Abstract
In Rhodobacter sphaeroides, MreB, MreC, MreD, PBP2, and RodA are encoded at the same locus. The localizations of PBP2, MreB, and MreC, which have all been implicated in the synthesis of the peptidoglycan layer, were investigated under different growth conditions to gain insight into the relationships between these proteins. Immunofluorescence microscopy showed that PBP2 localized to specific sites at the midcell of elongating cells under both aerobic and photoheterotrophic conditions. Visualizing PBP2 at different stages of the cell cycle showed that in elongating cells, PBP2 was found predominately at the midcell, with asymmetric foci and bands across the cell. PBP2 remained at midcell until the start of septation, after which it moved to midcell of the daughter cells. Deconvolution and three-dimensional reconstructions suggested that PBP2 forms a partial ring at the midcell of newly divided cells and elongated cells, while in septating cells, partial PBP2 rings were present at one-quarter and three-quarter positions. Due to the diffraction limits of light microscopy, these partial rings could represent unresolved helices. Colocalization studies showed that MreC always colocalized with PBP2, while MreB colocalized with PBP2 only during elongation; during septation, MreB remained at the septation site, whereas PBP2 relocalized to the one-quarter and three-quarter positions. These results suggest that PBP2 and MreC are involved in peptidoglycan synthesis during elongation and that this occurs at specific sites close to midcell in R. sphaeroides.
The peptidoglycan layer governs the cell shapes of most bacterial species. It is thought that two distinct phases of peptidoglycan synthesis take place in gram-negative bacterial species (27). Two penicillin binding proteins (PBPs), PBP2 and PBP3, appear to perform independent roles in peptidoglycan synthesis and to specify the phase of peptidoglycan synthesis. In Escherichia coli, PBP2 is thought to function during the elongation phase of the cell cycle in the expansion of the longitudinal axis of the cell (25, 27). This phase of peptidoglycan synthesis, known as the elongation-specific phase, deposits new material in a diffuse manner along the long axis of the cell (5). Once the cell length has doubled, the cell enters the septation phase of the cell cycle, and a switch is proposed to take place from PBP2- to PBP3 (more commonly known as FtsI)-mediated peptidoglycan synthesis (27). This septation-specific phase synthesizes peptidoglycan specifically at midcell or the site of septation, thereby generating the daughter cells (6).
In E. coli, the inhibition of PBP2 with amdinocillin results in a rounding of cell shape and eventually cell lysis, while temperature-sensitive pbp2 mutations produce round cell morphologies when shifted to the restrictive temperature (5). PBP2 localizes as discrete foci along the longitudinal axis of E. coli (4) and also forms band-like structures perpendicular to the lateral wall in the gram-negative Caulobacter crescentus (8). PBP2 function is thought to depend upon the integral membrane protein RodA; as the genes encoding these proteins form an operon (14), mutations in either gene produce similar phenotypic effects (5) and localization of the transpeptidase of similar transpeptidase-integral membrane protein pairs depends upon the integral membrane protein partner (15). It is thought that PBP2, in concert with RodA, contributes to the synthesis of the peptidoglycan layer, specifically functioning in the extension of the lateral wall (5).
MreB is an actin-like cytoplasmic protein thought to be involved in peptidoglycan synthesis. This protein is essential in E. coli (12), C. crescentus (8), and Rhodobacter sphaeroides (23), and impairment of MreB function is associated with morphological abnormalities in all these bacterial species (8, 12, 23). In E. coli, MreB forms a helix spanning the length of the cell, and two-dimensional (2D) localization images also suggest that the protein forms a ring (22). In C. crescentus, in newly formed and elongating cells, MreB forms a helix along the length of the cell, while in early predivisional cells, MreB forms a ring at the site of septation and remains at this location until the completion of septation (9). In R. sphaeroides, MreB localizes to the sites of peptidoglycan synthesis, and 3D reconstructions reveal an MreB ring at midcell, rather than a helix in this bacterium, at all stages of cell growth (23). In C. crescentus, the localization pattern of PBP2 depends upon the presence of MreB, suggesting that the specific role of MreB may involve the organization of PBP2-containing peptidoglycan-synthetic complexes (8).
The mre locus consists of mreB, mreC, and mreD in many bacterial species (1, 8, 28). In E. coli, depletion of MreC results in cell shape defects and ultimately cell death (12). In E. coli, MreC localizes to the cytoplasmic membrane and interacts with the cytoplasmic MreB and the integral membrane protein MreD (12), and in C. crescentus, MreC interacts with PBP2 (7). A complex consisting of MreB, MreC, MreD, PBP2, and RodA is proposed to function in the synthesis of the peptidoglycan layer in E. coli, specifically contributing to the expansion of the longitudinal axis (12).
