Abstract
The KdpD protein is a K+ sensor kinase located in the cytoplasmic membrane of Escherichia coli. It contains four transmembrane stretches and two short periplasmic loops of 4 and 10 amino acid residues, respectively. To determine which part of KdpD functions as a K+ sensor, genetic variants were constructed with truncations or altered arrangements of the transmembrane segments. All KdpD constructs were tested by complementation of an E. coli kdpD deletion strain for their ability to grow at a K+ concentration of 0.1 mM in the medium. A soluble protein composed of the C-terminal cytoplasmic domain was able to complement the kdpD deletion strain. In addition, analysis of the β-galactosidase activity of an E. coli strain which carries a transcriptional fusion of the upstream region of the kdpFABC operon and a promoterless lacZ gene revealed that this soluble KdpD mutant responds to changes in the K+ concentration in the extracellular medium. The results suggest that the sensing and response functions are both located in the C-terminal domain and might be modulated by the N-terminal domain as well as by membrane anchoring.
KdpD is an inner membrane protein of Escherichia coli. Together with KdpE, it is required for the transcriptional control of the kdpFABC operon, which encodes a high-affinity K+ uptake system to maintain intracellular osmolarity. Under K+-limiting growth conditions, KdpD autophosphorylates, most likely at His673 (25). Subsequently, the phosphoryl group is probably transferred to Asp52 of the cytoplasmic response regulator KdpE, a transcriptional activator of the kdpFABC operon (18, 23) which encodes the high-affinity K+ pump KdpFABC (6). KdpD is functionally related to other sensor kinases, like PhoR, EnvZ, and CreC, and shows moderate sequence homology to those proteins in parts of its C-terminal domain (27).
The nature of the environmental stimulus for the Kdp system has not been conclusively determined. It is not known how the K+ concentration of the growth medium is detected by the sensor, nor whether KdpD interacts directly with K+ ions. It is unlikely that the K+ signal is sensed in the cytoplasm, since the intracellular K+ concentration in E. coli is regulated by the osmolality of the medium and not by the external K+ concentration (26). On the other hand, it has been hypothesized that cell turgor generates the signal (13) by altering the outward pressure on the cytoplasmic membrane to change the stretch or tilt of the four transmembrane segments of KdpD (1). Mutations within transmembrane region 4 caused a loss of sensitivity to the K+ signal, but cells containing these mutant proteins were able to respond to high salt stress, suggesting that KdpD senses two signals: the K+ concentration and the physico-chemical state of the cytoplasmic membrane (24). Initially, the four transmembrane regions of KdpD were thought to be necessary for signal perception (18), until a mutant lacking all four transmembrane domains was found to be still able to respond to low K+ concentrations (10). These findings led to a new model, in which the sensing was suggested to involve communication between the N- and C-terminal domains of the protein in the cytoplasm (10).
In this study, we intended to investigate the communication between the N- and C-terminal domains of KdpD and asked whether membrane anchorage is important for the K+ sensing. KdpD was split into halves between helix 2 and 3 to generate fragments KdpD-N and KdpD-C. These fragments were expressed independently and were both found to insert into the cytoplasmic membrane (7). When plasmids encoding wild-type KdpD, KdpD-N, or KdpD-C were introduced into the kdpD deletion strain E. coli TKV2208, the N-terminal fragment (KdpD-N) alone did not allow the cells to grow at low K+ concentrations (0.1 mM). However, expression of the C-terminal fragment (KdpD-C) allowed growth at 0.1 mM K+ and showed a regulated β-galactosidase activity from a kdpFABC promoter-lacZ fusion. Furthermore, a soluble C-terminal fragment containing only the cytoplasmic domain of KdpD (C499-894) was also able to complement the growth of TKV2208 and to sense K+ concentrations as monitored in the kdpFABC promoter-lacZ fusion strains. We conclude that neither the N-terminal domain nor membrane anchoring is absolutely necessary for K+ sensing.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
KdpD and mutant derivatives thereof were expressed in E. coli strain MC1061 [kdpD+ hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi] (16). To study KdpD sensor activity and the ability to support growth in low-K+ medium, plasmids encoding KdpD constructs were introduced into strain HAK006 [ΔkdpABCD Δ(lac-pro) ara thi], which carries a kdpFABC promoter-lacZ fusion gene and is kdpE+. The deletion within kdpD spans residues 159 to 726 (19). The constructs were also introduced into the kdpD deletion strain TKV2208 (ΔkdpD trkA405 trkD1 nagA thi rha lacZ) (20), which still contains residues 1 to 127 of KdpD (P. Zimmann, personal communication). Since we could not exclude that the remaining N-terminal fragment in TKV2208 and HAK006 has some complementing effect, we performed a total deletion of kdpD in HAK006, resulting in HMK006 as described below.
