 mM phosphate buffer, pH 7.5, and concentrated 10 times in an Amicon 8200 system (Millipore). Active fractions were detected in all cases by their hydrolytic activity on 100 μM nitrocefin. After each purification step, samples were loaded on a 12% polyacrylamide gel and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (17), in order to assess their purification degree.
Determination of the N-terminal amino acid sequence.
The N-terminal amino acid sequence of the mature AmpC variants was characterized using a gas phase sequencer (Applied Biosystems) as previously described (6).
Determination of kinetic parameters.
Hydrolysis of β-lactam antibiotics by purified enzymes was monitored by following the absorbance variation, using an Uvikon 860 spectrophotometer equipped with thermostatically controlled cells. Cells with 0.2- to 1.0-cm path lengths were used, depending on the substrate concentration. Reactions were performed in a total volume of 500 μl at 30°C. For good substrates, the steady-state kinetic parameters (Km and kcat) were determined under initial rate conditions by using the Hanes-Woolf linearization of the Henri-Michaelis-Menten equation as described by De Meester et al. (8). Low Km values were determined as competitive inhibition constants, Ki, in the presence of nitrocefin as a good reporter substrate. When the β-lactam behaved as a poor substrate or inactivator, the residual activity of the enzyme in the presence of the drug was monitored using 100 μM nitrocefin as a reporter substrate. The pseudo-first-order inactivation constants, kis, were computed, and the different constants were calculated as previously reported (8). Tested antibiotics and inhibitors included benzylpenicillin (ΔɛM235 = −775 M−1 · cm−1), ampicillin (ΔɛM235 = −820 M−1 · cm−1), carbenicillin (ΔɛM235 = −780 M−1 · cm−1), piperacillin (ΔɛM235 = −820 M−1 · cm−1), nitrocefin (ΔɛM482 = 15,000 M−1 · cm−1), cephalothin (ΔɛM273 = −6,300 M−1 · cm−1), cefoxitin (ΔɛM260 = −6,600 M−1 · cm−1), cefotaxime (ΔɛM260 = −7,500 M−1 · cm−1), ceftazidime (ΔɛM260 = −9,000 M−1 · cm−1), cefepime (ΔɛM260 = −10,000 M−1 · cm−1), aztreonam (ΔɛM318 = −750 M−1 · cm−1), imipenem (ΔɛM300 = −9,000 M−1 · cm−1), and tazobactam (ΔɛM235 = 1,800 M−1 · cm−1).
Protein structure prediction and 3D modeling.
A prediction of the α-helix content of AmpC β-lactamases was performed with AGADIR software (25), and theoretical three-dimensional (3D) models for the enzymes were predicted by using the Swiss-Model tool (http://swissmodel.expasy.org) and constructed with SwissProt Pdb Viewer 3.7 (35).
Nucleotide sequence accession numbers.
Sequence data were deposited in the GenBank/EMBL nucleotide databases under accession no. AJ620115, AJ620362, and AJ620363.
RESULTS AND DISCUSSION
pI and in situ inhibition profile of AmpC variants.
IEF analysis of crude extracts obtained after induction of cultures with 50 μg/ml 6-APA showed the presence of a single band of β-lactamase activity with pI values of 6.6, 7.4, and 7.8 for M. morganii strains PP19, PP29, and PP37, respectively. No β-lactamase activity could be detected in the absence of an inducer. The enzymes were not inactivated or inhibited in the presence of 1 mM EDTA or 1 mM lithium clavulanate; on the other hand, 1 mM aztreonam behaved as a strong inactivator (data not shown).
In vitro selection and analysis of AmpC hyperproducers.
M. morganii mutants with derepressed expression of AmpC were obtained by incubating the bacteria in the presence of increasing concentrations of oxyimino-cephalosporins. Selection frequency ranked between 5.10−8 and 2.10−6 in accordance with that obtained for the in vivo selection of derepressed mutants of AmpC producers (i.e., Enterobacter spp.) during oxyimino-cephalosporin therapy (20). All mutants produced an enzyme with pIs identical to those of their parental wild-type AmpCs (data not shown).
Susceptibility testing of M. morganii strains.
