Abstract
Cleavage of the hepatitis C virus (HCV) polyprotein by the viral NS3 protease releases functional viral proteins essential for viral replication. Recent studies by Foy and coworkers strongly suggest that NS3-mediated cleavage of host factors may abrogate cellular response to alpha interferon (IFN-α) (E. Foy, K. Li, R. Sumpter, Jr., Y.-M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr., Proc. Natl. Acad. Sci. USA 102:2986-2991, 2005, and E. Foy, K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr., Science 300:1145-1148, 2003). Blockage of NS3 protease activity therefore is expected to inhibit HCV replication by both direct suppression of viral protein production as well as by restoring host responsiveness to IFN. Using structure-assisted design, a ketoamide inhibitor, SCH 503034, was generated which demonstrated potent (overall inhibition constant, 14 nM) time-dependent inhibition of the NS3 protease in cell-free enzyme assays as well as robust in vitro activity in the HCV replicon system, as monitored by immunofluorescence and real-time PCR analysis. Continuous exposure of replicon-bearing cell lines to six times the 90% effective concentration of SCH 503034 for 15 days resulted in a greater than 4-log reduction in replicon RNA. The combination of SCH 503034 with IFN was more effective in suppressing replicon synthesis than either compound alone, supporting the suggestion of Foy and coworkers that combinations of IFN with protease inhibitors would lead to enhanced therapeutic efficacy.
It is estimated that approximately 170 million people worldwide are infected with hepatitis C virus (HCV), the etiologic agent of non-A non-B hepatitis, identified by Choo and coworkers in the late 1980s (7, 41). Hepatitis C virus, in roughly 80% of the cases, leads to a chronic form of hepatitis which, without therapeutic intervention, can lead to morbidity in 10 to 20 years either through cirrhosis and hepatic failure or hepatocellular carcinoma (1). The current standard of care for chronic HCV infection is pegylated alpha interferon (IFN-α) alone or in combination with oral ribavirin (28). Although this combination therapy is reasonably successful (∼70 to 80% sustained virological response [SVR]) with the majority of genotypes (1, 2, 4, 5), its efficacy against genotype 1, the major genotype affecting North America, Europe, and Japan, is moderate at best, with only about 40% of treated patients showing SVR (16, 48). Lack of a complete response, relapse following therapy, and premature termination of therapy due to intolerability of side effects contribute to this poor eradication rate observed among genotype 1-infected individuals and underscore the need for new anti-HCV therapeutics.
Hepatitis C virus (HCV) is a member of the Flaviviridae, a family that includes other human pathogens such as Yellow Fever, Dengue, and West Nile viruses. Although it comprises its own genus (Hepacivirus), HCV, like the other family members, is an enveloped, positive-strand RNA virus of approximately 9.6 Kb. Upon entering a suitable host cell, HCV genomes serve as templates for cap-independent translation through their 5′ internal ribosome entry sites (IRESs). The resulting 3,000-amino-acid polyprotein undergoes both co- and posttranslational proteolytic maturation by host- and virally encoded proteases (for a review, see reference 35). The virus-encoded protease responsible for processing the nonstructural (NS) portion of the polyprotein is located in the N-terminal third of the NS3 protein (9, 10, 14, 20, 39). NS3 protease forms a heterodimeric complex with the NS4A protein, an essential cofactor that enhances enzymatic activity, and is believed to help anchor it to the endoplasmic reticulum (17, 21, 42).
Following autoproteolysis of the NS3-NS4A junction, the protease severs the NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions to release the downstream NS proteins that subsequently self assemble on the endoplasmic reticulum to generate the replicative complex or replisome (35). The replisome, using the viral genome as a template, generates negative-strand viral RNA intermediates, which in turn are used to synthesize new positive-strand (genomic) RNAs, which are either translated to yield more polyprotein or, later in the infection cycle, encapsidated to generate progeny virions.
The NS3 protease structure (8, 17, 25, 43) confirmed early homology modeling efforts (4, 13) and provided the necessary detailed insight to permit rational inhibitor design. The NS3 protease is a typical β-barrel serine protease, containing the signature canonic asp-his-ser catalytic triad (18, 25). Unique to the NS3 protease is an extended polydentate substrate binding cleft, which ensures substrate specificity (5). The major protease-substrate interactions, which span the protease domain, include hydrogen bonds with the substrate backbone and complementary electrostatic or hydrophobic contacts along the binding site. Drug design efforts targeting the active site resulted in the discovery of SCH 503034, a structurally novel ketoamide HCV NS3 protease inhibitor.
The lack of either a robust in vitro cell culture system or a small-animal model capable of supporting HCV replication and propagation has precluded traditional approaches to evaluating anti-HCV compounds, though recent development of an infection system by the laboratories of Lindenbach, Wakita, and Zhong (23, 40, 46) as well as the development of a human-mouse chimeric liver animal model (29) are encouraging. Prior to these recent developments, the HCV subgenomic replicon system developed by Lohmann and coworkers (24) has been critical in enabling preclinical evaluation of potential anti-HCV agents over the last few years. In spite of its inherent limitations, preclinical results utilizing the replicon system have been borne out in recent proof-of-concept clinical trials with the HCV protease inhibitors BILN-2061 and VX-950 (18, 36).