The pBD plasmid carrying the kdpD gene was kindly provided by K. Jung and K. Altendorf (11). The strains and plasmids used in this study are listed in Table 1. Media preparation and bacterial manipulations were performed according to standard methods (15). Where appropriate, ampicillin (100 μg/ml final concentration) or chloramphenicol (25 μg/ml final concentration) was added to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Name | Genotype or relevant characteristics | Source or reference |
|---|---|---|---|
| Strains | |||
| HAK006 | ΔkdpABCD Δ(lac-pro) ara thi kdpFABC promoter-lacZ+ fusion gene at the att λ site | 19 | |
| TKV2208 | ΔkdpD trkA405 trkD1 nagA thi rha lacZ | 20 | |
| MC1061 | kdpD+hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi | 16 | |
| HMK006 | ΔkdpD Δ(lac-pro) ara thi kdpFABC promoter-lacZ+ fusion gene at the att λ site | This study | |
| DY330 | W3110 ΔlacU169 gal490 (λcI857Δcro-broA) | 28 | |
| Plasmids | |||
| pBD | KdpD | Apr, pBAD18, kdpD | 11 |
| pSF1 | KdpD-N | Cmr, pBAD33, kdpDΔ449-894 | This study |
| pSF2 | KdpD-C | Apr, pBAD18, kdpDΔ1-443 | This study |
| pMR1 | N1-2C | Apr, pBAD18, kdpDΔ446-496 | This study |
| pSF3 | N3-4C | Apr, pBAD18, kdpDΔ398-446 | This study |
| pSF4 | N1-4C | Apr, pBAD18, kdpDΔ423-476 | This study |
| pMR2 | H4+C | Apr, pBAD18, kdpDΔ1-466 | This study |
| pMR3 | H4+C+RR | Apr, pBAD18, kdpDΔ1-466, (D474R, V475R) derivative of pMR2 | This study |
| pMR4 | C494-894 | Apr, pBAD18, kdpDΔ1-493 | This study |
| pMR5 | C499-894 | Apr, pBAD18, kdpDΔ1-498 | This study |
| pKD3 | Apr, Cmr, PCR template | 4 | |
| pCP20 | Apr, Cmr, ts FLP recombinase | 3 |
Construction of the HMK006 strain.
The E. coli strain HMK006 with a complete chromosomal deletion of kdpD was constructed from HAK006 using the recombination system described by Datsenko and Wanner (4). A linear PCR fragment encoding a chloramphenicol resistance (Cmr) cassette, cat, was obtained from the plasmid pKD3 (4) with primers matching the flanking regions of the kdpD gene on the chromosome. Primer sequences were 5′ GGATAA ACTTGATGAATAAGTGTAGGCTGGAGCTGCTTCGAAGT-TCCTATAC 3′ and 5′ CGTTTGTCACATATCCTCATGCATATGAATATCCTCCTTAGT-TCCTATTCCGAAGTTCCTATT 3′. Linear fragments were transformed into the RED strain DY330 (28). Recombinants were screened for Cmr, and P1 phage lysates were obtained from the respective candidates. The lysates were used to infect HAK006 as an acceptor strain. Cmr candidates were selected and tested for their K+-dependent β-galactosidase activity in a blue-white plate screen. The cat cassette was eliminated from the chromosome by transformation with plasmid pCP20 as described by Datsenko and Wanner (4). This plasmid contains a temperature-sensitive replication origin for temporal production of the Flp enzyme (3). The resulting strain, HMK006, has lost the kdpD gene completely, as verified by genomic PCR. HMK006 was tested for its K+-dependent growth as well as for its K+-dependent β-galactosidase activity.
Construction of KdpD deletion mutants.
To facilitate mutant construction, single restriction sites were introduced by site-directed mutagenesis using the QuikChange procedure (Stratagene). Restriction analysis and DNA sequencing confirmed the introduction of the new restriction sites. Plasmids encoding KdpD-N (i.e., encoding residues 1 to 448 of KdpD) and KdpD-C (i.e., encoding residues of 444 to 894 of KdpD) have been described previously (7). To construct plasmids pSF1 and pSF2, the sequences coding for KdpD-N and KdpD-C were cleaved with XbaI/HindIII and cloned into pBAD33 and pBAD18 (8), respectively.
To construct pMR1 (N1-2C), BamHI restriction sites were introduced at the codon position for amino acid 446 (corresponding to the end of helix 2) in pSF1 and at the codon position for 497 (corresponding to the end of helix 4) in pSF2. The sequence encoding the cytoplasmic domain of KdpD-C was then cleaved with BamHI/HindIII, and the resulting fragment was cloned into pSF1 to generate pMR1 (Fig. 1, N1-2C). An NdeI fragment encoding the cytoplasmic N-terminal domain of KdpD-N was joined in frame with KdpD-C to produce pSF3 (N3-4C). This construct was made using site-directed mutagenesis to introduce an NdeI restriction site at codon position 398 (before helix 1) in the plasmid coding for KdpD-N. To construct pSF4 (N1-4C), a DNA fragment encoding helix 1 and the cytoplasmic N-terminal domain of KdpD-N was cloned into the MunI restriction site between helices 3 and 4 (at the codon position for amino acid 477) of the plasmid coding for KdpD-C. To construct pMR2 (H4+C), an NdeI restriction site was inserted at codon position 466 between helix 3 and helix 4, and the NdeI/HindIII fragment encoding helix 4 and the cytoplasmic C-terminal domain of KdpD-C was subcloned into pBAD18 (8). Using site-directed mutagenesis, two arginine residues were introduced in pMR2 at codon positions 474 and 475 to generate pMR3 (H4+C+RR).
FIG. 1.
