Abstract
The secretion of PlcH and its homolog PlcN of Pseudomonas aeruginosa through the inner membrane depends upon a functional twin arginine translocase (Tat) system and a Tat signal sequence. Conserved twin arginine (Arg) residues within the Tat signal sequence consensus motif (S/TRRxFLK) are considered essential for the secretion of Tat substrates, but some exceptions (e.g., Lys and Arg) to the twin Arg residues in this motif have been noted. The roles of all three Arg residues within the PlcH RRRTFLK consensus motif were examined. Data are presented which indicate that Arg-9 and Arg-10 are essential for PlcH secretion across the inner membrane, but the mutation of Arg-8 (e.g., to Ala or Ser) had no observable effect on the localization of PlcH. In the signal sequence of PlcH and in all of its homologs in other bacteria, there are basic amino acid residues (Arg, Lys, and Gln) immediately adjacent to the signal peptidase cleavage site (Ala-X-Ala) that are not seen in Sec-dependent signal sequences. The mutation of these basic residues to Ala caused slightly decreased levels of extracellular PlcH, but normal localization was still observed. Deletion of the entire Tat signal sequence of PlcH not only resulted in the absence of detectable extracellular PlcH activity and protein but also caused a substantial decrease in the detectable level of plcH mRNA. Finally, data are presented which indicate that P. aeruginosa PlcH exhibits cross-species compatibility with the Escherichia coli Tat secretion machinery, but only when the E. coli Tat machinery is expressed in a P. aeruginosa host.
In bacteria, the translocation of proteins across membranes can proceed by several different routes. While the general secretory (Sec) pathway is responsible for the translocation of the majority of bacterial proteins, a second system for transport across the inner membrane, termed the twin arginine transport (Tat) system, was more recently discovered (43). Originally identified in the thylakoid membranes of chloroplasts (43), homologs of the Tat system have been subsequently characterized in numerous bacterial species (39, 52) as well as archaea (7, 25). Unlike the Sec system, which uses the energy of ATP hydrolysis to transport proteins in an unfolded conformation, the Tat system is a ΔpH dependent, ATP independent protein transport system operating in parallel to Sec in translocating folded proteins as well as protein complexes across the inner membrane (2, 16, 30, 37).
In Escherichia coli, three genes, tatA, tatB, and tatC, coding for components of the Tat translocon, have been shown to play a role in Tat translocation (2, 6, 40, 51). Proteins that are destined for transport via the Tat pathway are targeted to the translocon by an N-terminal signal peptide displaying the same tripartite organization as Sec signal sequences: a positively charged N region, followed by a hydrophobic H region and a polar C terminus preceding the signal peptidase cleavage site. In contrast to Sec leader sequences, Tat leaders possess two conserved arginine (Arg) residues in a consensus motif (S/T)-R-R-X-F-L-K within the positively charged N region (1).
The mutagenesis of these twin Arg residues either diminishes or abrogates the secretion of numerous Tat substrates (11, 15, 45). Recent studies, however, have shown that a conservative substitution of the first Arg for a lysine (Lys) still allowed transport through the Tat system (45), and a naturally occurring Tat signal peptide lacking one of the invariant twin Arg residues was recently found (20). This suggests that the Tat system is tolerant of some deviation from the twin Arg motif. The central H region of Tat signal peptides is less hydrophobic than the same region in Sec signal peptides, and this hydrophobicity was shown to play a role in the specificity of substrates for either the Tat or the Sec pathway of secretion (15). The more polar C terminus preceding the signal peptidase cleavage site contains basic residues which are absent from Sec signal sequences, and these have been shown to target Tat substrates exclusively to the Tat system, functioning as a “Sec avoidance” motif in thylakoids (5, 15). This may also be the case for bacterial Tat signal sequences, since the introduction of positively charged residues into the C-terminal region of Sec leaders slows down the transport of Sec substrates (45).
Most Tat substrates that have been examined to date are periplasmic (P) proteins that participate in redox processes as well as anaerobic growth (1). Recently, Voulhoux et al. (50) demonstrated that the opportunistic pathogen Pseudomonas aeruginosa possesses a functional Tat secretion system and that the heat-labile hemolytic phospholipase C (PlcH) and the nonhemolytic phospholipase C (PlcN), two known extracellular virulence factors of P. aeruginosa, (34, 48) depend upon the Tat system's transport of these extracellular enzymes across the inner membrane. This is the first report of extracellular proteins that rely on both the Tat secretion system and the extracellular secretion machinery of P. aeruginosa (Xcp) for transport.
The N-terminal leader sequence of PlcH in P. aeruginosa and its homologs in other bacteria (46) resemble those of most Tat-secreted proteins in that they are typically longer (35 amino acids) and more positively charged (3+ to 6+) than Sec-dependent signal sequences, and they contain the typical twin Arg residues within the so-called Tat consensus motif. However, in contrast to most Tat substrates, which contain either a serine (Ser) or a threonine (Thr) at the first position within the consensus motif (1), PlcH contains an additional Arg in this region (R-R-R-T-F-L-K). Furthermore, unlike typical Tat leader sequences, which contain only one positively charged residue prior to the Ala-x-Ala cleavage site, the PlcH leader sequence contains two Arg residues. These observations led us to examine the role of these multiple positively charged residues within the Tat signal peptide consensus motif and within the C-terminal region of the signal peptide. We also examined the localization of PlcH, which is devoid of either a Tat or a Sec signal sequence. Finally, we evaluated whether the signal peptide of PlcH in P. aeruginosa is compatible with a Tat secretion system from another bacterium, E. coli.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa PAO1 is from the collection of Michael Vasil, and it was originally obtained from Bruce Holloway and has been stored at −70°C continuously. Bacterial strains were grown in Luria Bertani (LB) medium, M9 minimal medium, or HEPES minimal medium. Casamino Acids medium was used for pyoverdine detection as previously described (33). For the overexpression of PlcH from the T7 overexpression system, bacterial strains were grown in LB medium at 37°C overnight. Cultures were passaged in M9 minimal medium to an optical density at 590 nm (OD590) of 0.5 and grown at 37°C for 1 h. For T7 induction, isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a 1 mM concentration. For the induction of chromosomal expression of PlcH, bacterial cultures were grown in HEPES minimal medium supplemented with 20 mM K2HPO4 and 0.2% (wt/vol) choline at 37°C for 12 h as previously described (14). Where needed, the following antibiotics were added at the indicated concentrations: ampicillin, 100 μg/ml; gentamicin, 15 μg/ml; and tetracycline, 100 μg/ml for E. coli and carbenicillin, 750 μg/ml; tetracycline, 100 μg/ml; gentamicin, 75 μg/ml; and irgasan, 25 μg/ml for P. aeruginosa.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototroph, chl-2 | 23 |