Recent studies have suggested that, in addition to its role in processing the NS region of the HCV polyprotein, NS3 acts directly against host cells, abrogating the IFN response (11, 12). In these studies it was demonstrated that phosphorylation and nuclear translocation of interferon response factor 3 (IRF3) was disrupted by the presence of either the subgenomic replicon or a full-length HCV genome. Further analysis confirmed that expression of a functional NS3/4A protein was sufficient to block IRF3 phosphorylation by disrupting intracellular RIG-1 signaling. NS3 protease inhibitors (SCH 6) could restore normal signaling.
In this report, studies with the HCV NS3 protease inhibitor SCH 503034 are presented that demonstrate the efficacy of this compound in cell-free enzyme and replicon assays and which confirm (22) the potential therapeutic advantage of combining NS3 protease inhibitors with IFN therapy using the replicon system.
MATERIALS AND METHODS
Synthesis of SCH 503034.
The details of the synthesis of SCH 503034 have been described previously (F. G. Njoroge et al., Abstr. 230th Am. Chem. Soc. Natl. Meet., abstr 252, 2005). The ketoamide moiety in SCH 503034 (Fig. 1) which is essential for the protease inhibitory activity leads to rapid racemization of the α carbon of the P1 residue under physiological conditions, and consequently all studies were performed on the racemic mixture. SCH 503034 used in these studies was purified by reverse-phase high-performance liquid chromatography, and the structure was confirmed by 1H and 13C nuclear magnetic resonance and high-resolution mass spectrometry (F. G. Njoroge et al., Abstr. 230th Am. Chem. Soc. Natl. Meet., abstr. 252, 2005).
FIG. 1.
SCH 503034 structure and chemical name.
NS3 enzyme assay.
The production and characterization of the single-chain NS3 protease domain, NS4A21-32-GSGS-NS33-181 (plasmid p24), has been previously described (38). Single-chain NS3 protease inhibition was evaluated using the chromogenic assay previously described (45). The assays were performed at 30°C in a 96-well microtiter plate. One-hundred microliters protease (3 to 5 nM nominally) was added to 100 μl of assay buffer (25 mM morpholinepropanesulfonic acid, pH 6.5, 20% glycerol, 0.3 M NaCl, 0.05% lauryl maltoside, 5 μM EDTA, 5 μM dithiothreitol) containing chromogenic substrate acetyl-DTEDVVP(norvaline)-O-phenylazophenol. The reactions were monitored at an interval of 30 s for 1 h for change in absorbance at 370 nm using a Spectromax Plus microtiter plate reader (Molecular Devices, Sunnyvale, CA). Progress curves were fitted to the two-step slow-binding inhibition model of Morrison and Walsh: P = vst + (v0 − vs)(1 − e−kt)/k (30) using SAS version 8.0 (Cary, NC). The overall inhibition constant, Ki*, was calculated from the estimated steady-state velocities {vs = VmaxS/[Km(1+I/Ki*)]}. Curve-fitting errors were propagated, and multiple independent estimates were used to generate a weighted average.
Generation of HCV replicon cells.
The establishment of HCV replicon cell lines has been previously described (6, 24). The replicon sequence used in the study was the same as that in reference 24 with the addition of the adaptive mutation S1179I identified by Blight et al. (6). Clone 16, bearing the bicistronic replicon analogous to the I377neo/NS3-3′/wt replicon, was used for evaluation of potency. Monocistronic and full-length replicons were obtained from Pietschmann et al. (34). All constructs were confirmed by sequencing. HCV replicon-bearing cell lines were selected and maintained in 0.5 mg/ml G418 (Geneticin; Invitrogen, Carlsbad, CA) for the bicistronic replicon and 25 μg/ml hygromycin for the monocistronic replicon in Dulbecco's modified minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS).
Immunofluorescence.
Rabbit polyclonal antiserum against full-length NS3 (residues 1 to 631) was from Sigma-Genosys (Woodland, TX). Clone 16 replicon cells were seeded at 30 to 50% confluence on a coverslip in a 6-well plate containing DMEM supplemented with G418 and FBS. Following overnight incubation, cells were treated with 400 nM SCH 503034 for 72 h. Medium and drug were refreshed every 24 h. At the end of the third day, cells were washed twice with phosphate-buffered saline (PBS) and fixed first with 4% paraformaldehyde for 15 min and then with 0.5% Triton X-100 (Sigma, St. Louis, MO) for 15 min. Cells were blocked with 1% bovine serum albumin in PBS for 30 min and incubated with a 1:500 dilution of the rabbit anti-NS3 serum in PBS with 0.5% normal goat serum for 2 h at room temperature. After three washes with PBS, the coverslips were incubated in a 1:200 dilution of goat-anti-rabbit immunoglobulin G-rhodamine antibody (IM0834; Beckman Coulter) for 1 h at room temperature. The coverslips were washed again with PBS, sealed in mounting medium (Vector Laboratories Inc., Burlingame, CA), and examined by fluorescent microscopy. Images were captured with a CCD camera.