R. sphaeroides is an α-subgroup purple nonsulfur photosynthetic coccobacillus capable of growth under a range of conditions, including aerobic and photoheterotrophic growth conditions (2). The cytoplasmic membrane of aerobic R. sphaeroides cells is uninvaginated, while under photoheterotrophic conditions, the inner membrane invaginates to accommodate the photosynthetic apparatus. Early studies showed that PBPs of R. sphaeroides localized to the uninvaginated regions of the membrane, suggesting that proteins are targeted to different regions of the continuous membrane (21). Previously, a putative mreB-mreC-mreD-pbp2-rodA operon was reported in R. sphaeroides (23). This genetic arrangement suggested a functional interaction between the encoded proteins. This study investigates whether PBP2 and the mre-encoded proteins in R. sphaeroides physically colocalize to regions of the cell during any or all phases of cell growth and therefore whether a functional relationship might exist between any or all of these proteins.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids are listed in Table 1. R. sphaeroides WS8N was cultured in succinate medium at 30°C either aerobically in the dark with shaking or anaerobically with 35 μM m−2 s−1 illumination. The amount of aeration received by the aerobic cultures was sufficient to completely prevent the formation of photosynthetic pigments. E. coli strain DH5α was used for all molecular cloning, strain S17-1 λpir was used for conjugal transfer into R. sphaeroides, and strain M15 pREP4 was used for protein expression. E. coli strains were cultured in Luria-Bertani medium at 37°C with shaking. Kanamycin and nalidixic acid were used at 25 μg ml−1, and ampicillin was at 100 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Characteristics | Source/reference |
|---|---|---|
| E. coli | ||
| DH5α | General cloning strain; allows blue-white screening during cloning | Gibco-BRL |
| S17-1λpir | Strain capable of mobilizing the suicide vector pK18mobsacB into R. sphaeroides; Smr | 17 |
| M15 | Protein expression host | Qiagen |
| R. sphaeroides | ||
| WS8N | Spontaneous nalidixic acid-resistant mutant of wild-type WS8 | 24 |
| Plasmids | ||
| pK18mobsacB | Allelio-exchange suicide vector mobilized by E. coli S17-1λpir; Kmr | 19 |
| pQE-80L | Ptac-based expression vector; introduces RGS(H)6 sequence at the N termini of expressed proteins; Apr | Qiagen |
| pREP4 | Plasmid carrying the lacIq gene; compatible with pQE80; reduces “leaky” expression from the tac promoter of pQE80; Kmr | Qiagen |
| pPAMK1 | Derivative of pK18mobsacB containing the region upstream of pbp2 | This study |
| pPAMK2 | Derivative of pK18mobsacB containing the regions upstream and downstream of pbp2 | This study |
| pBET1 | Derivative of pQE80 containing pbp2 | This study |
| pBINX1 | Derivative of pQE80 containing mreC | This study |
Molecular genetic techniques.
The cloning steps were performed as described by Sambrook and Russell (18). Sequencing-quality DNA was prepared using the WizardPlus kit (Promega), sequenced by the University of Oxford Biochemistry sequencing service, and analyzed with the GCG software package (University of Wisconsin). All primers were supplied by Genosys Biotechnologies Inc.
Deletion construct.
A derivative of pK18mobsacB was constructed for the generation of a pbp2 in-frame deletion strain as described below. The construct was sequenced to ensure that upstream and downstream regions were in frame and contained no errors. The construct was introduced into R. sphaeroides by allelic exchange as described previously (10, 19).
pbp2 in-frame deletion.
A 0.5-kb region immediately upstream of pbp2 was amplified by PCR using primers that encompassed the start codon and included 5′ EcoRI and 3′ BamHI sites. A 0.5-kb region that included the five 3′ codons and the downstream flanking DNA of pbp2 was amplified by PCR using primers that included 5′ BamHI and 3′ HindIII sites. The first PCR product was ligated into appropriately cut pK18mobsacB to produce pPAMK1. The second PCR product was ligated into appropriately cut pPAMK1 to generate the final construct, pPAMK2.
Inhibition studies.
Early-log-phase R. sphaeroides cells were cultured aerobically in the presence of amdinocillin at 25 μg ml−1 for 1 h at 30°C. Subsequently, cells were embedded in 1.2% agarose on microscope slides. DIC (differential interference contrast) images were acquired using a Nikon TE200 microscope and recorded with a cooled charge-coupled device camera (Hamamatsu). The images were processed with SimplePCI image analysis software (Digital Pixel).