| ADD1976 | Prototroph, chl-2 Tcr | 8 |
| ADD1976ΔtatC | ΔtatC::Gm | This study |
| ADD1976ΔtatC PlcH | ΔtatC::Gm::plcH | This study |
| ADD1976ΔtatC PlcH R34A | ΔtatC::Gm::plcHR34A | This study |
| ADD1976ΔtatC PlcH R35A | ΔtatC::Gm::plcHR35A | This study |
| ADD1976ΔtatC PlcH R34AR35A | ΔtatC::Gm::plcHR34AR35A | This study |
| PAO1 ΔHRN | ΔplcH ΔplcN ΔplcR1 ΔplcR2 | 44 |
| PAO1ΔHRN PlcH | ΔplcH ΔplcN ΔplcR1 ΔplcR2::plcH | This study |
| PAO1ΔHRN PlcH R9K | ΔplcH ΔplcN ΔplcR1 ΔplcR2::plcHR9K | This study |
| PAO1ΔHRN PlcH R10K | ΔplcH ΔplcN ΔplcR1 ΔplcR2::plcHR10K | This study |
| PAO1ΔtatC | ΔtatC::Gm | 33 |
| PAO1Δtat operon | ΔtatA ΔtatB ΔtatC ΔPA5071 | This study |
| E. coli | ||
| BL21-DE3(pLysS) | hsdS gal ompT rB− mB− (cIts857 ind1 Sam7 nin5 lacUV5 T7) | 47 |
| Plasmids | ||
| pUCP19 | Cbr pUC derivative, P. aeruginosa origin of replication | 8 |
| pUC18plcH | plcHR1R2 3.2-kb BamHI-SphI | 46 |
| pADD3268 | Broad host range, Apr Cbr T7 promoter | 8 |
| pADDplcH | plcHR 3.2-kb BamHI/HindIII | 46 |
| pADDplcHR8K | PlcH R8K point mutation | This study |
| pADDplcHR8S | PlcH R8S point mutation | This study |
| pADDplcHR8A | PlcH R8A point mutation | This study |
| pADDplcHR9K | PlcH R9K point mutation | 33 |
| pADDplcHR10K | PlcH R10K point mutation | This study |
| pADDplcHR34A | PlcH R34A point mutation | This study |
| pADDplcHR35A | PlcH R35A point mutation | This study |
| pADDplcHR34R35A34A35 | PlcH R34AR35A double mutation | This study |
| pADDplcHΔss | PlcH Δaa 1-35 signal sequence mutation | This study |
| pUCP19E.c tat | Cbr pUC derivative, Pseudomonas origin of replication 2-kb SpeI E. coli tatABC | This study |
| pUCP19P.a tat | Cbr pUC derivative, Pseudomonas origin of replication 4-kb PstI P. aeruginosa tatABC | This study |
General genetic procedures.
All PCRs were performed using Taq polymerase and custom-made primers (Integrated DNA Technologies) in a Genemate thermal cycler (ISC Bioexpress) with 30 cycles of denaturing (1 min, 95°C), annealing (1 min 10 sec, 55°C), and extension (2 min, 72°C), followed by a 10-min extension at 72°C. The PCR products were purified via the Marligen rapid gel extraction system (Marligen Bioscience Inc., Ijamsville, MD), cloned into pCR2.1 TOPO (Invitrogen), and sequenced with T7 and M13 primers by the University of Colorado Health Sciences sequencing facility.
Construction of signal sequence mutant overexpression strains.
All PlcH signal sequence mutants were constructed by site-directed mutagenesis using PCR and the overlap extension method (24) as previously described. For PCR amplification, plasmid pUC18plcH, which contains the 4-kb BamHI-SphI DNA fragment encompassing the plcHR1,2 operon, was used as the template. Primers used for the construction of each mutant are listed in Table 2. PCR products were cloned into pCR2.1 TOPO and verified for correct mutant construction by sequencing. Once confirmed, a 543-bp BamHI-NcoI fragment containing the mutation was ligated into the pUC18plcH construct digested with BamHI and NcoI, replacing the 543-bp wild-type fragment. The pUC18plcH signal sequence mutant clone was resequenced for final construct verification, subcloned into pADD3268, and transformed into ADD1976 for the overexpression from the lacUV5 IPTG inducible promoter (8, 14). Alternatively, the construct pADDplcH and the signal sequence mutant constructs pADDplcHR34A, pADDplcHR35A, and pADDplcHR34AR35A were transformed into the ADD1976ΔtatC strain, which was generated by the allelic exchange of an internal tatC gene fragment with a gentamicin cassette as previously described (22, 42).
TABLE 2.
Primers used in this study
| Primer | Primer sequence | Characteristics |
|---|---|---|
| PAMS F002 | 5′-AGG CAC CCC AGG CTT TAC AC-3′ | External sense primer for site-directed mutagenesis of all signal sequence mutants |
| PAMS R002 | 5′-GTA GAG GCG GTT CGG ATT GG-3′ | External antisense primer for site-directed mutagenesis of all signal sequence mutants, antisense primer for RT-PCR of plcHΔss mutant |
| PLCHR | 5′-CTT CAC TGA TCG CCT GGT TC-3′ | Reverse primer for qPCR studies |
| PLCHF | 5′-CAG TTG AAC ATC CGC AAT CTC-3′ | Forward primer for qPCR studies |
| R8K-1 | 5′-TGG AAA TTC AAG CGT CGA ACC-3′ | Sense primer to generate the plcHR8K mutant |
| R8K-2 | 5′-GGT TCG ACG CCT GAA TTT CCA-3′ | Antisense primer to generate the plcHR8K signal sequence mutant |
| R8S-1 | 5′-TGG AAA TTC TCG CGT CGA ACC-3′ | Sense primer to generate the plcHR8S signal sequence mutant |
| R8S-2 | 5′-GGT TCG ACG CGA GAA TTT CCA-3′ | Antisense primer to generate the plcHR8S signal sequence mutant |
| R8A-1 | 5′-TGG AAA TTC GCC CGT CGA ACC-3′ | Sense primer to generate the plcHR8A signal sequence mutant |
| R8A-2 | 5′-GGT TCG ACG GGC GAA TTT CCA-3′ | Antisense primer to generate the plcHR8A signal sequence mutant |
| R10K-1 | 5′-TTC CGC CGT AAG ACC TTT CTC-3′ | Sense primer to generate the plcHR10K signal sequence mutant |
| R10K-2 | 5′-GAG AAA GGT CTT ACG GCG GAA-3′ | Antisense primer to generate the plcHR10K signal sequence mutant |
| R34A-1 | 5′-GAG ACG CTC GCG CGC GCC CTG-3′ | Sense primer to generate the plcHR34A signal sequence mutant |
| R34A-2 | 5′-CAG GGC GCG CGC GAG CGT CTC-3′ | Antisense primer to generate the plcHR34A signal sequence mutant |
| R35A-1 | 5′-ACG CTC CGG GCG GCC CTG GCC-3′ | Sense primer to generate the plcHR35A signal sequence mutant |
| R35A-2 | 5′-GGC CAG GGC CGC CCG GAG CGT-3′ | Antisense primer to generate the plcHR10K signal sequence mutant |
| R3435A-1 | 5′-ACG CTC GCG GCG GCC CTG GCC-3′ | Sense primer to generate the plcHR34AR35A signal sequence mutant |
| R3435A-2 | 5′-GGC CAG GGC CGC CGC GAG CGT-3′ | Antisense primer to generate the plcHR34AR35A signal sequence mutant |
| PlcH ss-1 | 5′-CAT ATG GTC GAG CCG GAC ATC-3′ | Sense primer to generate the plcHΔss mutant (engineered NdeI site is underlined) |
| PlcH ss-2 | 5′-CAT ATG TTT ATT TCC CGG GTG-3′ | Antisense primer to generate the plcHΔss mutant (engineered NdeI site is underlined) |
| PAMS SEQ3 | 5′-ACG AGT TCT CCC CCT ATC AC-3′ | Sense primer for RT-PCR of plcHΔss mutant |
| OmlA-1 | 5′-TGA TAG ACC AGT TGC GTC CCG-3′ | Sense primer for RT-PCR of omlA |
| OmlA-2 | 5′-TGC TGC CCT CCT TGC CGA G-3′ | Antisense primer for RT-PCR of omlA |
| TAT 6518 | 5′-CGG CCT GAA CCT ACA CAT TGC-3′ | Sense primer to confirm Δtat mutant |
| TAT 8931 | 5′-CAT TAC TCC AGG CAG TGC GAC-3′ | Antisense primer to confirm Δtat mutant |
Generation of chromosomally expressed PlcH and PlcH signal sequence mutants.