In vitro potency.
To test the in vitro potency of SCH 503034, replicon cells were seeded at 4,000 cells/well in 96-well collagen I-coated plates (Biocoat; Becton Dickinson) containing DMEM supplemented with G418 and FBS. Twenty-four hours postseeding, SCH 503034 was added to the media in concentrations indicated in the figure legends. The final concentrations of dimethyl sulfoxide (DMSO), FBS, and G418 were 0.5%, 5%, and 500 μg/ml, respectively. Media and inhibitors were refreshed daily for both the standard 3-day and the extended studies. G418 was omitted from cultures that were dosed for extended periods (>3 days).
At harvest, cells were washed with PBS and lysed in 30 μl of 1× commercial cell lysis buffer (Ambion catalog no. 8721). One microliter of cell lysate was used directly as template for reverse transcription-PCR and real-time PCR (TaqMan) analysis. Replicon RNA level was measured using real-time PCR (TaqMan). The amplicon was located in NS5B. The PCR primers were the following: 5B.2F, ATGGACAGGCGCCCTGA; 5B.2R, TTGATGGGCAGCTTGGTTTC. The probe sequence was 6-carboxyfluorescein-labeled CACGCCATGCGCTGCGG. GAPDH RNA was used as an endogenous control and was amplified in the same reaction as NS5B (multiplex PCR) using primers and VIC-labeled probe recommended by the manufacturer (PE Applied Biosystems, Foster City, CA). The real-time reverse transcription-PCRs were run on the ABI PRISM 7900HT Sequence Detection System using the following program: 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The changes in cyclic threshold (ΔCT) values (CT5B-CTGAPDH) were plotted against drug concentration and fitted to the sigmoid dose response model using SAS version 8.0 (SAS Institute) or PRISM software (Graphpad Software Inc.). The 50% effective concentration (EC50) was the drug concentration necessary to achieve an increase of 1 in ΔCT over the estimated baseline. EC90 was the drug concentration necessary to achieve an increase of 3.3 over the baseline. All TaqMan reagents were from PE Applied Biosystems.
For extended incubation studies, replicon RNA was extracted from cell lysates using an RNeasy kit (QIAGEN), and the concentration was determined using RiboGreen (Molecular Probes, Carlsbad, CA). HCV-specific sequences were amplified utilizing real-time PCR, and copy number was estimated by direct comparison with a replicon cRNA standard. The normalized total RNA replicon copy number was used for analysis.
Cloning.
The cDNA fragment of the neomycin phosphotransferase gene was from the pCI-neo plasmid (Promega, Madison, WI), and the pHelix 1(+) cloning vector was from Boehringer Mannheim/Roche (Indianapolis, IN). Restriction enzymes were obtained from New England Biolabs (Beverly, MA), and the Neomycin Resistance Gene Rapid Ligation kit was from Roche.
To assemble a vector for producing strand-specific gene RNA probes, the pCI-neo and pHelix 1(+) plasmids were both restricted using PstI and SphI. From the digestion of the pCI-neo plasmid, a 352-bp fragment was purified and ligated into the linearized pHelix vector. Insert-positive vectors were characterized and designated pHelix-Neo.
Probe transcription.
To produce a neomycin resistance gene probe that detects positive-strand HCV replicon RNA, pHelix-Neo was linearized using NciI and transcribed using the T3 MAXIscript kit (Ambion, Austin, TX). For probe that detects negative-strand replicon RNA, pHelix-Neo was linearized using SphI and transcribed using the T7 MAXIscript kit. For probe RNA of either polarity, transcription reactions were supplemented with biotin-14-CTP (Life Technologies, Rockville, MD) to label the RNA for nonradioactive detection (60% biotin-CTP-40% CTP). An internal control probe detecting 18S rRNA was generated and biotin labeled in the same manner using the pTRI-RNA 18S-human antisense cDNA template (Ambion).
RNase protection assays (RPA).