Protein expression constructs.
pbp2 and mreC were amplified by PCR using primers that included 5′ BamHI and 3′ HindIII sites. The primers were designed to amplify the sequence encoding the soluble periplasmic regions of PBP2 and MreC. The PCR products were ligated into appropriately cut pQE-80L (QIAGEN) to produce pBET1 and pBINX1, respectively. The vector attaches an N-terminal tag containing six histidine residues to the expressed protein, thereby facilitating purification. The constructs were sequenced to ensure that the coding sequence contained no errors.
Protein purification and antibody production.
His-tagged, truncated PBP2 and MreC were expressed individually in E. coli M15 pREP4 cells containing pBET1 and pBINX1, respectively, and purified as described previously (13). Antibodies were raised against both of the truncated proteins in rabbits, and also against PBP2 in a guinea pig (Eurogentec).
Immunofluorescence staining.
Cells were fixed and permeabilized as described previously (11, 23). For individual protein localization studies, immunofluorescence staining was performed as described previously (23), except that the blocking solution contained a 1:7,500 dilution of either PBP2 or MreC rabbit polyclonal antibodies. For colocalization studies, immunofluorescence staining was performed as described previously (23) with the following modifications. Cells were incubated in a blocking solution containing a 1:7,500 dilution of guinea pig polyclonal anti-PBP2 antibodies and either a 1:10,000 dilution of rabbit polyclonal anti-MreB antibodies or a 1:7,500 dilution of rabbit polyclonal anti-MreC antibodies. After being washed, the cells were incubated with blocking solution containing Alexa Fluor 568-conjugated goat anti-guinea pig at 40 μg/ml (Molecular Probes) and a 3:1,000 dilution in blocking solution of fluorescein isothiocyanate-conjugated goat anti-rabbit antibodies (Sigma).
Fluorescence analysis.
DIC and fluorescence images were acquired using a Nikon TE200 microscope with either a fluorescein isothiocyanate filter set for the individual protein localization studies or a yellow fluorescent protein-DsRed filter set for the colocalization studies and were recorded with a cooled charge-coupled device camera (Hamamatsu). Z series were captured with a custom built piezo-driven (Physik Instruments) mechanical stage. The images were processed with SimplePCI image analysis software (Digital Pixel). Although all experiments were performed with both aerobically and photoheterotrophically grown cells, the DIC images were clearer and the fluorescence intensity was brightest in cells grown under photoheterotrophic conditions; therefore, unless otherwise stated, all images presented in this study were acquired from photoheterotrophically grown cells.
RESULTS
Localization of PBP2 in R. sphaeroides.
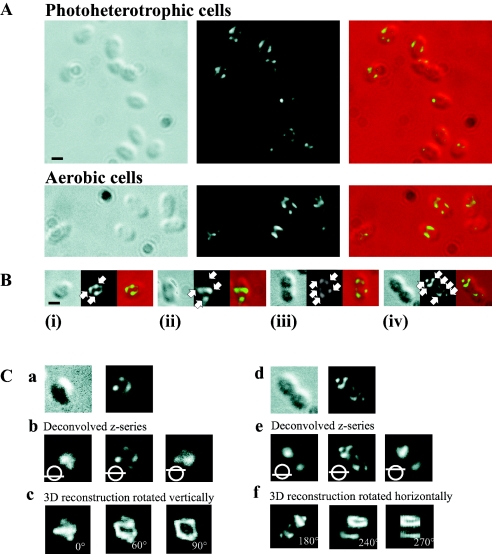
The localization of PBP2 was determined by immunofluorescence microscopy in populations of R. sphaeroides cells grown under either aerobic or photoheterotrophic growth conditions. Similar patterns of localization were observed under both growth conditions (Fig. 1A). A total of 150 cells were analyzed individually and then grouped according to their positions in the cycle as (i) newly formed, (ii) elongating, (iii) septation initiating, or (iv) septation nearing completion, depending upon their cell length and evidence of cellular invagination, as described previously (24). PBP2 localized as distinct punctate foci (Fig. 1B), and the number of foci present in cells increased as the cell cycle progressed (Fig. 2A). Many of these foci were arranged on opposing sides of the longitudinal axis, forming bands across the cell, while the remaining foci appeared disconnected from the surrounding foci. In newly formed and elongating cells, the bands were commonly observed either at or near midcell. However, in septating cells, the PBP2 bands were not present at midcell; instead, they were found at or near the one-quarter and three-quarter positions, i.e., at midcell of the forming daughter cells.
FIG. 1.