The P. aeruginosa ΔHRNplcH and ΔHRNplcH signal sequence mutants were constructed according to the method of Hoang et al. (22) by using a mini-CTX2 vector and the site-specific integration of the construct at the attB site on the chromosome of ΔHRN. Briefly, a 3,120-bp fragment encompassing the plcH operon with either the wild-type plcH gene or the signal sequence mutants was cloned into the BamHI/HindIII digested mini-CTX2 integration vector and constructs were transformed into E. coli SM10. Constructs were transferred into the recipient P. aeruginosa ΔHRN strain, a triple knockout strain lacking the plcH, plcR, and plcN genes via biparental mating (42). Generation of the unmarked mutant was achieved by Flp recombinase from the pFLP2 plasmid (21, 22). Constructs were confirmed by PCR.
Generation of the PlcHΔss mutant.
The pUC18plcH construct was used as a template for PCR amplification of a 230-bp plcH product with primers PlcH ss-1 and PAMS F002 (Table 2), engineering an NdeI restriction site at the 3′ end of the product. Primers PlcH ss-2 and PAMS R002 (Table 2) were used to generate a 438-bp plcH product with an NdeI site engineered at the 5′ end of the product. Both products were cloned into the pCR2.1 TOPO vector to generate pCR2.1-230 and pCR2.1-438, respectively, and confirmed by sequencing. The pCR2.1-230 construct was digested with BamHI and NdeI to release a 230-bp product, which was ligated into the NdeI/BamHI-restricted pCR2.1-438 construct, generating pCR2-1-1. This construct was confirmed by sequencing. The pCR2-1-1 construct was digested with BamHI and NcoI, and the 565-bp product was ligated into the BamHI/NcoI-digested pUC18plcH construct to generate pUC18plcHΔss. This PlcHΔss mutant lacks the first 38 amino acids of the N terminus of PlcH except for the initial Met residue. That is, the methionine residue is now adjacent to the Val residue of the mature PlcH (see Fig. 1). The pUC18plcHΔss construct was digested with BamHI and HindIII and ligated into BamHI/HindIII-digested pADD3268 to generate pADDplcHΔss. This construct was transformed into ADD1976 to generate the strain ADD1976 PlcHΔss.
FIG. 1.
Schematic of the signal sequence of P. aeruginosa wild-type PlcH and PlcH signal sequence mutant proteins used in this study. The twin Arg consensus motif is in boldface type. The C-terminal basic residues prior to the Ala-x-Ala cleavage site are underlined. The signal peptidase cleavage site is shown by an arrow.
Generation of the PAO1Δtat operon mutant.
Construction of the PAO1Δtat knockout mutant was performed by the gene replacement strategy of Schweizer and Hoang (42). A gentamicin (Gmr) resistance cassette obtained from pPS856 (22) replaced the tatA, tatB, tatC, and PA5071 genes within the pUCP19P.a tat construct, generating pUCP19tatGmR. This new construct was ligated into pEX100T (42), transformed into E. coli SM10, and used for an allelic exchange in a biparental mating with P. aeruginosa PAO1. Gene replacement was verified by PCR across the relevant region. The generation of the PAO1 tat operon unmarked mutant was achieved by Flp recombinase from the pFLP2 plasmid (21, 22, 42). Constructs were confirmed by PCR.
Preparation of bacterial fractions.
The fractionation of cells into the cytosolic (C), cytoplasmic membrane (inner membrane [IM]), P, and outer membrane (OM) fractions was done according to the method of Mizuno and Kageyama (29) with slight modifications. Bacterial cultures were grown in M9 minimal medium and induced by the addition of 1 mM IPTG for 1 to 3 h. After induction, cultures were collected by centrifugation at 10,000 rpm in a JA17 rotor (Beckman Instruments) for 15 min at 4°C. Culture supernatants were frozen. For chromosomal inductions of PlcH, supernatants were treated with trichloroacetic acid to precipitate PlcH. Cell pellets were washed with ice-cold 20% (wt/vol) sucrose and recollected by centrifugation. The washed cells were weighed and resuspended in 18 ml per 1.5 g of cells in ice-cold 20% (wt/vol) sucrose. Cells were kept on ice, and reagents were added, based on 1.5 g of cells, in the following order: 9 ml 2 M sucrose, 10 ml 0.1 M Tris-HCl (pH 7.8), 0.8 ml 15 M Na-EDTA (pH 7.0), and 1.8 ml fresh 0.5% (wt/vol) lysozyme. Cells were warmed to 30°C for 30 min. DNase was added to a concentration of 6 μg/ml, and cells were returned to 30°C for an additional 30 min. The resulting spheroplasts were collected by centrifugation at 13,000 rpm in a JA17 rotor (Beckman Instruments) for 20 min. The supernatant containing the OM and P fractions was ultracentrifuged at 35,000 rpm for 1 h at 4°C in a Ti70.1 fixed-angle rotor (Beckman Instruments). The supernatant contained the P fraction, while the pellet contained the OM fraction. The OM fraction was resuspended in phosphate-buffered saline (PBS) to 1:10 of the original pellet weight. Spheroplasts were washed in 20% (wt/vol) ice-cold sucrose and recentrifuged at 13,000 rpm in a Ti70.1 (Beckman Instruments) rotor for 10 min at 4°C. Spheroplasts were resuspended in PBS and boiled immediately, and an equal volume of 2× sample buffer was added to each spheroplast resuspension. Samples were boiled again, loaded, and run on a 6% sodium dodecyl sulfate-polyacrylamide electrophoresis gel (SDS-PAGE). Alternatively, spheroplasts were disrupted via the addition of 40 ml 5 mM MgCl2 per 1.5 ml of cells and the IM fraction was separated from the cytosolic fraction by centrifugation at 15,000 rpm in a JA17 rotor (Beckman Instruments) for 30 min at 4°C. The supernatant contained the C fraction, while the pellet contained IM fraction. The IM fraction was resuspended in PBS to 1:10 of the original pellet weight. Supernatants and bacterial fractions corresponding to an identical number of cells were subjected to SDS-PAGE and Western blotting using anti-PlcH monoclonal antibodies.
Enzyme assays.