Antimicrobial susceptibilities of both wild-type and mutant strains were determined (Table 1). A typical pattern was observed in wild-type strains, with resistance to aminopenicillins and in combination with clavulanate, cephalothin, and cefoxitin. Inhibition by tazobactam combinations was also observed, in agreement with previous reports (1). Mutants showed patterns that are similar to those of Morganella derepressed mutants: resistance to piperacillin and reduced susceptibility to oxyimino-cephalosporins, cefoxitin, and aztreonam. As expected, double-disk assays for the detection of inducible β-lactamases failed when these mutant strains were analyzed (data not shown).
TABLE 1.
MICs of β-lactams for M. morganii clinical isolates, derived AmpC hyperproducers, and E. coli strains harboring ampC from M. morganii
| Antibiotic(s) | MIC (μg/ml) for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild-type strains
|
AmpC hyperproducers
|
Escherichia coli strains
|
|||||||
| PP19 | PP29 | PP37 | PP19C | PP29II | PP37B | TCR192c | TC192-2d | Top10 F′ | |
| Ampicillin | 256 | 64 | 32 | 1,024 | 1,024 | 512 | 512 | 256 | 2 |
| Ampicillin-clavulanatea | 128/64 | 128/64 | 64/32 | 512/256 | 512/256 | 256/128 | 256/128 | 128/64 | 2/1 |
| Piperacillin | 16 | 0.25 | 0.25 | >64 | 32 | 16 | 64 | 32 | 1 |
| Piperacillin-tazobactamb | 1 | 0.063 | 0.063 | 32 | 8 | 4 | 16 | 8 | 0.5 |
| Cephalothin | >512 | >512 | >512 | >2,048 | >2,048 | >2,048 | >512 | 256 | 2 |
| Cefoxitin | 32 | 8 | 4 | 64 | 16 | 64 | 64 | 16 | 2 |
| Cefotaxime | ≤0.032 | ≤0.032 | ≤0.032 | 16 | 4 | 16 | 2 | 0.5 | ≤0.032 |
| Ceftazidime | 1 | 0.5 | 0.5 | 32 | 16 | 64 | 8 | 4 | 0.063 |
| Cefoperazone | 1 | 1 | 2 | 32 | 8 | 16 | 0.25 | 0.25 | ≤0.032 |
| Cefepime | ≤0.008 | ≤0.008 | ≤0.008 | ≤0.008 | ≤0.008 | ≤0.008 | 0.008 | 0.016 | 0.016 |
| Imipenem | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 0.063 |
| Aztreonam | ≤0.032 | ≤0.032 | ≤0.032 | 0.5 | 0.5 | 0.064 | 0.5 | 0.5 | 0.063 |
Used at a ratio of 2:1.
Fixed concentration of 4 μg/ml.
Strain harboring pMCR19-2 (ampC, ampR from M. morganii strain PP19).
Strain harboring pMC192-2 (ampC from M. morganii strain PP19).
Cloning of M. morganii ampC genes.
A 2.8-kb KpnI chromosomal DNA fragment from M. morganii strain PP19 was cloned into the pMCL210 vector to yield the pMC19-2 plasmid. The susceptibility profile of the E. coli recombinant clone TCR192 was similar to that for the M. morganii PP19C strain (Table 1). However, by double-disk tests, we were able to detect the production of an inducible β-lactamase in E. coli TCR192 (data not shown), suggesting that an AmpR-dependant regulation does occur, such as the induction observed in microorganisms harboring the DHA enzymes. Moreover, when E. coli TCR192 was grown in the presence of 50 μg/ml 6-APA, specific activity increased up to 240% compared to the basal level (no induction), reinforcing the hypothesis of AmpC induction. When ampC from M. morganii PP19 was subcloned without the ampR gene (pMC-192-2 recombinant plasmid), MICs for the recombinant clone TC192-2 were overall similar or slightly lower that those for TCR192 (Table 1), but a positive result was not observed with the double-disk tests for detection of inducible enzymes (data not shown), supporting the results that imply a negative regulation by AmpR.
Sequence of ampC genes and structural analysis of cephalosporinases.
The entire inserts from recombinant plasmids pMCR19-2, pGMC29-31, and pGMC37-43 (the latter were pGEM-T derivatives containing a PCR-amplified copy of the ampC genes from M. morganii strains PP29 and PP37, respectively) were sequenced in both strands. Analysis of these inserts for coding regions showed an open reading frame of 1,137 bp, encoding a deduced 379-amino-acid peptide, with at least 98% and 97% identities within them and with other AmpCs from M. morganii, respectively.