Localization of PBP2 in R. sphaeroides cells by immunofluorescence microscopy. (A) Localization in 2D. DIC images, fluorescence images, and an overlay of these images are shown from left to right. Bar, 1 μm. Representative fields of photoheterotrophic and aerobic cells. (B) Photoheterotrophic cells representing the different stages of the cell cycle: (i) newly formed, (ii) elongating, (iii) septation initiating, and (iv) septation nearing completion. The white arrows highlight the regions of intense fluorescence termed foci. In newly formed cells, two or three PBP2 foci were organized into bands across the cell. The number of PBP2 foci increased in elongating cells; two of these foci were commonly observed to form bands across the cell near midcell. As the cell cycle progressed to and through septation, PBP2 foci were observed to form bands across the cell near to the midcell sites of the forming daughter cells. The scale bar represents 1 μm. (C) 3D reconstruction of the PBP2 subcellular structure. (a) DIC and fluorescence images of a newly formed cell. (b) Optical sections through this cell; the bottom, middle, and top of the z series are shown. (c) 3D reconstruction of the z series at 0°, 60°, and 90°; the images are rotated vertically. (d) DIC and fluorescence images of a septating cell. (e) Optical sections through this cell; the bottom, middle, and top of the z series are shown. (f) 3D reconstruction of the z series at 180°, 240°, and 270°; the images are rotated horizontally. The cells in panel C are enlarged relative to those in panel A. No scale bar is presented for panel C, as the 3D reconstruction process can fractionally distorts the image parameters, and therefore, a scale bar might prove misleading in terms of the dimensions of the 3D reconstruction structure.
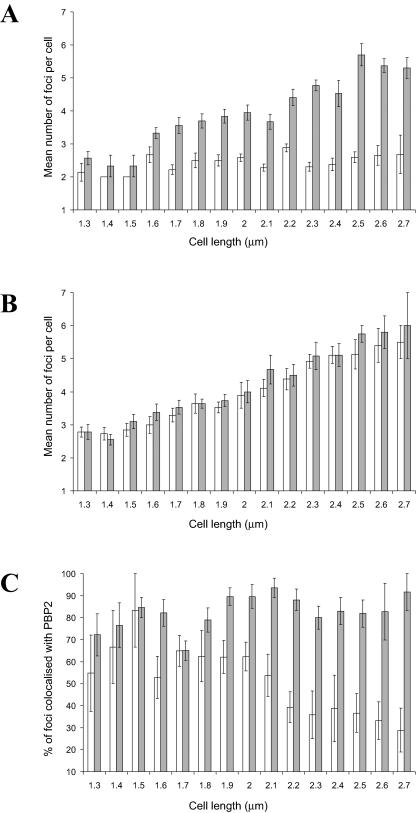
FIG. 2.
Colocalization studies of MreB and MreC with PBP2 in photoheterotrophic R. sphaeroides cells. (A) Graph showing the changes in the numbers of MreB and PBP2 foci per cell as the cell length increases throughout the cell cycle. White bars, MreB; gray bars, PBP2. A total of 153 cells were examined. (B) Graph showing the changes in the numbers of MreC and PBP2 foci per cell as the cell length increases throughout the cell cycle. White bars, MreC; gray bars, PBP2. A total of 162 cells were examined. (C) Graph showing the changes in the colocalization of MreB and MreC foci with PBP2 foci as the cell length increases throughout the cell cycle. White bars, MreB colocalized with PBP2; gray bars, MreC colocalized with PBP2. The error bars show the standard errors of the mean.
3D reconstruction of PBP2 localization.
The 2D fluorescent images of PBP2 could be explained by two possible subcellular arrangements of the protein: (i) a helix or (ii) a ring plus disconnected foci. Z series of fluorescent images were therefore captured with the aim of distinguishing between these two possibilities. A z series of a cell displaying the characteristic 2D localization of PBP2 in newly formed and elongating cells (Fig. 1C, a) was selected and subjected to deconvolution. The top and bottom images of the deconvolved z series contained a single focus, while the middle image contained multiple foci (Fig. 1C, b). 3D reconstruction and rotation revealed a partial ring plus disconnected foci (Fig. 1C, c) (see the supplemental material). This partial ring was positioned perpendicular to the longitudinal axis at or near midcell. In cells at the septation stage of the cell cycle (Fig. 1C, d), deconvolution of an appropriately selected z series revealed a single focus in the daughter cells at both the beginning and end of the z series and multiple foci in the middle image (Fig. 1C, e). 3D reconstruction and rotation revealed the formation of partial PBP2 rings in the forming daughter cells (Fig. 1C, f) (see the supplemental material). These partial rings were positioned approximately perpendicular to the longitudinal axis and at midcell of the forming daughter cells.