Purity of bacterial fractions was measured via the nitrocefin [3-(2,4-dinitrostyryl)-(6R,7R)-7-(2-thienylacetamido)-ceph-3-em-4-carboxylic acid, E-isomer] assay (Calbiochem, San Diego, CA). Outer membrane, periplasmic, inner membrane, and cytosolic fractions were mixed with nitrocefin (dissolved in dimethyl sulfoxide and 0.1 M phosphate buffer [pH 7.0]) and 0.1 M phosphate buffer. An OD490 of each sample was taken at 5 min. β-Lactamase activity is presented as percent activity in each fraction of the total β-lactamase activity in all the bacterial fractions. For PlcH enzymatic assays, the artificial substrate p-nitrophenyl-phosphorylcholine (NPPC), purchased from Sigma, was used as previously described (27). Reactions were carried out in 100 mM Tris-HCl (pH 6.8), 25% (vol/vol) glycerol with 3.1 mg/ml NPPC at 37°C (46). Values were measured as relative units to wild-type PlcH activity, which was set at 1 or as relative units = 100 × ΔOD405/Δtime and recorded as the percentage of total NPPC activity overall for the bacterial fractions and supernatant. The assay for NADH oxidase activity was performed essentially as described by Cheng et al. (13). Briefly, NADH oxidase activity was measured as a decrease in absorbance at 340 nm of each bacterial fraction sample mixed with 4 × 10−4 NADH (wt/vol) in 0.05 M Tris-HCl buffer (pH 7.6). Activity was recorded as change in absorbance over time and expressed as percentage of total activity for all the bacterial fractions.
Gel electrophoresis and Western immunoblot conditions.
SDS-PAGE was carried out according to the method of Laemmli (28). Supernatant samples and all bacterial fractions except inner membranes were loaded and run on 12% SDS-PAGE gels. Inner membrane fractions and spheroplasts were loaded and run on 6% SDS-PAGE gels. Protein samples were then transferred onto nitrocellulose (Optitran; Schleicher and Schuell, Inc.) and probed with PlcH monoclonal antibody at a 1:500 dilution. Horseradish peroxidase-conjugated goat anti-mouse antibody was used as the secondary antibody at a 1:25,000 dilution, and bands were detected using the ECL Plus Western blot detection kit (Amersham Pharmacia Biotech). For quantitative analysis, films were scanned and imported into Adobe Photoshop for the adjustment of brightness and contrast (Adobe Systems, Inc., San Jose, Calif.). Quantitative analysis was performed with NIH ImageJ software, version 1.55. A standard curve of purified PlcH was generated and used to calculate the amount of secreted PlcH in the PlcH signal sequence mutants. For the confirmation of E. coli Tat protein expression, rabbit polyclonal sera against E. coli TatA, E. coli TatB, and E. coli TatC, provided by Timothy Yahr, were used at a 1:1,000 dilution. Goat anti-rabbit secondary antibody was used at a 1: 25,000 dilution.
Complementation of the PAO1Δtat mutant with E. coli tat operon.
The E. coli pBADtatABC construct (53) was digested with NcoI and XbaI to obtain the 2-kb tatABC fragment, which was ligated into the pUCP19 plasmid, generating the pUCP19tatABC construct. This construct was transformed into the PAO1 Δtat mutant strain as well as the previously constructed PAO1 ΔtatC mutant (33).
Preparation of RNA and real-time quantitative RT-PCR.
Total bacterial RNA was isolated from P. aeruginosa ADD1976 and the signal sequence mutant grown in M9 minimal medium. The wild type and the mutant were grown to an OD590 of 1.0 and induced with IPTG. Samples of the wild-type culture and the mutant culture were taken just before induction and at 1, 7, 15, and 30 min after induction and immediately treated with RNAlater (Ambion, Inc.), and RNA was isolated by the QIAGEN RNeasy mini kit according to the manufacturer's instructions. The RNA preparations were treated with RNase-free DNase (Promega) for 1 h, and samples were treated with the QIAGEN RNeasy mini kit. RNAs were used as a template in reverse transcription (RT)-PCRs by utilizing the SuperScript one-step RT-PCR system (Invitrogen) generally according to the manufacturer's instructions. Real-time quantitative RT-PCR (qPCR) was performed by the TaqMan assay in a Cephid Smart Cycler II (Sunnyvale, CA) by using the plcH probe 5′/56-6-carboxyfluorescein/ACGGTG ACG CCG AAC CCG GCC TA/BBHQ_1/-3′ that was synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The constitutively expressed omlA gene was measured in all wild-type and mutant samples to confirm that the same amount of RNA was used in each reaction for the qPCR experiments (Table 3). Predictions of the plcH mRNA signal sequence secondary structure were done using the M-fold program (54).
TABLE 3.
Real-time quantitative RT-PCR analysis of plcH wild type and the plcHΔss mutant
| Time after IPTG induction (min) | Relative abundancea of:
|
|
|---|---|---|
| plcH wild-type signal sequence | plcHΔss mutant | |
| 0 | 362 | .007 |
| 1 | 371 | .012 |
| 7 | 881 | .013 |
| 15 | 1,028 | .031 |
| 30 | 1,256 | .036 |
Values are expressed as relative abundance of plcH wild-type or plcHΔss transcripts compared to that for the omlA transcripts. omlA is a constitutively expressed gene.
RESULTS
PlcH R10K signal sequence mutant is mislocalized to the inner membrane.
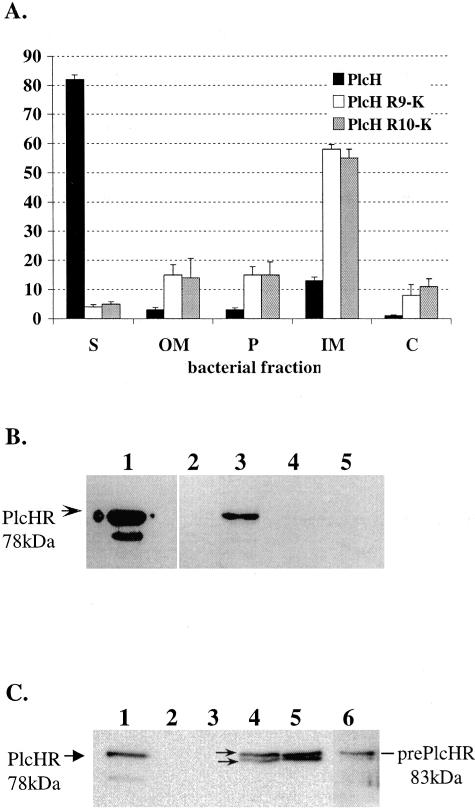
As mentioned above, PlcH has three tandem Arg residues instead of the typical twin Arg consensus motif of Tat substrates. Consequently, it is possible that any or all of these Arg residues are required for the secretion of PlcH through the inner membrane. To begin, the third Arg residue (Arg-10) within the Tat consensus motif of PlcH was mutated to Lys in order to generate the PlcH R10K mutant (Fig. 1). The wild type, the previously generated R9K mutant, and the newly constructed R10K mutant were expressed in the P. aeruginosa PAO1-derived ADD1976 T7 overexpression system from the IPTG-inducible T7 promoter of plasmid pADD3268. All constructs also expressed the PlcR1 and PlcR2 chaperone proteins that were previously shown to be important in PlcH secretion (14). Cultures were grown in M9 minimal medium and induced with IPTG, and supernatants and bacterial fractions were examined for PlcH by activity on the synthetic substrate NPPC and by immunoblot analysis as described in Materials and Methods. In this expression system, no PlcH activity or protein was detected from the ADD1976 with vector alone (Fig. 2B, lane 2). The induction of wild-type PlcH in the ADD1976 expression system resulted in high levels of extracellular PlcH, as shown by NPPC activity (Fig. 2A). Immunoblot analysis revealed a single 78-kDa form of the PlcH protein (Fig. 2B, lane 3), corresponding to the processed PlcH that was observed for the positive control (purified PlcH enzyme) (Fig. 2B, lane 1). The smaller band in the positive control lane of this and all subsequent immunoblots represents nonspecific degradation products of PlcH. In contrast, the PlcH R10K mutant showed almost no NPPC activity in culture supernatants (Fig. 2A) and PlcH protein was undetectable in that fraction by immunoblot analysis (Fig. 2A and B, lane 5). As shown previously, the PlcH R9K mutant also showed almost no extracellular PlcH activity (Fig. 2A) or PlcH protein (Fig. 2B, lane 4). These results indicate that Arg-10 within the twin Arg motif of the PlcH signal sequence is also critical for PlcH secretion.