Cell pellets recovered from lysates tested for luciferase activity were processed using the Direct Protect Lysate kit (Ambion) per the manufacturer's instructions. Briefly, cell pellets were resuspended into 100 μl of lysis buffer (containing guanidine thiocyanate). A 30-μl aliquot of cell lysate from each sample was mixed with 200 pg of strand-specific Neo probe and 500 pg of 18S rRNA gene probe and incubated overnight at 37°C. Treatment of the samples with RNases A and T1, followed by proteinase K, was performed per the manufacturer's instructions. Protected RNA fragments recovered from isopropanol precipitation were resuspended in 10 μl of denaturing gel loading buffer, and 5 μl was loaded, alongside undigested probes and an RNA Century size ladder (Ambion), on Tris-borate-EDTA-5% polyacrylamide gels containing 6 M urea (Bio-Rad). Gel-separated fragments were then transferred by semidry electroblotting to positively charged nylon membranes (BrightStar-Plus nylon membranes; Ambion) and processed for detection of the biotinylated RNA probe using the BrightStar BioDetect kit (Ambion) per the manufacturer's instructions. Chemiluminescent signals on the membranes were visualized using X-ray film and quantitated by densitometry using ImageQuant software (Molecular Dynamics). Measurements of the 18S rRNA signal were used to normalize cell material loading for an accurate assessment of changes in replicon RNA levels.
Western blot analysis.
Whole-cell lysates were separated on 4 to 12% NuPAGE gels (Novex; Invitrogen), transferred to polyvinylidene difluoride membranes, and probed with a monoclonal antibody against NS5A (1126) (1:500) that was kindly provided by Johnson Lau. Bands were visualized using the ECF Western blot detection system (Amersham Pharmacia, Piscataway, NJ).
Cellular toxicity.
A 10 mM stock of SCH 503034 in DMSO was used to prepare serial twofold dilutions in DMSO. A volume of 2.5 μl of these 200× solutions was transferred into 500 μl of diluent (complete DMEM plus 5 to 10% heat-inactivated fetal bovine serum) for final concentrations ranging from 50 μM to 0.1 μM. Eight-thousand replicon cells/well were seeded onto 96-well plates and incubated in the presence of compound for 72 h. All reagents and media were refreshed daily. The MTS assay was performed at 72 h according to the manufacturer's protocol (catalog no. G3580; Promega).
Cellular uptake of SCH 503034.
Primary human hepatocytes (T3, Inc.) were incubated with 10 μM [14C]SCH 503034 (synthesized in house) in DMEM supplemented with 10% FBS. At each time point (2, 40, and 240 min), cells were harvested, washed, and pelleted. Radioactivity in the supernatant and the cell pellets was counted on a Packard Tri-carb 2700 TR liquid scintillation counter (Perkin Elmer, Dewers Grove, IL). Counts were normalized based upon packed cell volume and used to calculate the cell/medium ratios.
Effect of SCH 503034 on interferon potency.
Escalating doses of SCH 503034 (10 nM to 5 mM) were added to a standard titration study of IFN-α-2b (0.1 to 40 IU/ml; Schering-Plough Corp., Kenilworth, NJ) to generate a 10 by 10 matrix of concentrations ranging from below EC50 to well above EC90 for both drugs. At 72 h, replicon RNA levels were estimated using the standard single-tube multiplex assay described above. Statistical analysis was performed using a nonparametric spline fit (32).
RESULTS
Inhibition of NS3 protease.
SCH 503034 (Fig. 1) showed time-dependent inhibition of the single-chain NS3 protease characteristic of ketoamide inhibitors of serine proteases (Fig. 2) (19, 33, 37, 44). As crystallographic analysis had indicated the formation of a covalent adduct with the active-site serine (Fig. 3A), the progress curves were fitted to a two-step paradigm assuming a fast initial binding followed by a slow formation of the covalent adduct (30). The overall binding constant for the formation of adduct, Ki* (30), was estimated to be 14 ± 1 nM (n = 12). Other derived kinetic parameters (±standard errors) include a Ki (initial inhibition constant) of 7.8 ± 2.5 μM, a half life (t1/2) for reactivation of the inhibited enzyme of 23 ± 4 h, k5 (forward rate constant for isomerization of the EI to EI* “tight” complex) of 0.43 ± 0.13 min−1, and k6 (backward rate constant for the isomerization of the EI* to EI complex) of 0.00084 ± 0.0001 min−1. Activity against human leukocyte elastase (HLE), a serine protease which also shows a preference for small hydrophobic residues in the P1 residue (i.e., the residue immediately adjacent to the scissile bond toward the amino terminus of the substrate), did not show time dependence and was very weak (Ki > 25 μM).
FIG. 2.
Progress curve of peptide hydrolysis by the single-chain HCV protease domain (genotype 1b) showing time-dependent inhibition by SCH 503034. SCH 503034 was incubated with the single-chain genotype 1b NS3 protease (see Materials and Methods). Reactions were initiated by addition of enzyme and absorbance (abs) was monitored for 120 min. Data were fitted to the two-step “slow-binding” inhibition model to obtain kinetic parameters.
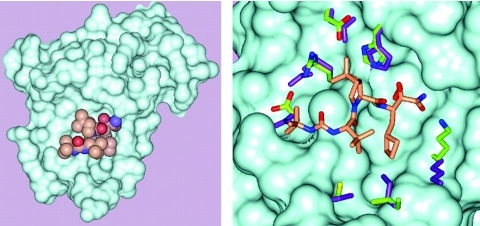
FIG. 3.