The N-terminal sequence of mature β-lactamase was Ala-Asp-Asn-Val-Ala-Ala-Val. Therefore, the pre-β-lactamase contained a 23-amino-acid signal peptide and mature AmpC β-lactamases consisted of 356-amino-acid proteins. These results are in agreement with the theoretical determination of the signal peptide by SignalP V2.0 (30).
By comparing the sequences of mature AmpCs from the three M. morganii isolates with the other available sequences within the species, we could describe three new variants of AmpC β-lactamases (Table 2). AmpC from M. morganii strain PP37 (AmpC M37) seems to be related to GUI-1 (3, 4, 11, 31), which is identical to plasmid-borne DHA-1 (3, 4, 11, 31), displaying a single amino acid change, Met79Thr (99.7% identity). On the other hand, AmpCs from M. morganii strains PP29 and PP19 (AmpC M29 and M19) appear to be more related to the enzyme from M. morganii strain SLM01 (3), presenting one mutation, Glu167Asp (M29, 99.7% identity), and two mutations, Arg138Trp and Ala239Thr (M19, 99.4% identity), respectively.
TABLE 2.
Amino acid substitutions of AmpC β-lactamases from Morganella morganiia
| AmpC variant | pI | Amino acid position
|
Amino acid sequence identity (%)
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 44 | 79 | 80 | 86 | 110 | 123 | 126 | 129 | 138 | 167 | 239 | 253 | GUI-1/DHA-1 | SLM01 | DHA-2 | M19 | M29 | M37 | ||
| GUI-1/DHA-1 | 7.6 | I | M | A | A | A | S | D | N | R | E | A | T | 100 | |||||
| SLM01 | 7.4 | V | T | E | H | 98.9 | 100 | ||||||||||||
| DHA-2 | 7.8 | V | M | E | T | N | E | H | 98.0 | 99.2 | 100 | ||||||||
| M19 | 6.6 | V | T | E | H | W | T | 98.3 | 99.4 | 98.6 | 100 | ||||||||
| M29 | 7.4 | V | T | E | H | D | 98.6 | 99.7 | 98.9 | 99.2 | 100 | ||||||||
| M37 | 7.6 | T | 99.7 | 98.6 | 97.7 | 98.0 | 98.3 | 100 | |||||||||||
In order to determine if some of the amino acid substitutions could be associated with residues involved in the catalytic activity or the structure of the enzyme, hypothetical α-helix domains and theoretical 3D models were built using the X-ray structure of the Escherichia coli AmpCs, which is the closest enzyme available (65.7% similarity) (35). Resulting models obtained for the three variants gave similar results, showing a strict correspondence in the position of the class C conserved motifs and domains with known crystals. Figure 1 shows the predicted 3D model of AmpC from M. morganii strain PP29. Mutations occurring in our variants seem to be located on the surface of the molecule and not associated with any catalytically important residue. Therefore, we expected a similar kinetic behavior between all these AmpC variants and probably compared to other cephalosporinases. The Arg138Trp mutation in AmpC M19 could lead to a pI decrease from 7.4 to 6.6 when compared to AmpC SLM01 (Table 2).
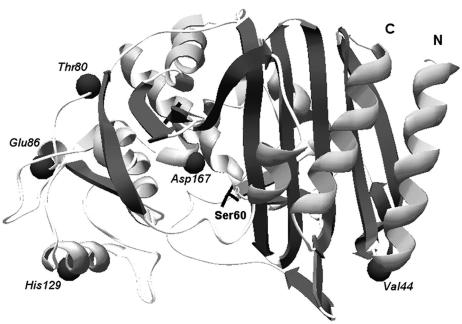
FIG. 1.
Predicted 3D model of the AmpC β-lactamase from M. morganii strain PP29, constructed using Swiss-Model and the Deep-View/Swiss-Pdb Viewer v3.7. The following amino acid shifts, compared to AmpC GUI-1 (AF055067), are shown as dark spheres: Ile44Val, Ala80Thr, Ala86Glu, Asn129His, and Glu167Asp. The active-site serine residue (Ser60) is also shown. N, amino terminal; C, carboxy terminal.
Sequence of ampR from M. morganii strain PP19 and analysis of intercistronic region.