FIG. 2.
The PlcH R9K and PlcH R10K signal sequence mutants mislocalize to the inner membrane. (A) PlcH activity in supernatants (S) and bacterial fractions. Error bars indicate standard errors of the means of three independent experiments. (B and C) Total PlcH protein in supernatants (B) and inner membrane fractions (C). Lane 1, purified PlcH enzyme, used as size marker for mature PlcH (the lower band represents nonspecific degradation product); lane 2, ADD1976 with the vector; lane 3, ADD1976 with wild-type PlcH; lane 4, ADD1976 with PlcH R9K mutant; lane 5, ADD1976 with PlcH R10K mutant; lane 6, ADD1976 with PlcH R9K spheroplast. The arrows and arrowhead in lanes 1 (B and C) and 4 (C) show the different forms of PlcH.
Since the extracellular secretion of both the PlcH R9K and PlcH R10K mutants was arrested within the bacterial cell, we examined other bacterial compartments to determine PlcH localization. Bacterial pellets were fractionated into outer membrane, periplasm, inner membrane, and cytosolic fractions, and the location of PlcH was evaluated by the detection of NPPC activity and immunoblot analysis. The purity of bacterial fractions for all fractionation protocols was assessed by measuring the activity of the control proteins β-lactamase, a periplasmic protein, and NADH oxidase, an inner membrane protein, for all of the bacterial fractions. Control protein localization was determined by a nitrocefin assay (for lactamase) and an NADH oxidase assay as described in Materials and Methods. An analysis of these enzymes indicated that the separation of the cellular compartments (e.g., periplasm and inner and outer membranes) was accomplished as previously described (data not shown). An examination of the bacterial compartments for the presence of PlcH activity and PlcH protein revealed that the majority of PlcH produced by the PlcH R9K and PlcH R10K mutants localized to the inner membrane (Fig. 2A and C). Immunoblot analysis of the inner membrane fraction demonstrated two forms of the PlcH protein present in this fraction, the smaller band corresponding to the faster migrating, 78-kDa mature protein lacking the signal peptide and the slower band corresponding to the 83-kDa prePlcH precursor form, which likely still contained the twin Arg signal peptide (Fig. 2C, lanes 4 and 5).
The presence of both forms of PlcH was somewhat surprising, since it suggested that the PlcH R9K and PlcH R10K mutants were being translocated across or into the inner membrane and were partially processed by a signal peptidase. On the other hand, the observed PlcH processing in the inner membrane fraction could be an artifact of the fractionation protocol. To distinguish between these two possibilities, we expressed the PlcH R9K mutant and isolated bacterial spheroplasts containing both the inner membrane and cytosolic fractions as described in Materials and Methods. Spheroplasts were immediately boiled to allow lysis and minimize any artificial processing of PlcH by the signal peptidase. Boiled samples were loaded immediately on an SDS-PAGE gel, followed by Western immunoblotting. Since the cytosolic fraction generated through the fractionation protocol contained negligible amounts of PlcH, as shown by both detection of NPPC activity and Western blot analysis (Fig. 2A and data not shown), any PlcH detected in the spheroplasts would presumably be localized in the inner membrane. An examination of the PlcH R9K spheroplasts for PlcH revealed mainly the unprocessed, 83-kDa form, with very little of the processed 78-kDa form (Fig. 2C, lane 6). This suggests that PlcH is not processed in the spheroplast, but instead, processing from the 83-kDa form to the 78-kDa form occurs only when the spheroplasts are further fractionated into the inner membrane and cytosolic fractions (Fig. 2C, lanes 4 and 5) and is a consequence of the fractionation protocol. Taken together, these results suggest that, unlike most Tat substrates not secreted through Tat that are found in the cytoplasm, the PlcH R9K and PlcH R10K mutants associate with the inner membrane and this association is independent of either Arg residue within the Tat leader peptide. This finding contrasts with results found for most E. coli Tat substrates impaired in Tat secretion, which localize to the cytoplasm (1).
Overexpression from the P. aeruginosa T7 system does not affect PlcH localization.
One caveat to the protein expression from the ADD1976 T7 overexpression system is that induction leads to very high levels of protein, potentially leading to artificial mislocalization of PlcH, due to the blockage of the Tat secretion system. We complemented a previously generated PlcH mutant strain (ΔHRN) with either the wild-type PlcH or the PlcH R9K or PlcH R10K mutants in single copy under the native PlcH promoter in the P. aeruginosa chromosome to determine whether the overexpression of PlcH from the ADD19767 T7 expression system affects the localization of the PlcH protein. PlcH localization of the wild type and mutants reflected that found with the overexpression data, with the majority of wild-type PlcH localized to the supernatant and the majority of PlcH R9K and PlcH R10K localized to the inner membrane (data not shown). These results indicate that overexpression of either the wild-type PlcH or the PlcH R9K and PlcH R10K mutants from the T7 promoter does not substantially affect the final localization of PlcH.
The Arg-8 residue does not play a role in the localization of PlcH.