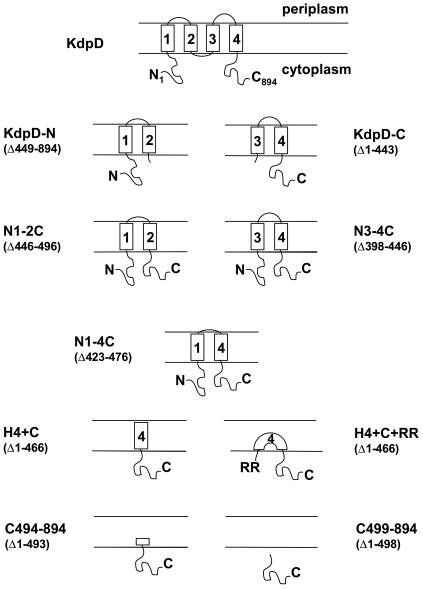
Schematic representation of KdpD and variants showing the predicted membrane topologies of the proteins used in this study. The numbered boxes represent the transmembrane domains. KdpD (wild type), KdpD-N, KdpD-C, N1-2C, N3-4C, and N1-4C are shown with their associated cytoplasmic regions. H4+C contains transmembrane helix 4 and the C-terminal cytoplasmic region, and H4+C+RR contains two arginine residues introduced N-terminally to helix 4. The C494-894 and C499-894 proteins have a truncation or complete deletion of helix 4, respectively.
Two PaeI restriction sites were inserted into pMR2 at codon positions 469 and 494 by site-directed mutagenesis. Cleavage with PaeI and subsequent religation created pMR4 (C494-894), which contains the last four amino residues of helix 4 and the cytoplasmic C-terminal domain. To construct pMR5 (C499-894), two PaeI restriction sites were inserted into pMR2 at codon positions for residues 469 and 499. All the kdpD constructs were verified by DNA sequencing.
Assay of β-galactosidase activity.
Assays for β-galactosidase were performed in strain HAK006 (19) and HMK006 according to the method described by Miller (17), with minor modifications. Cells transformed with pBD or plasmids expressing the KdpD mutant proteins (Table 1) were grown in minimal medium (6) containing KCl at various concentrations (0.1 to 10 mM). Samples (100 μl) were taken at the times indicated and added to Z-buffer (100 mM sodium phosphate, pH 7.0, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). The cells were permeabilized by the addition of chloroform (45 μl) and 0.1% sodium dodecyl sulfate (30 μl) and incubated at 28°C for 5 min before addition of 200 μl of ο-nitrophenyl-β-d-galactopyranoside (4 mg/ml). The reactions were stopped after 10 min by the addition of 1 M Na2CO3 (0.5 M final concentration) and centrifuged for 10 min to remove cell debris. The amount of ο-nitrophenol released per cell mass (optical density minus wavelength value) was measured by determining the absorbance at 420 nm of the supernatant. Assays were done in triplicate. All activities are expressed in Miller units (17).
Complementation of the kdpD deletion strain TKV2208.
The function of the mutant proteins was tested in the kdpD deletion strain TKV2208 (20) containing pBD or the plasmids expressing the KdpD variants (Table 1). The ability to grow under K+-limiting conditions was used to test for complementation. The plasmid-bearing cells were grown overnight in minimal medium containing 0.1 mM KCl (6) with glucose as sole carbon source.
Sodium carbonate extraction.
Cell fractionation experiments were performed with sodium carbonate to determine whether the KdpD variants partition into the membrane (5). Cultures of strain MC1061 (16) bearing pBD or the plasmids expressing the KdpD mutant proteins were grown in LB medium at 37°C. Overnight cultures were diluted 1:100 into fresh LB medium, grown to an optical density at 600 nm (OD600) of 0.2, and induced with l-arabinose (0.2%) for 3 hours. Cells (1 ml) were then removed, chilled on ice, and centrifuged. Pellets were resuspended in 300 μl spheroplast buffer (33 mM Tris, pH 8.0, 40% sucrose). Lysozyme (5 μg/ml final concentration) and EDTA (1 mM final concentration) were then added. After 15 min, 400 μl H2O and 700 μl sodium carbonate (pH 11.5; 0.2 M final concentration) were added, and after 30 min the samples were vortexed vigorously to lyse the cells. These samples were centrifuged (110,000 × g) for 30 min at 4°C to pellet the membrane fractions. The supernatant and membrane fractions were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with antiserum against KdpD.
RESULTS
Construction of truncated KdpD variants.
To investigate which parts of the KdpD protein are required for sensor function, the deletion constructs shown in Fig. 1 were analyzed. Plasmid pSF1 (KdpD-N) encodes the N-terminal cytoplasmic domain and transmembrane regions 1 and 2 (amino acid residues 1 to 448). Plasmid pSF2 (KdpD-C) encodes transmembrane regions 3 and 4 and the entire C-terminal cytoplasmic domain (residues 444 to 894). Variants containing both cytoplasmic regions but only two membrane spans were constructed and were designated as N1-2C, N3-4C, and N1-4C (Fig. 1). The H4+C construct contains only the fourth transmembrane region and the cytoplasmic C-terminal domain, and H4+C+RR was designed to block insertion into the membrane by introducing two arginine residues preceding the fourth helix. Mutants without any transmembrane helices were named C494-894 and C499-894. All mutant proteins were tested for expression and membrane anchoring.