MreB colocalizes with PBP2 in a cell cycle-dependent manner.
In C. crescentus, the localization of PBP2 has been shown to depend upon MreB (8). Since the mreB and pbp2 genes of R. sphaeroides are found in the same operon (23), we reasoned that PBP2 and MreB may be functionally related. Therefore, in R. sphaeroides, we determined the localization of MreB and PBP2 simultaneously in the same cells using immunofluorescence microscopy. If MreB and PBP2 colocalized, it would suggest that the two proteins are functionally related.
The rate at which the number of PBP2 foci per cell increased as the cell cycle progressed differed from that of the MreB foci (Fig. 2A). In newly formed cells, there were similar numbers of PBP2 and MreB foci. However, the number of PBP2 foci increased as the cell cycle progressed, while the number of MreB foci remained relatively constant.
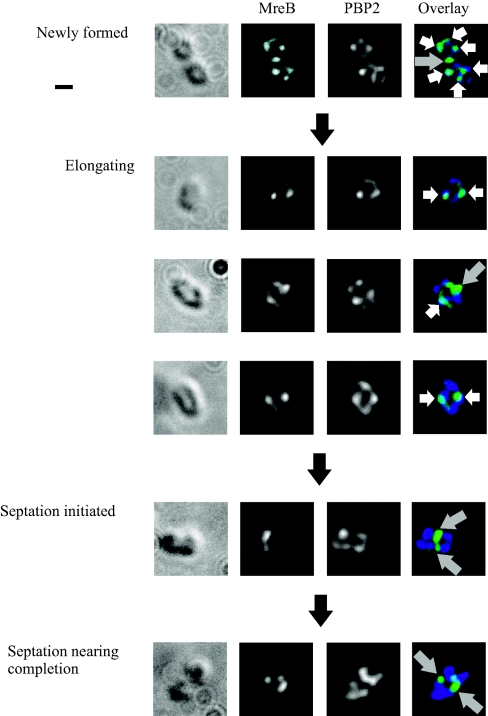
MreB localized with PBP2, but only at certain stages of the cell cycle (Fig. 2C and Fig. 3). In newly formed and elongating cells (cell length, <2.1 μm), 60% ± 3% (standard error of the mean [SEM]) of MreB foci localized with PBP2; however, in septating cells (cell length, ≥2.1 μm), MreB remained at the site of septation while PBP2 localized to the midcell sites of the forming daughter cells, with the number of colocalizing foci decreasing to 35% ± 4% (SEM). These data suggest that while most of the MreB and PBP2 molecules colocalize during elongation, this is not the case during septation.
FIG. 3.
Colocalization images of MreB and PBP2 in photoheterotrophic R. sphaeroides cells by immunofluorescence microscopy. Cells representing the different stages of the cell cycle are shown. DIC images, fluorescence images of MreB and PBP2, and an overlay of these fluorescent images are shown from left to right. The white arrows highlight MreB foci colocalizing with PBP2 foci; the gray arrows highlight MreB foci that are not colocalized with PBP2. In newly formed and elongating cells, MreB and PBP2 foci were frequently colocalized. However, in septating cells, MreB remained at the site of septation, while PBP2 localized in the forming daughter cells away from the site of septation. The scale bar represents 1 μm.
MreC colocalizes with PBP2 throughout the cell cycle.
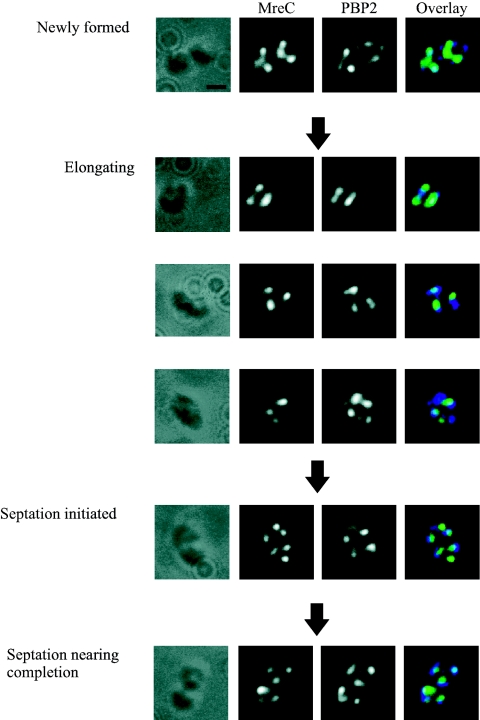
The number of MreC and PBP2 foci increased throughout the cell cycle; the ratio of the number of MreC to PBP2 foci was maintained at approximately 1 for the entire cell cycle (Fig. 2B). The majority of MreC foci (81% ± 2% [SEM]) were colocalized with PBP2 foci at all stages of the cell cycle (Fig. 2C).