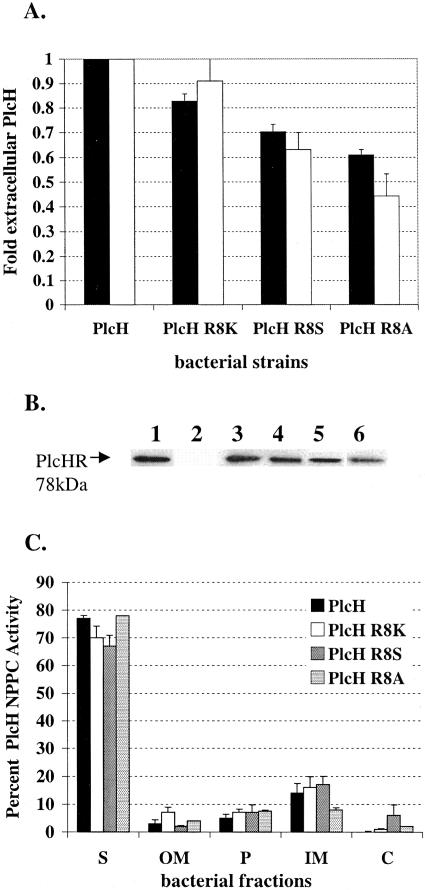
The (S/T)-R-R-x-F-L-K consensus motif of a typical Tat leader sequence in E. coli and several other bacterial species contains a serine or threonine in the position just prior to the twin Arg. This is not the case in the P. aeruginosa PlcH signal sequence, where the position just prior to the twin Arg residues is occupied by another Arg, which is a much larger, charged amino acid (Fig. 1). We substituted this residue for a Lys, a Ser, or an Ala by the overlap extension method, generating the PlcH R8K, PlcH R8S, and PlcH R8A signal sequence mutants to assess whether this Arg plays a role in the secretion of PlcH. PlcH R8K is a conserved substitution; the PlcH R8S substitution converts the consensus motif of PlcH to a more typical E. coli Tat consensus motif (S/TRRxFLK) and PlcH R8A is a nonconserved substitution. As shown in Fig. 3A, the induction of the wild-type PlcH and the R8K, R8S, and R8A mutants in the ADD1976 overexpression system resulted in high levels of extracellular PlcH NPPC activity for the wild-type strain. In contrast, all R8 mutants showed decreased NPPC PlcH activity in the supernatants relative to the wild type. That is, secretion of the R8K mutant was ∼80% of that of the wild-type PlcH activity, the PlcH R8S mutant was ∼70% of that of the wild-type activity, and the relative activity of R8A mutant was ∼60% of that of the wild-type PlcH (Fig. 3A). Immunoblot analysis of the supernatants followed by quantitation of the each of the secreted PlcH proteins (Fig. 3A) reflected these results when PlcH activity was measured. That is, the protein level differences observed between the wild-type and mutant extracellular PlcH closely correlated with the NPPC activity differences observed (Fig. 3A, black bars versus white bars), suggesting that the enzymatic activities of these mutants were not affected by the substitutions.
FIG. 3.
The PlcH R8K, PlcH R8S, and PlcH R8A signal sequence mutants are not affected in secretion but produce less total PlcH. (A) PlcH in supernatants of ADD1976 expressing wild-type PlcH or the PlcH signal sequence mutants was measured by detecting NPPC activity (black bars) and by quantitation of total extracellular PlcH protein by immunoblot analysis (white bars). Wild-type PlcH NPPC activity (calculated as 100 × ΔOD590/Δtime) and total wild-type extracellular PlcH protein concentration were set to 1, and the PlcH activity and PlcH protein concentration detected in the supernatants of each mutant were compared to this standard. Error bars indicate standard errors of the means of three independent experiments. (B) Total PlcH protein in supernatants by immunoblot analysis. Lane 1, purified PlcH control; lane 2, ADD1976 with the vector; lane 3, ADD1976 with wild-type PlcH; lane 4, ADD1976 with PlcH R8K mutant; lane 5, ADD1976 with PlcH R8S mutant; lane 6, ADD1976 with PlcH R8A mutant. The arrow in lane 1 shows the twin Arg signal peptide. (C) Distribution of bacteria-associated PlcH activity. Bacterial pellets of ADD1976 expressing wild-type PlcH or PlcH signal sequence mutants were fractionated into OM, P, IM, and C as described in Materials and Methods. Error bars indicate standard errors of the means of three independent experiments. S, supernatant.
Since the PlcH R8 mutants demonstrated lower PlcH activity and protein levels in the supernatants, we examined other bacterial compartments to determine whether any PlcH was arrested within the bacterial cell. Bacterial pellets were fractionated into the outer membrane, periplasm, inner membrane, and cytosolic fractions as previously described, and the location of PlcH was evaluated by the detection of NPPC activity and immunoblot analysis. An evaluation of the percent distribution of total PlcH activity in the wild type and mutants revealed that pellet-associated PlcH activity constituted only a small percentage of the overall total PlcH activity for the wild type as well as the mutants, and no difference in the amount of PlcH activity that was detected in each fraction was observed between the wild type and mutants (Fig. 3C). These data suggest that the differences in extracellular PlcH that were observed between the mutants and the wild type are not likely due to an arrest of the protein in its translocation through the cellular compartments. The actual mechanism by which these levels of these Tat signal sequence mutants are affected will have to await further experimental analysis.
The C-terminal Arg residues (R34 and R35) do not play a role in localization of PlcH.
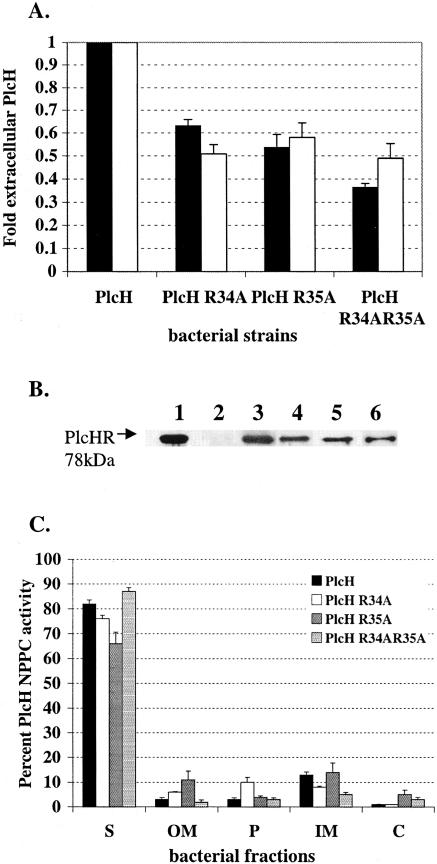
Residues outside the conserved twin Arg motif have been shown in previous studies to play a role in the proper localization of Tat-transported substrates. Of note, basic residues within the C-terminal region of the leader sequence have been implicated in the “Sec avoidance” phenomenon observed in thylakoids (5) and, more recently, in bacteria (3). Numerous E. coli Tat-secreted proteins contain a basic residue just prior to the Ala-x-Ala cleavage site within the leader sequence. In addition, an examination of the leader sequences of Tat-secreted phospholipase homologs from the PLC superfamily in P. aeruginosa PlcH indicates that they all contain either one or two highly conserved basic residues at the location just prior to the cleavage site. PlcH contains two basic residues, which are both Arg, at this position. We generated single PlcH R34A and PlcH R35A mutants as well as a double PlcH R34AR35A mutant via site-directed mutagenesis to examine the possible contribution of these basic residues to PlcH secretion. Mutants PlcH R34A and PlcH R35A maintain a single basic residue at the position prior to the cleavage site, while the double mutation PlcH R34AR35A abrogated both basic residues. Wild-type and mutant PlcH were expressed in the ADD1976 T7 overexpression system, and the supernatants were collected and tested for the presence of PlcH by the detection of NPPC activity and immunoblot analysis. As shown in Fig. 4A, the amount of PlcH activity detected in the mutants was lower than the amount of wild-type PlcH activity. The PlcH R34A and PlcH R35A mutants had ∼50 to 60% of the PlcH activity detected in the wild type, while the double PlcH R34AR35A mutant had ∼40 to 50% of wild-type PlcH activity in the culture supernatant. This finding was further confirmed by Western immunoblot analysis (Fig. 4B). Quantitation of the extracellular PlcH protein demonstrated that the supernatant of PlcH R34A mutant contained ∼50% of the PlcH seen in the wild type. The PlcH R35A mutant had ∼55% of the PlcH seen in the wild type, and the PlcH R34AR35A double mutant had ∼50% of the PlcH seen in the wild type (Fig. 4A). The protein level differences observed between the wild-type and mutant extracellular PlcH correlated with the NPPC activity differences that were observed (Fig. 4A, black bars versus white bars), indicating that the substitutions did not affect the enzymatic PlcH activity.