Effects of the deletions on membrane anchoring.
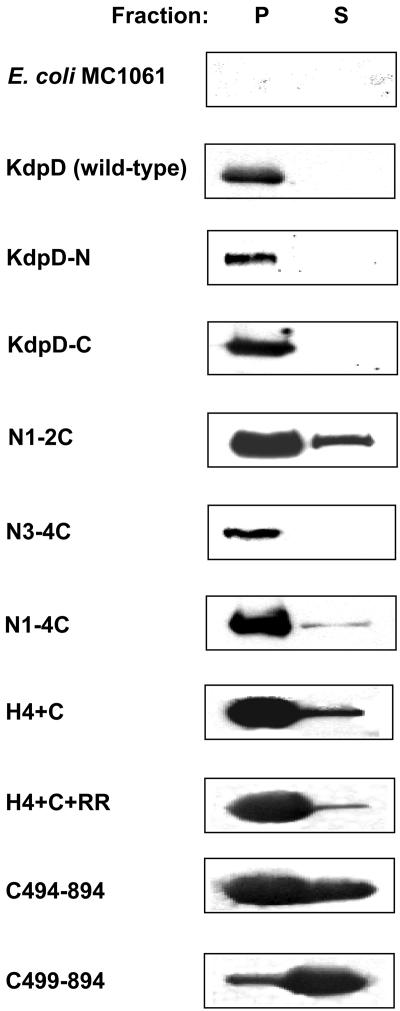
The expression levels of the mutant proteins and membrane partitioning were examined by sodium carbonate extraction (5) and Western blotting (Fig. 2). MC1061 cells transformed with plasmid pBD (wild-type KdpD) or the plasmids expressing the KdpD mutants (Table 1) were grown to exponential phase and incubated with arabinose for 3 hours to induce expression of the plasmid-borne polypeptides. Proteins were extracted from the cells with 0.2 M sodium carbonate and subjected to ultracentrifugation to separate the membrane (pellet) and cytosolic (supernatant) fractions. All mutant proteins were expressed like wild-type KdpD, except for KdpD-N and N3-4C, which were present at reduced levels, possibly due to protein instability. All proteins with a transmembrane region were found mainly in the pellet fraction and about 5% of N1-2C was found in the supernatant, suggesting that all the mutants with an intact transmembrane region had integrated into the membrane. The H4+C and H4+C+RR proteins were also found in the pellet fraction, since they probably interact hydrophobically with the membrane. The hydrophobic interaction was weakened for C494-894, which was found both in the pellet and supernatant. Finally, the C499-894 protein was found mainly in the supernatant, suggesting that it is a soluble, cytoplasmic protein.
FIG. 2.
KdpD mutants with transmembrane segments are found in the membrane fraction. Plasmids carrying the different engineered kdpD genes were transformed into E. coli MC1061. Cultures were grown in LB medium to an OD600 of 0.2 and then induced with 0.2% arabinose for 3 h. The supernatant (S) and pellet (P) fractions after sodium carbonate extraction were prepared as described in Materials and Methods.
Complementation by KdpD mutant proteins.
Cells expressing the KdpD mutant proteins (Table 1) were tested for growth under K+-limiting conditions (0.1 mM) in the kdpD deletion strain TKV2208. Although strain TKV2208 alone did not grow under these conditions, the presence of a plasmid encoding wild-type KdpD or any of the variants with the C-terminal cytoplasmic domain (H4+C, H4+C+RR, C494-894, or C499-894) allowed normal growth, whereas the mutants N3-4C and N1-2C were retarded in their growth rate (Table 2). In contrast, cells bearing the plasmid expressing only the N-terminal fragment (KdpD-N) did not grow.
TABLE 2.
Growth of KdpD constructs in E. coli TKV2208 under K+-limiting conditions
| KdpD construct | Cell densitya (OD600) | Growth rateb (h−1) | Colony formationc |
|---|---|---|---|
| KdpD | 0.89 ± 0.07 | 0.45 | + |
| KdpD-N | 0.06 ± 0.09 | 0.00 | − |
| N1-2C | 0.82 ± 0.11 | 0.18 | + |
| KdpD-C | 0.94 ± 0.12 | 0.45 | + |
| N3-4C | 0.89 ± 0.08 | 0.27 | + |
| N1-4C | 0.95 ± 0.07 | 0.33 | + |
| H4+C | 0.99 ± 0.06 | 0.39 | + |
| H4+C+RR | 0.93 ± 0.03 | 0.40 | + |
| C494-894 | 0.88 ± 0.10 | 0.43 | + |
| C499-894 | 0.95 ± 0.11 | 0.44 | + |
Cells were grown overnight in minimal medium containing 0.1 mM KCl.
Overnight cultures were grown in KML complex medium (1% KCl, 1% tryptone, 0.5% yeast extract), washed in minimal medium without K+, and diluted 1:50 in minimal medium containing 0.1 mM KCl.
Cells were streaked on minimal agar plates containing 0.1 mM KCl.
Responses mediated by KdpD variants at different K+ concentrations.