The AmpR-encoding gene from M. morganii strain PP19, cloned in the pMCR192-2 recombinant plasmid, was also sequenced in both strands. The deduced AmpR protein, consisting of 291 amino acids (ca. 32,000 Da), seems to have the characteristics of a LysR family regulator, showing a putative DNA-binding domain named helix-turn-helix, located at the N terminus of M. morganii AmpR between Phe23 and Ala43, where the highest homology with other LysR-regulators occurs (5, 10). By comparison with other LysR regulators from enterobacteria, AmpR from M. morganii strain PP19 has 99% identity with available AmpR sequences from M. morganii isolates, showing so far a high degree of conservation within the species, and shares 51 to 63% of its amino acidic sequence with other AmpRs, being closer to those from E. coli, Enterobacter spp., Citrobacter freundii, and Yersinia enterocolitica and with other regulators of class A β-lactamases such as NMC-R and SmeR, suggesting similar functions and probably a common origin (26, 27).
A DNA intercistronic sequence between the divergently transcribed ampR and ampC genes is also found in M. morganii strain PP19, showing the characteristics of the hypothetical AmpR-binding site (5). As in other Morganella strains, the intercistronic region is shorter (110 bp) than in other species, such as C. freundii (131 bp), Enterobacter cloacae (132 bp), and Y. enterocolitica (134 bp).
Purification and kinetic parameters of AmpC variants.
AmpC M19, AmpC M29, and AmpC M37 were purified; 1.54 to 2.1 mg of purified enzyme per liter of culture was obtained after purification, with a final yield of 22 to 34%.
The main kinetic parameters for AmpC M29 (taken as representative) are shown in Tables 3 and 4, and some differences with other AmpC enzymes have been addressed previously (13, 14, 21).
TABLE 3.
Kinetic parameters of AmpC M29 from Morganella morganii strain PP29 with different substrates
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|
| Benzylpenicillina | 0.25 ± 0.007 | 0.0074 ± 0.0004 | 0.03 ± 0.002 |
| Ampicillina | 0.13 ± 0.009 | 0.065 ± 0.003 | 0.5 ± 0.02 |
| Piperacillina | 0.17 ± 0.005 | 1.5 ± 0.08 | 9 ± 0.4 |
| Oxacillina | 0.003 ± 0.0003 | 0.022 ± 0.001 | 7 ± 0.3 |
| Imipenema | 2 ± 0.3 | 0.07 ± 0.003 | 0.035 ± 0.002 |
| Nitrocefin | 25 ± 4 | 93 ± 8 | 3.8 ± 0.3 |
| Cephalothin | 148 ± 10 | 140 ± 35 | 1 ± 0.2 |
| Cefoxitina | 0.018 ± 0.0015 | 0.038 ± 0.0018 | 2.1 ± 0.1 |
| Cefotaximea | 0.02 ± 0.0016 | 0.032 ± 0.002 | 1.6 ± 0.1 |
| Ceftazidimea | 0.9 ± 0.04 | 0.04 ± 0.002 | 0.045 ± 0.002 |
| Cefepimea | 95 ± 9 | 0.008 ± 0.0007 | 8 × 10−5 ± 7 × 10−6 |
For these substrates, the Km value was determined as a Ki value using nitrocefin as a reporter substrate.
TABLE 4.
Kinetic parameters of AmpC M29 from Morganella morganii strain PP29 with different inactivators
| Inactivatora | k+2 (s−1) | K (μM) | k+2/K (μM−1 · s−1) | k+3 (s−1) |
|---|---|---|---|---|
| Carbenicillin | 0.35 ± 0.02 | 0.4 ± 0.02 | 0.87 ± 0.12 | 0.004 ± 0.0002 |
| Aztreonam | ND | ND | 1 ± 0.05 | 0.004 ± 0.0003 |
| Tazobactam | ND | ND | 0.014 ± 0.001 | 0.017 ± 0.0003 |
For these drugs, the different constants were calculated from the computed pseudo-first-order inactivation constant ki using nitrocefin as a reporter substrate. ND, not determined.
Interaction with penicillins was generally characterized by low values of Km and kcat, yielding high kcat/Km values (catalytic efficiencies). All three variants showed good affinities against a wide variety of antibiotics, supporting the idea of their wide hydrolytic spectrum compared to class A enzymes.