FIG. 4.
The PlcH R34A, PlcH R35AS, and PlcH R34R35A34A35 signal sequence mutants are not affected in secretion but produce less total PlcH. (A) PlcH in supernatants of ADD1976 expressing wild-type PlcH or the PlcH signal sequence mutants was measured by the detection of NPPC activity (black bars) and by the quantitation of total extracellular PlcH protein by immunoblot analysis (white bars). Wild-type PlcH NPPC activity (calculated as 100 × ΔOD590/Δtime) and total wild-type extracellular PlcH protein concentration were set to 1, and the PlcH activity and PlcH protein concentration detected in the supernatants of each mutant were compared to this standard. Error bars indicate standard errors of the means of three independent experiments. (B) Total PlcH protein in supernatants was detected by immunoblot analysis. Lane 1, purified PlcH enzyme (used as size marker); lane 2, ADD1976 with vector; lane 3, ADD1976 with wild-type PlcH; lane 4, ADD1976 with PlcH R34A; lane 5, ADD1976 with PlcH R35A; lane 6, ADD1976 with PlcH R34AR35A. The arrow and lane 1 show the twin Arg signal peptide. (C) Distribution of bacterium-associated PlcH activity. Bacterial pellets of ADD1976 expressing wild-type PlcH or the PlcH signal sequence mutants were fractionated into OM, P, IM, and C as described in Materials and Methods. Error bars indicate standard errors of the means of three independent experiments. S, supernatant.
To determine whether this decrease in extracellular PlcH was due to incomplete secretion from the bacterial cell, bacterial pellets were fractionated into the outer membrane, periplasm, inner membrane, and cytosolic fractions and each fraction was examined for PlcH by detection of NPPC activity and immunoblot analysis as described previously. As shown in Fig. 4C, an examination of the overall PlcH activity produced by the wild type and mutants indicated that the majority of PlcH produced was localized to the supernatant for both the wild type and mutant strains, with very little PlcH activity localized to any of the other bacterial fractions. This suggests that the lower extracellular PlcH levels that were observed in the mutants are due to lower total PlcH protein.
Previous studies have suggested that the removal of the charged residues within the C-terminal region of a Tat signal peptide eliminates the “Sec avoidance” motif and allows the recognition and secretion of the substrate by the Sec secretion machinery. To determine whether the secretion of PlcH in the C-terminal Arg mutants was being coordinated by the Sec secretion machinery instead of the Tat secretion machinery, we generated an ADD1976ΔtatC mutant that lacked a functional Tat secretion system but maintained a functional Sec secretion system and transformed this strain with the individual C-terminal Arg mutant pADD PlcHR34A, PlcHR35A, and PlcHR34AR35A constructs. Wild-type and mutant PlcH were expressed from the T7 overexpression system, and supernatants were examined for the presence of PlcH by detection of the NPPC activity and Western immunoblot analysis. No PlcH activity was detected in the supernatants of any of the C-terminal mutants (data not shown), indicating that the replacement of the Arg-34 and Arg-35 residues within the signal peptide does not reroute PlcH secretion to the Sec pathway.
Deletion of the PlcH signal sequence coding region affects plcH transcript levels.
A current model of Tat secretion based on previous studies in E. coli (9) proposes that Tat substrates associate with the inner membrane prior to their recognition and translocation by the Tat secretion apparatus. This association occurs by interaction of the Tat signal sequence with the inner membrane but is independent of the residues within the signal sequence that are required for Tat recognition, i.e., the twin Arg residues. In this study, we have shown that the membrane association of PlcH is independent of either Arg residue within the signal sequence, as PlcH R9K and R10K mutants are still able to associate with the inner membrane but are not secreted through the Tat secretion system (Fig. 2). In order to determine whether the entire PlcH signal sequence is required for the association with the inner membrane, we generated a PlcH mutant lacking the Tat signal peptide. Wild-type PlcH and the PlcH signal sequence mutant were expressed in the ADD1976 T7 overexpression system, and bacterial pellets were fractionated into the outer membrane, periplasm, inner membrane, and cytosolic fractions to determine the localization of PlcH. As shown in Table 4, wild-type PlcH activity was localized primarily to the supernatant, with some residual activity being detected in the other bacterial fractions, most likely due to protein overexpression causing inefficient translocation. In contrast, in the mutant devoid of the entire Tat signal sequence (see Materials and Methods) (Fig. 1), no PLC activity was detected in the supernatant. Moreover, an examination of the bacterial fractions (e.g., the periplasm and outer membrane) from this mutant for PLC activity or PlcH protein (e.g., immunoblot) revealed that neither was detectable in these compartments as well (data not shown and Table 4).
TABLE 4.
PlcH NPPC activity in bacterial fractionsa of the PlcH Δsignal sequence mutant
| P. aeruginosa strain | Activityb for:
|
|||||
|---|---|---|---|---|---|---|
| S | OM | P | IM | C | Total unitsc | |
| Vector control | 0.04 | 0.10 | 0.12 | 0.12 | 0.06 | 0.44 |
| PlcH | 7.24 | 0.54 | 0.80 | 2.08 | 0.16 | 10.82 |
| PlcH Δss | 0.04 | 0.08 | 0.07 | 0.08 | 0.14 | 0.40 |
All cell fractions were adjusted based on total bacterial pellet weight for each sample obtained prior to fractionation. S, supernatant.
Assayed by hydrolysis of the synthetic substrate NPPC and expressed as activity units ΔOD415/Δtime ×100.
Total PlcH NPPC activity units for each construct represent the sum total activity of all the fractions.
To determine whether the deletion of the entire Tat signal sequence affected plcH transcript levels, P. aeruginosa expressing wild-type PlcH or the signal sequence mutant PlcHΔSS were grown in M9 minimal medium and induced for PlcH expression with IPTG. Bacterial pellets from the wild-type and the PlcHΔSS mutant cultures were collected before induction (time zero) and at 1, 7, 15, and 30 min and 2 h postinduction, and bacterial RNA was extracted as described in Materials and Methods. The level of plcH transcript was determined by RT-PCR with primers that were internal to the plcH transcript. The constitutively expressed omlA (PA4765) gene was used as a control for the quality and amount of RNA between samples. With nonquantitative RT-PCR, a 259-bp plcH product indicative of a plcH transcript was detected at 30 min and 2 h after IPTG induction, which was indicative of robust wild-type plcH mRNA levels. In contrast, relative to wild-type PlcH, no product and a barely detectable product were observed by this method at 30 min and 2 h postinduction, respectively, for the signal sequence mutant (data not shown). Equivalent levels of omlA product were observed between samples. To confirm and extend these data, the RNA samples from before induction (time zero) and at 1, 7, 15, and 30 min were analyzed by qPCR as described in Materials and Methods. These data are presented in Table 3. Using this more-sensitive method, at the earlier time points, we could detect transcripts from the signal sequence mutant but the relative levels of the transcripts in the signal sequence mutant were >10,000 times (i.e., 33,161 times higher in the strain expressing wild-type PlcH than in the strain expressing PlcH without its signal sequence at 15 min) lower than the levels detected in the strain expressing wild-type PlcH. Collectively, these data indicate that the deletion of the signal sequence coding region severely affects the transcription of plcH since the levels of the mutant transcript were considerably reduced in comparison to that of the wild type at all time points. Nevertheless, it is clear that the transcription of the strain expressing the mutant transcript (plcHΔss) increased over time upon induction with IPTG, suggesting considerable instability of the mutant mRNA.