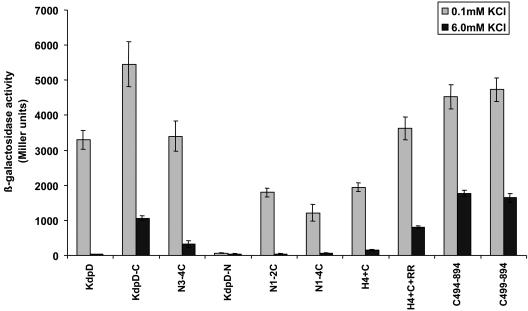
To compare the activities of the KdpD variants, expression of a kdpFABC promoter-lacZ fusion was analyzed in strain HAK006, which carries an internal in-frame kdpD deletion (19) (see Materials and Methods). Because the amount of a regulatory protein is critical in signal transduction, plasmids were designed to express the KdpD variants under control of the araBAD promoter. When cells were grown in minimal medium with 0.1 mM KCl in the absence of arabinose and with glucose present, the amount of wild-type KdpD produced was still sufficient to complement a kdpD null strain (10). Cells expressing different KdpD variants were grown in minimal medium with 0.1 or 6 mM KCl. When cultures reached a cell density (OD600) of 0.5, β-galactosidase activity was measured (Fig. 3). Cells producing wild-type KdpD had a high β-galactosidase activity in low-K+ medium (0.1 mM). At 6 mM K+, very little β-galactosidase activity was detected. There was no β-galactosidase activity measurable in cells expressing KdpD-N at 0.1 mM K+, but the cytoplasmic C-terminal domain (N1-2C) restored activity to 55% compared to wild-type KdpD. Activation of the kdpFABC promoter-lacZ fusion by KdpD-C responded to the different K+ concentrations, but the basal level of β-galactosidase activity was higher than with wild-type KdpD. Activities comparable to those of the wild type were observed when the N-terminal domain was present (N3-4C). The N1-2C and N1-4C constructs also mediated responses to K+ concentration, but they did not restore full activity at 0.1 mM K+. In addition, the mutant proteins with only one transmembrane segment (H4+C and H4+C+RR) supported considerable activity. Truncation or deletion of the fourth transmembrane segment (variants C494-894 and C499-894, respectively) produced higher levels of β-galactosidase activity than wild-type KdpD, but the activity was not fully repressed under 6 mM K+ conditions, like KdpD-C and H4+C+RR (Fig. 3). The C494-894 and C499-894 constructs at 6 mM KCl had a remaining activity of 30 to 40% of the levels seen at 0.1 mM KCl and 50% of the level of fully induced wild-type KdpD. Thus, the inactivation of the sensor system is affected in the deletion mutants. This suggests that the cytoplasmic N-terminal domain may be important in preventing constitutive expression at 6 mM K+.
FIG. 3.
The cytoplasmic C-terminal domain is sufficient to sense changes in external K+ concentration. Strain HAK006 was transformed with plasmids encoding the various KdpD variants. Cells were grown to mid-logarithmic phase in minimal medium containing 0.1 mM KCl or 6 mM KCl. The β-galactosidase activity was determined as described in Materials and Methods and is given in Miller units (17). The data represent the means of triplicate determinations.
KdpD variants are activated by shifts to a low K+ concentration.
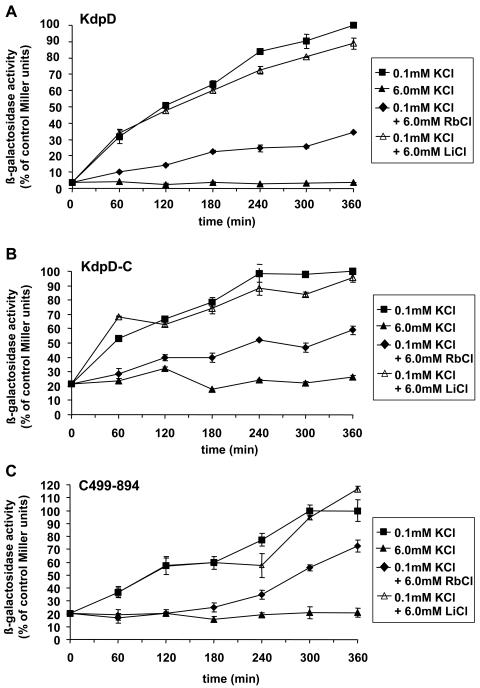
Cells were grown in 6 mM KCl medium to exponential phase and shifted to a growth medium with 0.1 mM KCl (Fig. 4). HAK006 cells producing wild-type KdpD (Fig. 4A) showed an immediate increase in the β-galactosidase activity. When 6 mM LiCl was present together with 0.1 mM K+, the sensor was fully activated, whereas 6 mM RbCl led to an intermediate level of activation. These results demonstrate that the K+ concentration in the medium modulates sensor activity and that Li+ does not inactivate the sensor. Cells expressing KdpD-C (Fig. 4B) or C499-894 (Fig. 4C) showed an elevated basal activity (at t0) but responded like cells expressing wild-type KdpD when the different salts were present. KdpD-C and C499-894 were still able to discriminate between K+, Rb+, and Li+, like wild-type KdpD.
FIG. 4.