The cellular positions of the colocalizing MreC and PBP2 foci were similar to the pattern observed for PBP2 (Fig. 4). In newly formed and elongating cells, the foci formed bands across the cell, which were predominately located at or near midcell. In cells undergoing septation, the foci were not observed at the site of septation but were positioned close to the midcell of the nascent/forming daughter cells. The colocalization data suggest that MreC and PBP2 colocalize throughout the cell cycle.
FIG. 4.
Colocalization images of MreC and PBP2 in photoheterotrophic R. sphaeroides cells by immunofluorescence microscopy. Cells representing the different stages of the cell cycle are shown. DIC images, fluorescence images of MreC and PBP2, and an overlay of these fluorescent images are shown from left to right. MreC colocalized with PBP2 throughout the cell cycle. The scale bar represents 1 μm.
PBP2 performs an essential role in R. sphaeroides.
To investigate whether PBP2 is essential for viability, we tried to delete or interrupt the pbp2 gene. Attempts at mutating R. sphaeroides pbp2 using an in-frame deletion strategy were unsuccessful. The strategy failed to produce mutants under either aerobic or photoheterotrophic growth conditions. The in-frame deletion strategy employed has been routinely used for the mutation of nonessential genes in R. sphaeroides; therefore, the failure to produce pbp2 mutants suggests indirectly that PBP2 performs an essential role in this bacterium.
PBP2 is the target of amdinocillin, a penicillin class antibiotic, in E. coli and other gram-negative bacterial species (26). The R. sphaeroides PBP2 homologue possesses 36% sequence identity to E. coli PBP2 (23), and therefore, R. sphaeroides cells were treated with amdinocillin in an attempt at targeting PBP2. This strategy has also been successfully used in C. crescentus (20), whose PBP2 homologue is 39% identical to R. sphaeroides PBP2.
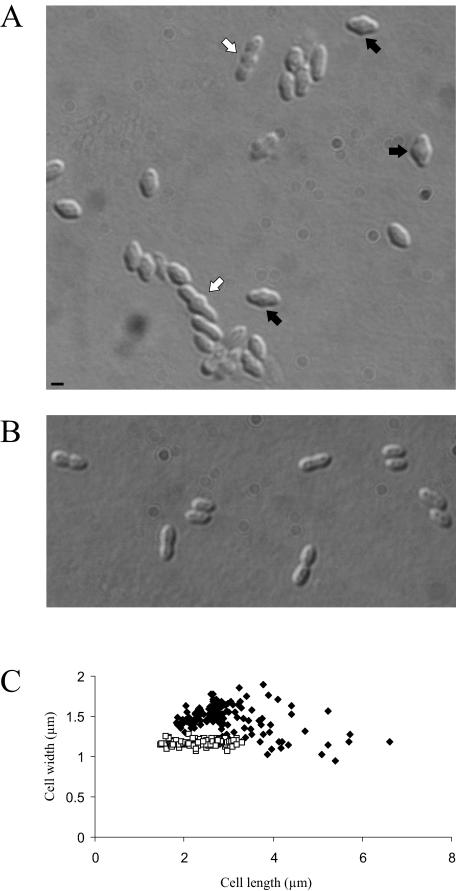
The treatment of R. sphaeroides with amdinocillin caused swelling at midcell and an increase in cell width and cell length (Fig. 5A and C), which was followed by cell lysis (measured as a decrease in culture optical density) (data not shown). These data suggest that, if the target of amdinocillin in R. sphaeroides is PBP2, then functional PBP2 is both essential for viability and required for the determination of the correct cell shape.
FIG.5.
Treatment of R. sphaeroides populations with amdinocillin results in phenotypic effects on cell shape prior to cell lysis. (A) DIC image of a population of R. sphaeroides incubated with amdinocillin for 1 h. The black arrows highlight cells that displayed increased cell widths; the white arrows highlight cells with increased cell lengths. The scale bar represents 1 μm. (B) A population of R. sphaeroides without amdinocillin. (C) Comparison of the lengths and widths of cells treated with amdinocillin and untreated. □, untreated cells; ⧫, cells treated with amdinocillin. A total of 247 cells were analyzed. Amdinocillin treatment increased the mean cell width from 1.17 ± 0.04 μm to 1.46 ± 0.01 μm, while the mean cell length was increased from 2.39 ± 0.05 μm to 2.95 ± 0.07 μm. The errors in the cell length/width refer to the standard errors of the mean.