SCH 503034 complexed with the HCV NS3 protease. A. Connolly surface for the NS3 protease. SCH 503034 is rendered in CPK format (Corey-Pauling-Koltun space-filling model): gold, carbon; red, oxygen; and blue, nitrogen. B. A close up of the NS3:SCH 503034 complex showing side chains that are perturbed upon binding of SCH 503034. Color code for SCH 503034 is the same as that for panel A. Side chains of the complex are colored as follows: carbon, green; nitrogen, blue; and oxygen, red. Side chains of the apoenzyme structure are shown in purple.
Structural analysis.
In addition to the formation of an adduct between the active-site serine (S139) and the carbonyl carbon of the ketoamide, crystallographic analysis confirmed that SCH 503034 binds to the active site of the protease in a manner analogous to the C terminus of the NS3 helicase as previously described (39) (Fig. 3A). Compared with the uninhibited enzyme structure, the side chains of the complex appear to move only slightly to accommodate SCH 503034 (Fig. 3B).
Anti-replicon activity.
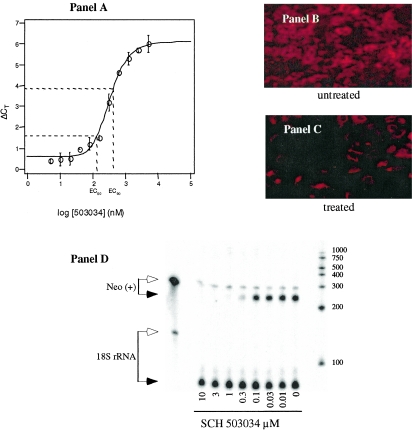
To estimate SCH 503034 antiviral potency, replicon-bearing cell monolayers were cultured for 72 h with compound as described above. Ten-point dose-response curves were generated, and drug concentrations yielding a twofold (1 CT above baseline) or a 10-fold (3.3 CT above baseline) reduction in replicon RNA were estimated using the grid search method NLIN (SAS, version 8.0). EC50 and EC90 for SCH 503034 were determined to be 200 nM (95% predictive interval, 70 to 500 nM; n = 23) and 400 nM (95% predictive interval, 200 to 700 nM; n = 23), respectively (Fig. 4A). In addition, three independent replicon clones (all bearing the same adaptive mutation, i.e., S1179I) as well as a monocistronic replicon (HCV IRES only) and a full-length replicon (expressing the structural proteins) yielded comparable results in dose-response studies (data not shown). Immunofluorescent staining of clone 16 replicon cells treated with SCH 503034 at the calculated EC90 (400 nM) showed barely detectable levels of NS3 protein relative to vehicle-treated control at the end of 72 h (Fig. 4B and C). RPA analysis of positive-strand replicon RNA showed comparable dose response inhibition of replicon RNA synthesis by SCH 503034 (Fig. 4D). Similar results were observed with negative-strand replicon RNA (data not shown).
FIG. 4.
Effect of SCH 503034 on HCV replicon RNA synthesis in Huh-7 cells. A. Real-time PCR (Taqman) analysis of the HCV replicon RNA level relative to the GAPDH internal control (ΔCT). Huh-7 cells were treated with increasing concentrations of SCH 503034 for 72 h in the presence of G418. Increases in ΔCT (y axis) indicate decreasing replicon RNA levels; each ΔCT reflects a twofold change in RNA level from baseline. A representative set of data is shown. Immunofluorescent detection of NS3 protein expression following exposure to SCH 503034. HCV replicon-bearing Huh-7 cells were treated with vehicle alone (0.5% DMSO) (B) or with SCH 503034 (1× EC90) (C) for 72 h and then probed with anti-NS3 polyclonal antibodies and visualized by staining with an immunofluorescently labeled secondary antibody. A comparable number of cells are present in each field. D. Detection of replicon RNA from drug-treated cells by RNase protection assay. Clone 16 cell cultures were treated with SCH 503034 for 3 days as described in Materials and Methods. Cell monolayers were recovered in cell lysis buffer and processed for RPAs detecting positive-strand replicon genomic RNA (via the Neo gene) and 18S rRNA (internal control). RPA products were separated on a TBE-5% polyacrylamide gel electrophoresis gel containing urea. Arrows show the positions of digested (filled) and undigested (open) riboprobe. Similar results were obtained in several experiments using other replicon cell lines (data not shown).
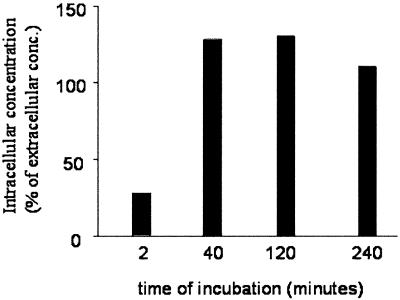
Kinetic studies which followed the change in replicon RNA level ascertained by TaqMan (data not shown) and RNase protection analyses (data not shown), as well as NS protein level as determined by Western blot analysis (Fig. 5), suggested that SCH 503034 acted rapidly, with effects becoming visible in as little as 1 h. These data, which support the rapid entry of compound into the cell with equilibrium concentrations attained in minutes, were confirmed using radiolabeled SCH 503034 (Fig. 6). [14C]SCH 503034 association with primary human hepatocytes was quantifiable within 2 min of initial exposure to compound and was maximal by 40 min.