Activation of KdpD, KdpD-C, and C499-894 is specific for K+. HAK006 cells producing KdpD (A), KdpD-C (B), or C499-894 (C) were grown to mid-logarithmic phase in minimal medium containing 6 mM KCl. Cells were washed with minimal medium containing 0.1 mM KCl and transferred into fresh medium containing 0.1 mM KCl. Then, 6 mM RbCl or 6 mM LiCl was added. A control culture was resuspended in medium containing 6 mM KCl. The β-galactosidase activity was determined at the indicated time points and normalized to the maximum β-galactosidase activity (100% of control Miller units) of the KdpD+ culture growing in medium containing 0.1 mM KCl.
KdpD variants are inactivated by high K+.
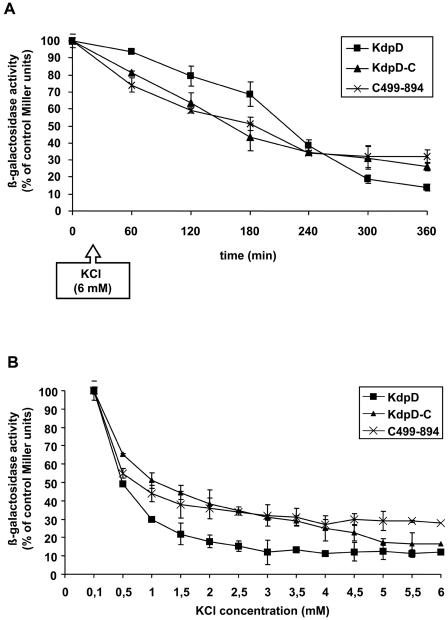
To confirm that the KdpD-C and the C499-894 proteins are functional K+ sensors, inactivation kinetics were analyzed in HAK006 cells expressing these proteins. Cells were grown in the presence of 0.1 mM KCl, and β-galactosidase activity was measured at different intervals after the addition of 6 mM KCl to the medium (Fig. 5A). The β-galactosidase activity slowly decreased over a period of 5 h in all three cases (Fig. 5A).
FIG. 5.
KdpD-C and C499-894 have inactivation kinetics like KdpD. (A) HAK006 cells expressing KdpD, KdpD-C, or C499-894 were grown to mid-logarithmic phase in minimal medium containing 0.1 mM KCl. Samples were taken at the indicated time points after addition of 6 mM KCl and assayed for β-galactosidase activity. (B) HAK006 cells expressing KdpD, KdpD-C, or C499-894 were grown to mid-logarithmic phase in minimal medium containing 0.1 mM KCl and divided into aliquots. The indicated amounts of KCl were added to the cultures, and growth was continued for 6 h. The β-galactosidase activity was determined and normalized to the value of the initial culture for each sample.
HAK006 cells containing the same three plasmids were also grown overnight in low-K+ medium (0.1 mM KCl) and back-diluted into media containing various K+ concentrations. Growth was continued for 6 h, and β-galactosidase activity was then measured (Fig. 5B). The β-galactosidase activity in all three transformed strains gradually decreased at higher K+ concentrations. Three millimolar K+ was sufficient to inactivate KdpD. The same was true for cells expressing KdpD-C and C499-894, but the basal activity was still higher than for the wild type.
Response of KdpD to changes in osmotic strength of the medium.
KdpD-dependent β-galactosidase activity was measured in HAK006 cells growing in media of various osmolarities and K+ concentrations (Table 3). KdpD was able to sense the change from low to high K+ concentrations in the presence of 100 mM NaCl or 100 mM LiCl. Under these conditions, it seems that the changes of osmolarities in the medium have no effect on the K+ sensing and the resulting activity of KdpD.
TABLE 3.
β-Galactosidase activity supported by wild-type KdpD in high-osmolarity medium with different KCl concentrations
| KCl (mM) | Added salt (100 mM) | Activity (% of initial Miller units) |
|---|---|---|
| 0.1 | 100 | |
| 6.0 | 1.4 | |
| 100 | 0.6 | |
| 0.1 | NaCl | 112 |
| 6.0 | NaCl | 0.7 |
| 0.1 | LiCl | 150 |
| 6.0 | LiCl | 0.8 |
Regulation of sensor activity in kdpD deletion strain HMK006.
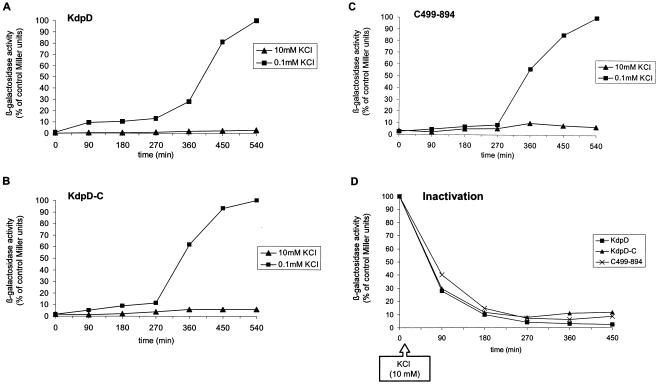
Since we could not exclude that the remaining kdpD fragment in HAK006 could lead to a complementing sensor activity, the total kdpD gene was removed in HMK006 by the method of Datsenko and Wanner (4). To analyze the sensor activation of wild-type KdpD, KdpD-C, and C499-894, the HMK cells were grown overnight in the presence of 10 mM KCl and back-diluted 1:100 into medium containing 0.1 mM KCl (Fig. 6). After a lag time of about 4 h, the β-galactosidase activity increased with a similar kinetics for KdpD (Fig. 6A), KdpD-C (Fig. 6B), and C499-894 (Fig. 6C). In addition, when cells were grown to the exponential phase in the presence of 0.1 mM KCl and then supplemented with KCl to 10 mM, the β-galactosidase activity decreased for KdpD, KdpD-C, and C499-894 (Fig. 6D). Remarkably, we did not observe an elevated constitutive activity for the mutants KdpD-C and C499-894, as was apparent in the HAK006 cells (Fig. 3, 4, and 5A). Taken together, the data clearly show that the C-terminal soluble fragment C499-894 retains the sensor properties of the wild-type KdpD.