DISCUSSION
The current model of the bacterial cell cycle includes two distinct phases of peptidoglycan synthesis (27). During elongation, it is proposed that a PBP2-specific peptidoglycan-synthetic complex drives the expansion of the longitudinal axis by depositing new peptidoglycan material in a diffuse manner along the length of the cell. During septation, it is proposed that PBP3/FtsI replaces PBP2 in the peptidoglycan-synthetic complex to specifically target synthesis at the site of septation.
During elongation, PBP2 localized as punctate foci organized as bands across the cell; the majority of these bands were found around the midcell of R. sphaeroides. 3D reconstructions showed that these fluorescent images represented partial rings plus disconnected foci, which, because of the diffraction limits of light microscopy, may represent unresolved helices. This localization suggests that PBP2 is involved in the elongation-specific phase of peptidoglycan synthesis but that it then occurs at distinct regions of the cell rather than diffusely throughout the lateral wall and that during this phase new material is deposited in a ringlike or helical manner. During septation, PBP2 was no longer visible at midcell but instead localized as bands across the cell at the midcell positions of the forming daughter cells. It is probable that an R. sphaeroides PBP3/FtsI homologue then performed peptidoglycan synthesis at the septum. These data suggest that peptidoglycan is being deposited, under the control of PBP2, at midcell until just before septation begins, when PBP2 relocalizes to the midcell positions of the daughter cells. The presence of PBP2 at midcell of the forming daughter cells suggests that, if localization reflects activity, elongation of the longitudinal axis occurs at the same time as septation-specific peptidoglycan synthesis. This possibility is supported by previous data for R. sphaeroides, which demonstrated that elongation continues as cells progress from the initiation of septation through to the completion of septation (23). The suggestion that elongation-specific peptidoglycan synthesis can occur at the same time as septation-specific peptidoglycan synthesis is also supported by studies of the gram-positive bacterium Streptococcus pneumoniae: in septating cells, the PBP2 homologue localizes in the forming daughter cells while the PBP3/FtsI homologue localizes at midcell (16). In addition, the staining of sites of nascent peptidoglycan synthesis in S. pneumoniae reflects this localization pattern: fluorescence is observed both at midcell and in the longitudinal cell walls of the forming daughter cells (3).
MreB, MreC, and PBP2 have all been implicated in the synthesis of the peptidoglycan layer (12), and in R. sphaeroides, they are encoded by the same operon (23); therefore, the colocalization of MreC and MreB with PBP2 was investigated. Colocalization is not necessarily indicative of related function, although given that these proteins are encoded by the same operon, it is likely that they will have similar functions. MreC and PBP2 colocalized throughout the cell cycle. In contrast, MreB localized with PBP2 only at specific stages of the cell cycle: the highest percentage of colocalization was observed during elongation, while the level of colocalization fell as septation occurred. These colocalization studies suggest either that these proteins directly interact and work together in the synthesis of peptidoglycan or that the proteins function independently at the same site in the formation of the peptidoglycan layer. The proteins whose functions involve peptidoglycan synthesis therefore appear to cluster in specific positions of the R. sphaeroides cell and to function, presumably cooperatively, in the deposition of new material. The constant colocalization of MreC and PBP2, if it reflects a functional relationship, suggests that these proteins may function in concert. The colocalization data on MreB and PBP2, again if they reflect a functional relationship, suggest that the two proteins interact during elongation but are involved in different cellular roles during septation.
A modified model of peptidoglycan synthesis in R. sphaeroides is proposed, based upon the localization data reflecting both functionality and functional relationships. (i) In elongating cells, peptidoglycan is synthesized in a helical or ringlike manner at or near midcell. This process is driven by a peptidoglycan-synthetic complex containing PBP2 and is supported by a range of proteins, including MreB and MreC. (ii) As the cell cycle progresses from elongation to septation, FtsI replaces PBP2 in the peptidoglycan-synthetic complex to drive synthesis of the peptidoglycan layer. MreB associates with this FtsI-containing peptidoglycan-synthetic complex and contributes to the formation of peptidoglycan at the site of septation. PBP2 and MreC relocalize in the nascent daughter cells to a site between the quarter and midcell positions and form additional peptidoglycan-synthetic complexes. (iii) During septation, peptidoglycan is synthesized in different regions of the cell by the FtsI- and PBP2-containing peptidoglycan-synthetic complexes. A bias toward the FtsI-containing peptidoglycan-synthetic complex results in a septation-specific dominance of peptidoglycan synthesis.