FIG. 5.
Inhibition of polyprotein processing in HCV replicon cells. Replicon cells were treated with SCH 503034 (0 to 100 μM, 1:3 serial dilution). Cells were harvested at various time points as indicated, and cell lysates were analyzed by Western blot for NS5A as described in Materials and Methods. The position of the high-molecular-weight processing intermediate (possibly NS3-NS5B) is indicated by an arrow. Additional processing intermediates, detected at 6 h posttreatment, are indicated by a bracket.
FIG. 6.
Uptake of [14C]SCH 503034 by replicon-bearing cells. Primary human hepatocytes were incubated in the presence of 10 μM radiolabeled SCH 503034 for various time intervals as indicated. Cells were harvested, washed, and counted. The intracellular concentration of SCH 503034 was estimated and compared with the initial concentration in the media. Results are expressed as percentages of medium concentration (conc.).
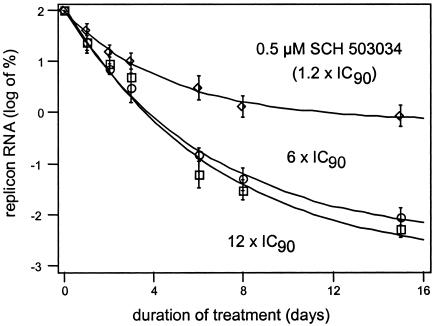
Loss of replicon RNA, reflecting the inhibition of newly synthesized protease and the concomitant reduction of new replisomes, appears to be roughly first order with respect to time of treatment, suggesting the compound is present in molar excess over protease (Fig. 7). Dosing with 6× EC90 (2.5 μM) or 12× EC90 (5 μM) SCH 503034 gave comparable results (Fig. 7). Treatment with either drug concentration for 72 h resulted in a 1.5- to 2-log drop in replicon RNA. Prolonged exposure, in the absence of neomycin selection, resulted in a 3.5- to 4-log decrease in RNA levels, or less than one copy per cell, by day 15. For these long-term studies, during which HCV replicon cells were cultured in the absence of G418 selection, copy number was normalized to day zero based upon studies by Pietschmann et al. (34) demonstrating that replicon copy number in the absence of G418 remains stable for several months if cells do not become confluent.
FIG. 7.
Effect of treatment duration and inhibitor concentration on replicon RNA level. Replicon-bearing Huh-7 cells (clone 16) were incubated for 14 days in media (without G418 selection) containing 0.5 μM (1.2 × 90% inhibitory concentration), 2.5 μM (6 × EC90) SCH 503034, or 5 μM (12 × EC90) SCH 503034. Compound and medium were refreshed daily. HCV replicon RNA level was determined by TaqMan as described in Materials and Methods and normalized to time zero. Experiments were performed in triplicate, and error bars represent the standard deviations.
As replicon level has been shown to be sensitive to cell division, the impact of SCH 503034 on cell growth and viability was measured. SCH 503034 showed no toxicity by MTS (26) on either replicon-bearing or parental Huh-7 cell lines at 50 μM after 96 h. Primary human hepatocytes likewise showed no toxic effects from exposure to ∼10× EC90 (4 μM) levels after 1 week compared with the no-drug control (n = 3, different donors) (data not shown). In a second study, the effect on doubling time of prolonged exposure to SCH 503034 was examined. Huh-7 cells were cultured in the presence of ∼10× EC90 (4 μM) for 3 weeks. No change in doubling time was observed over the course of the study, and growth rates were indistinguishable from those of control cells without compound (data not shown).
Effect of SCH 503034 on alpha interferon potency.
To assess the impact of SCH 503034 on alpha interferon potency, escalating doses of SCH 503034 were added to a standard dose-response study of IFN to generate a 10 by 10 matrix of concentrations varying from below EC50 to well above EC90 for both drugs. At 72 h, replicon RNA levels were estimated using the standard single-tube multiplex assay. Parametric evaluation methods proved unsuccessful, necessitating the use of a nonparametric spline fit (32) (Fig. 8A). Evaluation at the 90% suppression level indicated no statistically significant deviation of the isobole from the line of additivity (Fig. 8B).
FIG. 8.
Effect of SCH 503034 on alpha interferon (IFN) potency in vitro. Replicon cells were treated simultaneously with SCH 503034 and IFN as described in Materials and Methods. A. Nonparametric (spline-fit) analysis of efficacy as measured by the reduction in HCV replicon RNA. Axes are normalized to maximum dose. B. The isobole at the 90% suppression level. The line of additivity is shown. Although the isobole trends below the line of additivity, the deflection is within the error of the assay and therefore indicates simple additivity of alpha interferon and SCH 503034. IC90, 90% inhibitory concentration.