FIG. 6.
Activation (A to C) and inactivation kinetics (D) of KdpD, KdpD-C, and C499-894 in HMK006. HMK006 cells expressing KdpD (A), KdpD-C (B), or C499-894 (C) were grown overnight in minimal medium containing 10 mM KCl. Cells were washed with minimal medium containing 0.1 mM KCl and diluted 1:100 into fresh minimal medium containing 0.1 mM KCl. A control culture was resuspended in medium containing 10 mM KCl. The β-galactosidase activity was determined at the indicated time points and normalized to the maximum β-galactosidase activity (100% of control Miller units) of the KdpD+ culture growing in medium containing 0.1 mM KCl. (D) HMK006 cells expressing KdpD, KdpD-C, or C499-894 were grown in minimal medium containing 0.1 mM KCl to mid-logarithmic phase. Then, 10 mM KCl was added. Samples were taken at the indicated time points after addition of 10 mM KCl and assayed for β-galactosidase activity.
DISCUSSION
The present study investigated the minimal portion of the KdpD protein that can function as a K+ sensor. We found that the second half of the protein, KdpD-C, which includes transmembrane helices 3 and 4 and the C-terminal cytoplasmic domain, constitutes a sensor kinase that mediates responses to K+ in the medium (Fig. 3 to 6). A plasmid encoding KdpD-C complements the defect of a kdpD deletion strain to allow growth under K+-limiting conditions.
A KdpD mutant lacking all four transmembrane domains had shown a reduced level of both kinase and phosphatase activities but was still able to detect low K+ concentrations (10). This observation that the transmembrane portion of KdpD is dispensable for normal regulation of activity raises the question of whether membrane association is required for sensor activity. We examined this issue by systematically removing the membrane-spanning helices of KdpD-C. A construct with only one such helix (H4+C) exhibited only slightly decreased (60% of the wild type) sensor activity (Fig. 3). When two arginine residues were introduced at the N terminus of H4+C (H4+C+RR) to prevent a normal membrane insertion, the protein was still able to sense K+ limitation. However, this protein was still strongly associated with the membrane based on the results of a sodium carbonate extraction (Fig. 2). Membrane association was decreased when the fourth helix was truncated (the C494-894 construct) or deleted completely (C499-894). Surprisingly, the C499-894 protein still functioned as a K+ sensor (Fig. 3), and it also complemented a kdpD deletion strain to permit growth at 0.1 mM KCl. We conclude that C499-894 is a functional K+ sensor with the ability to sense K+ limitation. This finding does not support the model proposed by Heermann and coworkers (10), in which signal transduction relies on an interaction of the cytoplasmic but membrane-associated N- and C-terminal domains of KdpD.
If only the C-terminal half of KdpD is essential, what is the function of the N-terminal half? One possibility is that additional parameters may modulate the sensor activity. Recently, it has been suggested that the N-terminal cytoplasmic domain responds to intracellular ATP levels (12). Indeed, the N-terminal region contains two putative ATP-binding cassettes that could account for such a regulatory mechanism (11). Direct binding of the N-terminal to the C-terminal domain has been demonstrated (10), and our data also support that the basal sensor activity is controlled by this interaction. It has also been suggested that the N-terminal domain of KdpD has a stabilizing effect on the binding of KdpE∼P to its corresponding DNA-binding site (9).
The K+ concentrations at which expression of KdpFABC is turned off vary depending on the strain (13). In some strains, external K+ concentrations greater than 2 mM are sufficient to cause inhibition of Kdp (13, 21). At external KCl concentrations of 3 mM and above, transcription from the kdpFABC promoter was reduced in HAK006 cells expressing KdpD, KdpD-C, and C499-894 (Fig. 5B). Also, KdpD-C and C499-894 could be activated (Fig. 4 and 6) and inactivated (Fig. 5 and 6) with kinetics like those of the wild-type KdpD. In the HAK006 cells, the two mutants did not fully repress transcription, which explains the constitutive level that was observed in Fig. 3.