The deletion strategies routinely used for the mutation of nonessential genes in R. sphaeroides failed to produce mutant genotypes for pbp2. The addition of amdinocillin, the inhibitor of PBP2 in E. coli (5), to R. sphaeroides populations produced abnormalities in cell width and cell length and ultimately resulted in cell lysis. Similar phenotypic effects are also seen in C. crescentus populations treated with amdinocillin (20). These observations suggest that if amdinocillin is inhibiting PBP2, in both R. sphaeroides and C. crescentus, then this protein performs a role in cell shape that is essential for viability.
The cytoplasmic membrane of R. sphaeroides invaginates during photoheterotrophic growth; the resulting intracytoplasmic membrane contains the photosynthetic apparatus.The peptidoglycan synthesis enzymes are also embedded in the cytoplasmic membrane; it would be undesirable for peptidoglycan synthesis to occur in the invaginated folds of the membrane; therefore, a mechanism must exist for preventing peptidoglycan synthesis in these regions. Previous work demonstrated that the PBPs of R. sphaeroides are localized exclusively to the noninvaginated regions of the cytoplasmic membrane (21), i.e., to the portions of the cytoplasmic membrane that are adjacent to the peptidoglycan layer; thus, inappropriate peptidoglycan synthesis in the invaginated membrane folds is prevented. An intriguing question remains: how are the PBP proteins excluded from the invaginated region of the membrane? In this study, PBP2 has been shown to be organized into a helical or ringlike subcellular structure under both aerobic and photoheterotrophic conditions. One possible role for this structure is to keep the PBP2 out of the invaginated membrane regions. Since aerobic cells have fewer membrane invaginations than photoheterotrophic cells, the observation that the PBP2 organization is the same under both conditions suggests that this arrangement of PBP2 is capable of accommodating variable levels of membrane invagination.
Supplementary Material
Acknowledgments
This work was funded by the BBSRC.
Thanks to G. H. Wadhams for continued assistance throughout this study and to Mark Leaver, Alex Formstone, and Simon Townsend for their help.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:1053-1060. [DOI] [PubMed] [Google Scholar]
- 2.Chory, J., T. J. Donohue, A. R. Varga, L. A. Staehelin, and S. Kaplan. 1984. Induction of the photosynthetic membranes of Rhodopseudomonas sphaeroides: biochemical and morphological studies. J. Bacteriol. 159:540-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 4.den Blaauwen, T., M. E. G. Aarsman, N. E. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 5.De Pedro, M. A., W. D. Donachie, J. V. Holtje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divakaruni, A. V., R. R. Loo, Y. Xie, J. A. Loo, and J. W. Coober. 2005. The cell shape protein MreC interacts with extracytoplasmic protein including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 102:18602-18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 9.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamblin, P. A., N. A. Bourne, and J. P. Armitage. 1997. Characterization of the chemotaxis protein CheW from Rhodobacter sphaeroides and its effect on the behaviour of Escherichia coli. Mol. Microbiol. 24:41-51. [DOI] [PubMed] [Google Scholar]
- 11.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene-expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2004. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 13.Martin, A. C., G. H. Wadhams, D. S. H. Shah, S. L. Porter, J. C. Mantotta, T. J. Craig, P. H. Verdult, H. Jones, and J. P. Armitage. 2001. CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides. J. Bacteriol. 183:7135-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzawa, H., S. Asoh, K. Kunai, K. Muraiso, A. Takasuga, and T. Ohta. 1989. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J. Bacteriol. 171:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer, K. L. N., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 17.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19—selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 20.Seitz, L. C., and Y. V. Brun. 1998. Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J. Bacteriol. 180:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepherd, W. D., S. Kaplan, and J. T. Park. 1981. Penicillin-binding proteins of Rhodopseudomonas sphaeroides and their membrane localization. J. Bacteriol. 147:354-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slovak, P. M., G. H. Wadhams, and J. P. Armitage. 2005. Localization of MreB in Rhodobacter sphaeroides under conditions causing changes in cell shape and membrane structure. J. Bacteriol. 187:54-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sockett, R. E., J. C. A. Foster, and J. P. Armitage. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473-479. [Google Scholar]
- 25.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spratt, B. G. 1977. The mechanism of action of mecillinam. J. Antimicrob. Chemother. 3(Suppl. B):13-19. [DOI] [PubMed] [Google Scholar]
- 27.Vollmer, W., and J. V. Holtje. 2001. Morphogenesis of Escherichia coli. Curr. Opin. Microbiol. 4:625-633. [DOI] [PubMed] [Google Scholar]
- 28.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.