DISCUSSION
Since its first identification in 1993, the NS3 protease has been considered a potential target for therapeutic intervention in chronic hepatitis C infection (3, 14, 15, 27, 39). The subsequent elucidation of the NS3 atomic structure in 1996 revealed a classical serine protease with a relatively shallow, hydrophobic, and featureless active site (18, 25). Consequently, priority was given to identifying a mechanism-based inhibitor predicated on natural peptide substrates, a strategy well precedented in the literature to yield potent and selective molecules for serine proteases (47). After exploring many potential electrophilic moieties with only modest success, a potent ketoamide derivative of the NS5A-NS5B cleavage junction was identified which exhibited the slow-binding inhibition kinetics suggestive of true mechanism-based inhibition: nucleophilic attack by the active-site serine. This initial inhibitor, though unsuitable for therapeutic use, provided the starting point for an iterative design effort, which after several stepwise modifications yielded the current therapeutic candidate, SCH 503034.
SCH 503034, although roughly a third the size of the initial lead, retained greater than 90% of the potency in the cell-free enzyme assay. Detailed kinetic analysis of progress curves yielded kinetic parameters consistent with other reported ketoamide inhibitors of serine proteases (19, 33, 37, 44). Comparison of the t1/2 of reactivation (∼23 h) with the t1/2 of the NS3 replisome degradation (∼12 h) suggests that SCH 503034 may be effectively irreversible as cellular turnover of NS3 would be faster than dissociation of the inhibitor (34). Unlike the initial lead, SCH 503034 demonstrated efficacy in the HCV replicon system with virtually no cellular toxicity and has consequently been advanced for clinical evaluation.
As reported for other NS3 inhibitors, SCH 503034 acts rapidly to suppress proteolytic activity and prevent formation of new replisomes. The ongoing degradation of extant replisomes and replicon RNA leads to a first-order decline in replicon RNA with an apparent half-life of approximately 12 h. Proof-of-concept clinical studies with the protease inhibitors BILN 2016 and VX-950 have demonstrated that NS3 protease inhibition results in a biphasic drop in HCV viral load (31). The kinetics of NS3 protease inhibition observed with SCH 503034 in the replicon system (Fig. 7) is consistent with the initial rapid decline in HCV RNA observed in the clinic with BILN 2016 and VX-950 (i.e., initial viral load reductions occurring within 48 h). Initial biphasic pharmacodynamics have been observed with SCH 503034 (B. A. Malcolm, S. Zeuzem, M. Laughlin, S. Gupta, J. Zhang, and A. S. Perelson, Modeling of HCV dynamics during combination therapy with peginterferon alfa-2b and the NS3 protease inhibitor SCH 503034, Abstr. 56th Ann. Meet. Am. Assoc. Study Liver Dis., abstr. 1266, 2005).
Gale and coworkers have presented data which strongly suggest that NS3 protease activity suppresses the innate intracellular immune response, although the complete mechanism and specific cellular targets remain to be elucidated and identified (11, 12). Nevertheless, as IFN-α is the current standard of care for HCV, the potential of protease inhibition to enhance IFN therapy has attracted much attention. Although a reasonable expectation based on the theories of Gale and coworkers, to date no studies, neither those reported here nor those reported recently by Lin et al. (22), are sufficiently accurate to allow a statistically rigorous distinction between simple additivity and a true enhancement of IFN potency, i.e., synergy, by the coadministration of protease inhibitors. Whether this is purely a limitation of resolution (i.e., the accuracy of TaqMan-based arrays) or a reflection of the possibility that only a few active NS3 protease molecules are sufficient to abort the innate cellular response will require further investigation. In the interim, the pleiotropic effects of IFN-α on both the infected hepatocytes as well as the immune system suggest that combination therapy with protease inhibitors will likely remain preferable to simple monotherapy to effect a rapid sustained viral response and minimize the potential emergence of protease resistance.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Bain, V. G. 2001. Effect of HCV viral dynamics on treatment design: lessons learned from HIV. Am. J. Gastroenterol. 96:2818-2828. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazan, J. F., and R. J. Fletterick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, A., and I. Schechter. 1970. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 257:249-264. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-364. [DOI] [PubMed] [Google Scholar]
- 8.De Francesco, R. P., and C. Steinkuhler. 1998. The hepatitis C virus NS3 proteinase: structure and function of a zinc-containing serine proteinase. Antivir. Ther. 3:99-109. [PubMed] [Google Scholar]