Morganella AmpC showed a higher catalytic efficiency for ampicillin (0.5 μM−1 · s−1) than benzylpenicillin (0.03 μM−1 · s−1), whereas for enzymes from other species, the opposite behavior usually takes place (14, 21). A better environment for the stabilization of amino-substituted penicillins probably exists in the catalytic site of Morganella AmpCs, but only the determination of the crystallographic structure of these enzymes will finally elucidate some of the presented results.
The high catalytic efficiency of piperacillin and oxacillin compared with that of other penicillins was noteworthy. As generally accepted, oxacillin and its chlorinated derivatives usually behave as poor substrates or even transient inactivators of this class of enzymes (14). In this case, oxacillin showed a high catalytic efficiency due to very low Km values (Table 3). Probably in Morganella variants, a higher efficiency for releasing the hydrolyzed molecule (high k+3 values) also occurs, leading to the increased kcat/Km values that we observed.
The catalytic efficiency against imipenem was significantly higher than that observed for class A β-lactamases (21). Furthermore, for some other AmpC β-lactamases, imipenem behaves as a transient inactivator instead (13). Carbapenems seem to be more efficiently hydrolyzed by Morganella AmpC than other variants, and this could (if associated with a lower permeability of this drug) be giving the high MICs observed in the isolates under study (Table 1).
Like other chromosome- and plasmid-encoded class C β-lactamases, Morganella AmpC displayed high catalytic efficiencies toward most of the tested cephalosporins. For nitrocefin and cephalothin, the high kcat/Km values apparently demonstrate that even with a high Km value, a fast turnover of the active site (higher kcat) occurs. This could be equivalent to the behavior of some class A β-lactamases against penicillins (13, 23).
Cefoxitin seems to be efficiently hydrolyzed by AmpCs from Morganella. Whereas cephamycins are efficiently hydrolyzed by class C enzymes, class A β-lactamases lack an efficient hydrolysis of these compounds, probably because of a displacement of the water molecule involved in the hydrolysis step by the cephamycin's 7α-methoxy moiety (22). The kcat/Km values toward oxyimino-cephalosporins, especially cefotaxime, are in good agreement with those reported for other Enterobacteriaceae (13). Cefepime was, on the contrary, hydrolyzed more than 1,000-fold less efficiently than other cephalosporins, explaining the remarkable stability of these compounds against Morganella AmpC β-lactamases, even when they are overproduced by derepression.
It was noteworthy that carbenicillin (often a good substrate for many class A β-lactamases) and aztreonam behaved as apparent poor substrates of AmpC (Table 4). The interaction between AmpC M29 and these compounds is characterized by a high acylation efficiency (k+2/K ≈ 1 μM−1 · s−1) and a slow deacylation of the acyl-enzyme complex (low k+3), making these compounds behave as transient inactivators. For carbenicillin, k+2 (acylation rate constant) is almost 90-fold higher than k+3 (deacylation rate constant), in agreement with the behavior of poor substrates or even transient inactivators (14). Further studies are necessary in order to explain this unusual behavior.
Morganella morganii AmpC M29 is better inhibited by tazobactam than other class C enzymes. This compound displayed a relatively fast acylation step (k+2/K = 1.4 × 10−2 μM−1 · s−1) but a slow deacylation step (k+3 = 0.017 s−1). Taking into account the apparently high catalytic efficiency of Morganella AmpC toward piperacillin and their inactivation by tazobactam, we assume that the latter could slightly protect the penicillin due to a rapid acylation of the active site by tazobactam. MICs support this idea of a partial protection of piperacillin by tazobactam, which was not observed in other AmpC-producing enterobacterial species.
All three variants of AmpC from Morganella showed similar kinetic parameters, with the most relevant similarities and differences presented in Table 5. The main differences, especially in their catalytic efficiencies, were observed within the cephalosporins, though these drugs seem to be better substrates than penicillins. The AmpC from M. morganii strain PP19 seems to hydrolyze cefotaxime 10-fold less efficiently than the other two variants, but the variant from M. morganii strain PP37 appears to be 5- to 10-fold less efficient in hydrolyzing cephalothin.
TABLE 5.