Presumably, the signal sensed by KdpD is K+ depletion in the external environment. If the signal perceived by KdpD is in the periplasm, how is it transmitted to the cytoplasmic domain of KdpD that controls activity? Since the mutant C499-894 is in the cytoplasm, accessory proteins might transmit the signal from the periplasm to the interior of the cell to modulate kdpFABC expression. Perhaps an additional component in the membrane senses the signal and communicates with KdpD. In Mycobacterium tuberculosis, a direct communication between KdpD and the membrane proteins LprF and LprJ was found. The C-terminal histidine kinase domain of KdpD participates in the formation of a ternary complex with these lipoproteins and the N-terminal domain of KdpD (22). However, no homologues of the Lpr proteins have been found in E. coli. Another possibility is that the K+ depletion in the environment has secondary effects that are sensed inside by the cells.
The idea that turgor alone could be the signal has been challenged by several findings (2, 11, 24). It had been proposed that cell turgor regulates KdpD (14), but experiments in which osmolarity was modulated showed weaker responses compared to changes in K+ concentrations (10). Also, Gowrishankar and coworkers observed that accumulation of intracellular solutes, such as trehalose or glycine betaine, did not affect KdpD activity, indicating that turgor pressure in general does not control kdpFABC transcription (2). Our experiments, by simply increasing the osmotic strength in the medium with NaCl or LiCl, showed regulated sensor activity in different osmotic media (Table 3). Under these conditions, wild-type KdpD still reacts to different K+ concentrations in the medium. Our results support the view that turgor is not the crucial signal to control kdpFABC transcription.
Taken together, we have shown that the C-terminal hydrophilic domain of KdpD is a K+ sensor that controls the transcription of kdpFABC, whereas the N-terminal region most likely modulates the activity by reducing the constitutive basal response.
Acknowledgments
We thank K. Jung and K. Altendorf for generously providing us with the initial plasmid, pBD, the E. coli strains HAK006 and TKV2208, and the KdpD antiserum.
This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 495.
REFERENCES
- 1.Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes, vol. 5. Jai Press Ltd., London, England. [Google Scholar]
- 2.Asha, H., and J. Gowrishankar. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J. Bacteriol. 175:4528-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, N. G., and P. Model. 1985. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell 41:607-614. [DOI] [PubMed] [Google Scholar]
- 6.Epstein, W., and M. Davis. 1970. Potassium-dependent mutants of Escherichia coli. J. Bacteriol. 101:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facey, S. J., and A. Kuhn. 2003. The sensor protein KdpD inserts into the E. coli membrane independent of the Sec translocase and YidC. Eur. J. Biochem. 270:1724-1734. [DOI] [PubMed] [Google Scholar]
- 8.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 51:51277-51284. [DOI] [PubMed] [Google Scholar]
- 10.Heermann, R., A. Fohrmann, K. Altendorf, and K. Jung. 2003. The transmembrane domains of the sensor kinase KdpD of Escherichia coli are not essential for sensing K+ limitation. Mol. Microbiol. 47:839-848. [DOI] [PubMed] [Google Scholar]
- 11.Jung, K., and K. Altendorf. 1998. Truncation of amino acids 12-128 causes deregulation of the phosphatase activity of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 273:17406-17410. [DOI] [PubMed] [Google Scholar]
- 12.Jung, K., and K. Altendorf. 2002. Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:223-228. [PubMed] [Google Scholar]
- 13.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA. 78:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Meissner, P. S., W. P. Sisk, and M. L. Berman. 1987. Bacteriophage λ cloning system for the construction of directional cDNA libraries. Proc. Natl. Acad. Sci. USA 84:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Nakashima, K., A. Sugiura, K. Kanamaru, and T. Mizuno. 1993. Signal transduction between the two regulatory components involved in the regulation of the kdpABC operon in Escherichia coli: phosphorylation-dependent functioning of the positive regulator, KdpE. Mol. Microbiol. 7:109-116. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima, K., A. Sugiura, and T. Mizuno. 1993. Functional reconstitution of the putative Escherichia coli osmosensor, KdpD, into liposomes. J. Biochem. 114:615-621. [DOI] [PubMed] [Google Scholar]
- 20.Puppe, W., P. Zimmann, K. Jung, M. Lucassen, and K. Altendorf. 1996. Characterization of truncated forms of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. J. Biol. Chem. 271:25027-25034. [DOI] [PubMed] [Google Scholar]
- 21.Roe, A. J., D. McLaggan, C. P. O'Byrne, and I. R. Booth. 2000. Rapid inactivation of the Escherichia coli Kdp K+ uptake system by high potassium concentrations. Mol. Microbiol. 35:1235-1243. [DOI] [PubMed] [Google Scholar]
- 22.Steyn, A. J., J. Joseph, and B. R. Bloom. 2003. Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 47:1075-1089. [DOI] [PubMed] [Google Scholar]
- 23.Sugiura, A., K. Nakashima, K. Tanaka, and T. Mizuno. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6:1769-1776. [DOI] [PubMed] [Google Scholar]
- 24.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14:929-938. [DOI] [PubMed] [Google Scholar]
- 25.Voelkner, P., W. Puppe, and K. Altendorf. 1993. Characterization of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. Eur. J. Biochem. 217:1019-1026. [DOI] [PubMed] [Google Scholar]
- 26.Walderhaug, M. O., D. C. Dosch, and W. Epstein. 1987. Potassium transport in bacteria, p. 85-130. In B. Rosen and S. Silver (ed.), Ion transport in prokaryotes. Academic Press, Inc., New York, N.Y.
- 27.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]