- 9.D'Souza, E. D., E. O'Sullivan, E. M. Amphlett, D. J. Rowlands, D. V. Sangar, and B. E. Clarke. 1994. Analysis of NS3-mediated processing of the hepatitis C virus non-structural region in vitro. J. Gen. Virol. 75:3469-3476. [DOI] [PubMed] [Google Scholar]
- 10.Failla, C. M., E. Pizzi, R. De Francesco, and A. Tramontano. 1996. Redesigning the substrate specificity of the hepatitis C virus NS3 protease. Struct. Folding Design 1:35-42. [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, R. Sumpter, Jr., Y.-M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, K. Q., J. M. Vierling, and A. G. Redeker. 2001. Viral, host and interferon-related factors modulating the effect of interferon therapy for hepatitis C virus infection. J. Viral Hepat. 8:1-18. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. (Erratum, 89:159.) [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A.-M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M.-A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C.-L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, S. D., B. J. Lucas, S. F. Brady, J. T. Sisko, K. J. Cutrona, P. E. J. Sanderson, R. M. Freidinger, S.-S. Mao, S. J. Gardell, and J. A. Shafer. 1998. Characterization of the two-step pathway for inhibition of thrombin by alpha-ketoamide transition state analogs. J. Biol. Chem. 273:4843-4854. [DOI] [PubMed] [Google Scholar]
- 20.Lin, C., B. M. Pragai, A. Grakoui, J. Xu, and C. M. Rice. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, K., A. D. Kwong, and C. Lin. 2004. Combination of a hepatitis C virus NS3-NS4A protease inhibitor and alpha interferon synergistically inhibits viral RNA replication and facilitates viral RNA clearance in replicon cells. Antimicrob. Agents Chemother. 48:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 25.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 26.Malich, G., B. Markovic, and C. Winder. 1997. The sensitivity and specificity of the MTS tetrazolium assay for detecting the in vitro cytotoxicity of 20 chemicals using human cell lines. Toxicology 124:179-192. [DOI] [PubMed] [Google Scholar]
- 27.Manabe, S., I. Fuke, O. Tanishita, C. Kaji, Y. Gomi, S. Yoshida, C. Mori, A. Takamizawa, I. Yosida, and H. Okayama. 1994. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology 198:636-644. [DOI] [PubMed] [Google Scholar]
- 28.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 29.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927-933. [DOI] [PubMed] [Google Scholar]
- 30.Morrison, J. F., and C. T. Walsh. 1988. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 61:201-301. [DOI] [PubMed] [Google Scholar]
- 31.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 32.O'Connell, M. A., and R. D. Wolfinger. 1997. Spatial regression models, response surfaces, and process optimization. J. Comput. Graph. Stat. 6:224-241. [Google Scholar]
- 33.Perni, R. B., J. Pitlik, S. D. Britt, J. J. Court, L. F. Courtney, D. D. Deininger, L. J. Farmer, C. A. Gates, S. L. Harbeson, and R. B. Levin. 2004. Inhibitors of hepatitis C virus NS3 · 4A protease 2. Warhead SAR and optimization. Bioorg. Med. Chem. Lett. 14:1441-1446. [DOI] [PubMed] [Google Scholar]
- 34.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Topics Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 36.Resnick, H. W., S. Zeuzem, A. van Vliet, L. McNair, S. Purdy, H-M. Chu, and P. L. Jansen. 2005. Initial results of a phase 1b, multiple-dose study of VX-950, a hepatitis C virus protease inhibitor, abstr. 527. Digestive Disease Week, 14 to 19 May 2005, Chicago, Ill.
- 37.St. Charles, R., J. H. Matthews, E. Zhang, and A. Tulinsky. 1999. Bound structures of novel P3-P1′ beta-strand mimetic inhibitors of thrombin. J. Med. Chem. 42:1376-1383. [DOI] [PubMed] [Google Scholar]
- 38.Taremi, S., B. M. Beyer, M. Maher, N. Yao, W. Prosise, P. C. Weber, and B. A. Malcolm. 1998. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 7:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomei, L., C. Failla, E. Santolini, R. De Francesco, and N. La Monica. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 42.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Struct. Folding Design 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 44.Yip, Y., F. Victor, J. Lamar, R. Johnson, Q. M. Wang, D. Barket, J. Glass, L. Jin, L. Liu, and D. Venable. 2004. Discovery of a novel bicycloproline P2 bearing peptidyl α-ketoamide LY514962 as HCV protease inhibitor. Bioorg. Med. Chem. Lett. 14:251-256. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, R., B. M. Beyer, J. Durkin, R. Ingram, F. G. Njoroge, W. T. Windsor, and B. A. Malcolm. 1999. A continuous spectrophotometric assay for the hepatitis C virus serine protease. Anal. Biochem. 270:268-275. [DOI] [PubMed] [Google Scholar]
- 46.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, J., and W. C. Groutas. 2004. Recent developments in the design of mechanism-based and alternate substrate inhibitors of serine proteases. Curr. Top. Med. Chem. 4:1203-1216. [DOI] [PubMed] [Google Scholar]
- 48.Zylberberg, H., M. L. Chaix, and C. Brechot. 2000. Infection with hepatitis C virus genotype 4 is associated with a poor response to interferon-alpha. Ann. Intern. Med. 132:845-846. [DOI] [PubMed] [Google Scholar]