Main kinetic similarities and differences among the three variants of AmpC from M. morganii
| Antibiotic | AmpC enzyme | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|---|
| Ampicillin | M19 | 0.06 ± 0.003 | 0.08 ± 0.004 | 0.75 ± 0.04 |
| M29 | 0.06 ± 0.003 | 0.13 ± 0.009 | 0.5 ± 0.02 | |
| M37 | 0.06 ± 0.003 | 0.06 ± 0.006 | 1 ± 0.05 | |
| Imipenem | M19 | 0.11 ± 0.006 | 4 ± 0.1 | 0.027 ± 0.002 |
| M29 | 0.07 ± 0.003 | 2 ± 0.3 | 0.035 ± 0.002 | |
| M37 | 0.07 ± 0.004 | 1 ± 0.06 | 0.07 ± 0.004 | |
| Cephalothin | M19 | 14 ± 2 | 46 ± 13 | 0.3 ± 0.04 |
| M29 | 140 ± 35 | 148 ± 10 | 1 ± 0.2 | |
| M37 | 6 ± 0.8 | 106 ± 10 | 0.06 ± 0.007 | |
| Cefotaxime | M19 | ≤0.07 ± 0.004 | 0.5 ± 0.06 | 0.14 ± 0.008 |
| M29 | 0.032 ± 0.002 | 0.02 ± 0.0016 | 1.6 ± 0.1 | |
| M37 | ≤0.09 ± 0.003 | 0.09 ± 0.01 | 1 ± 0.03 |
Conclusions.
Based on structural and catalytic properties, it was reported that class C β-lactamases have a higher homogeneity than class A enzymes (21). Our results show a slight variability in the kinetic properties of AmpCs from Morganella, with values that are more heterogeneous than those for AmpCs from other Enterobacteriaceae, both chromosome and plasmid encoded. More detailed studies on structural properties of the Morganella variants, by crystallographic and mutagenesis studies, are intended in order to determine if a relationship between the structural and kinetic properties does actually occur.
Acknowledgments
This work was supported in part by grants from the FP6 project LSHM-CT-503335 of the European Union to J.A.A., from the European Commission (Targeted Project COBRA) to M.G., and from Secretaría de Ciencia y Tecnología (PICT 2004/14234), Universidad de Buenos Aires and Ministerio de Salud (Beca Oñativia-Carrillo), Argentina, to G.G. Part of this work was also supported by an FNRS-SECyT agreement between Belgium and Argentina.
G.G. is a member of Carrera del Investigador Científico (CONICET).
REFERENCES
- 1.Akova, M., Y. Yang, and D. M. Livermore. 1990. Interactions of tazobactam and clavulanate with inducibly- and constitutively-expressed class I β-lactamases. J. Antimicrob. Chemother. 25:199-208. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, M., J. H. Tran, N. Chow, and G. A. Jacoby. 2004. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 48:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnaud, G., G. Arlet, C. Danglot, and A. Philippon. 1997. Cloning and sequencing of the gene encoding the AmpC β-lactamase of Morganella morganii. FEMS Microbiol. Lett. 148:15-20. [DOI] [PubMed] [Google Scholar]
- 4.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartowsky, E., and S. Normark. 1993. Interactions of wild-type and mutant AmpR of Citrobacter freundii with target DNA. Mol. Microbiol. 10:555-565. [DOI] [PubMed] [Google Scholar]
- 6.Bekal, S., J. van Beeumen, B. Samyn, D. Garmyn, S. Henini, C. Diviès, and H. Prévost. 1998. Purification of Leuconostoc mesenteroides citrate lyase and cloning and characterization of the citCDEFG gene cluster. J. Bacteriol. 180:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Meester, F., B. Joris, G. Reckinger, C. Bellefroid-Bourguignon, J. M. Frère, and S. G. Waley. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 36:2393-2403. [DOI] [PubMed] [Google Scholar]
- 9.Eliopoulos, G. M. 1988. Induction of β-lactamases. J. Antimicrob. Chemother. 22:37-44. [DOI] [PubMed] [Google Scholar]
- 10.Everett, M., T. Walsh, G. Guay, and P. Bennett. 1995. GcvA, a LysR-type transcriptional regulator protein, activates expression of the cloned Citrobacter freundii ampC β-lactamase gene in Escherichia coli: cross-talk between DNA-binding proteins. Microbiology 141:419-430. [DOI] [PubMed] [Google Scholar]
- 11.Fortineau, N., L. Poirel, and P. Nordmann. 2001. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 12.Gaillot, O., C. Clement, M. Simonet, and A. Philippon. 1997. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J. Antimicrob. Chemother. 39:85-87. [DOI] [PubMed] [Google Scholar]
- 13.Galleni, M., G. Amicosante, and J.-M. Frère. 1988. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem. J. 255:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galleni, M., and J.-M. Frère. 1988. A survey of the kinetic parameters of class C β-lactamases. Penicillins. Biochem. J. 255:119-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg. 1994. Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
- 16.Ghuysen, J. M. 1991. Serine β-lactamases and penicillin-binding-proteins. Annu. Rev. Microbiol. 45:37-67. [DOI] [PubMed] [Google Scholar]
- 17.Hames, B. D. 1981. An introduction to polyacrylamide gel electrophoresis. In B. D. Hames and D. Rickwood (ed.), Gel electrophoresis of proteins: a practical approach. IRL Press, Oxford, United Kingdom.
- 18.Jacobs, C., J.-M. Frère, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 19.Liebana, E., M. Batchelor, F. A. Clifton-Hadley, R. H. Davies, K. L. Hopkins, and E. J. Threlfall. 2004. First report of Salmonella isolates with the DHA-1 AmpC β-lactamase in the United Kingdom. Antimicrob. Agents Chemother. 48:4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matagne, A., A. Dubus, M. Galleni, and J.-M. Frère. 1999. The β-lactamase cycle: a tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 16:1-19. [DOI] [PubMed] [Google Scholar]
- 22.Matagne, A., J. Lamotte-Brasseur, G. Dive, J. R. Knox, and J. M. Frère. 1993. Interactions between active-site serine β-lactamases and compounds bearing a methoxy side chain on the α-face of the β-lactam ring: kinetic and molecular modelling studies. Biochem. J. 293:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matagne, A., J. Lamotte-Brasseur, and J.-M. Frère. 1998. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 330: 581-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 25.Muñoz, V., and L. Serrano. 1997. Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41: 495-509. [DOI] [PubMed] [Google Scholar]
- 26.Naas, T., D. M. Livermore, and P. Nordmann. 1995. Characterization of an LysR family protein, SmeR from Serratia marcescens S6, its effect on expression of the carbapenem-hydrolyzing β-lactamase Sme-1, and comparison of this regulator with other β-lactamase regulators. Antimicrob. Agents Chemother. 39:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas, T., and P. Nordmann. 1994. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc. Natl. Acad. Sci. USA 91:7693-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. 20. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Poirel, L., M. Guibert, D. Girlich, T. Naas, and P. Nordmann. 1999. Cloning, sequence analysis, expression, and distribution of ampC-ampR from Morganella morganii. Antimicrob Agents Chemother. 43:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Power, P., M. Radice, C. Barberis, C. de Mier, M. Mollerach, M. Maltagliatti, C. Vay, A. Famiglietti, and G. Gutkind. 1999. Cefotaxime-hydrolysing β-lactamases in Morganella morganii. Eur. J. Clin. Microbiol. Infect. Dis. 18:743-747. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fristch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sanger, I., S. Nicklen, and A. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31: 3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toda, M., M. Inoue, and S. Mitsuhashi. 1981. Properties of cephalosporinase from Proteus morganii. J. Antibiot. 34:1469-1475. [DOI] [PubMed] [Google Scholar]
- 37.Yan, J.-J., W.-C. Ko, Y.-C. Jung, C.-L. Chuang, and J.-J. Wu. 2002. Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 β-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40:3121-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, Y., and D. M. Livermore. 1988. Chromosomal β-lactamase expression and resistance to β-lactam antibiotics in Proteus vulgaris and Morganella morganii. Antimicrob. Agents Chemother. 32:1385-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yong, D., Y. Lim, W. Song, Y. S. Choi, D.-Y. Park, H. Lee, J. H. Yum, K. Lee, J. M. Kim, and Y. Chong. 2005. Plasmid-mediated, inducible AmpC β-lactamase (DHA-1)-producing Enterobacteriaceae at a Korean hospital: wide dissemination in Klebsiella pneumoniae and Klebsiella oxytoca and emergence in Proteus mirabilis. Diagn. Microbiol. Infect. Dis. 53:65-70. [DOI] [PubMed] [Google Scholar]