PlcH Tat signal sequence compatibility with the E. coli Tat secretion machinery is host dependent.
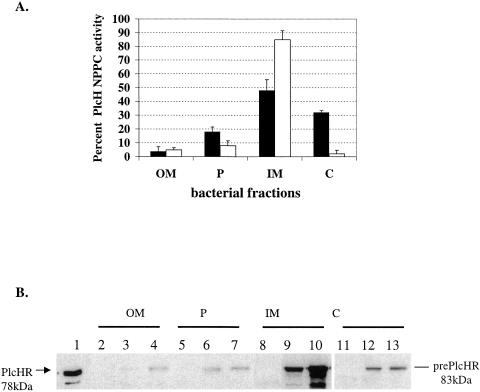
Several studies examining the compatibility between Tat signal sequences from heterologous hosts and the E. coli Tat transport system have been carried out. Examples have been found for full recognition of the heterologous signal sequence (32), leading to Tat transport of the substrate as well as noncompatibility between the signal sequence and E. coli Tat, leading to a sequestration of the Tat substrate in the cytosol (4, 35). Since our findings suggest that the PlcH Tat signal sequence resembles a typical E. coli Tat signal sequence, we tested whether the E. coli Tat translocation system can recognize and translocate the P. aeruginosa PlcH protein. Previous results have shown that PlcH is not secreted to the extracellular space when expressed in BL21 E. coli (DE3) cells (14). This result is not surprising because the E. coli strains used in that study lacked a functional Xcp homolog and were thus unable to transport PlcH through the outer membrane. We therefore hypothesized that, if the E. coli Tat translocation machinery were compatible with the PlcH Tat signal sequence, the protein would be transported through the inner membrane and localize to the periplasm. On the other hand, if the E. coli Tat machinery were incompatible with the PlcH signal sequence, PlcH would localize to either the inner membrane or the cytosol or both. We expressed wild-type PlcH as well as a ΔPlcR mutant (see below) that was previously shown to be deficient in secretion in BL21 (DE3) cells (14), fractionated the bacterial pellets into the outer membrane, periplasm, inner membrane, and cytosolic fractions, and localized PlcH by detection of NPPC activity and immunoblot analysis. As a control for PlcH localization, the BL21 ΔPlcR mutant, which contains the wild-type plcH gene but is deleted for the plcR1,2 genes, was used. Cota-Gomez et al. (14) previously showed that PlcH expressed in the absence of the PlcR chaperones formed an insoluble complex that was associated with the inner membrane. The PlcH delta R mutant strain was therefore included in this study as a control for the localization of a secretion-deficient PlcH when expressed in E. coli. The majority of wild-type PlcH activity (Fig. 5A) and PlcH protein (Fig. 5B) were localized to the inner membrane and cytosolic fractions, with a small percentage of activity and protein found in the periplasm and the outer membrane. PlcH delta R localized to the inner membrane as expected. Immunoblot analysis of the bacterial fractions detected only the larger 83-kDa unprocessed form of PlcH for both the wild-type and mutant fractions (Fig. 5B), suggesting that PlcH is not processed by the signal peptidase and that any localization to the periplasm and outer membrane is an artifact. These data indicate that PlcH expressed in E. coli is not transported through the Tat secretion machinery when overexpressed in E. coli and suggest that the E. coli Tat secretion system may not be compatible with PlcH secretion. However, it is possible that additional factors in addition to the Tat proteins are required for PlcH translocation and that these are absent when PlcH is expressed in E. coli. To examine this hypothesis in more detail, we asked whether PlcH could be recognized and transported by the E. coli Tat secretion machinery in the context of a P. aeruginosa host. Thus, any additional factors that are required for PlcH secretion would be provided by the host strain.
FIG. 5.
PlcH overexpressed in E. coli mislocalizes to the inner membrane and cytosol. BL21 (DE3) E. coli isolates overexpressing the P. aeruginosa PlcH were grown in LB medium and induced with 1 mM IPTG, and bacterial pellets were fractionated into the OM, P, IM, and C as described in Materials and Methods. (A) PlcH activity in bacterial fractions of E. coli expressing wild-type PlcH (black bars) or the PlcH delta R secretion-deficient mutant (white bars) was measured by the detection of NPPC activity and is expressed as the percentage of total PlcH activity measured in the bacterial pellet of each sample. Error bars indicate standard errors of the means of three independent experiments. (B) Immunoblot analysis of bacterial fractions. Lane 1, purified PlcH enzyme used as a size marker for the mature PlcH protein; lanes 2, 5, 8, and 11, E. coli expressing pGEM vector alone; lanes 3, 6, 9, and 12, E. coli expressing wild-type PlcH; lanes 4, 7, 10, and 13, E. coli expressing PlcH without the PlcR chaperones.
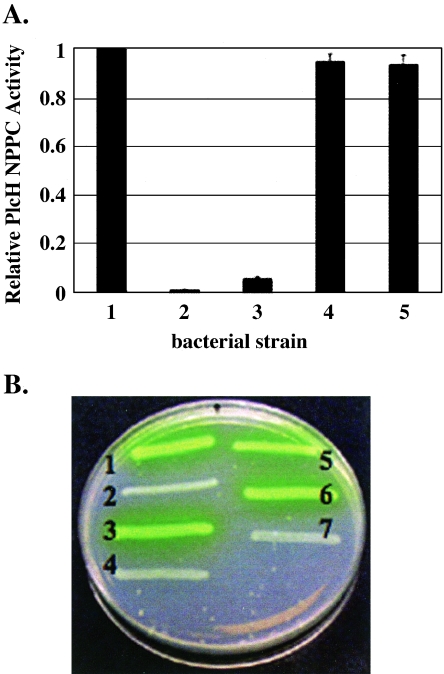
We generated a PAO1Δtat mutant that lacked all three tat genes and complemented this mutant with a plasmid expressing the tatA, tatB, and tatC genes of E. coli or the tat operon of P. aeruginosa. The expression of the tat genes of both E. coli and P. aeruginosa was driven from their native promoters to ensure that, although the Tat proteins would be overexpressed, the stoichiometry of the Tat proteins would be maintained. E. coli Tat protein expression was confirmed by Western analysis of bacterial pellets with rabbit serum directed against the E. coli TatA, TatB, and TatC proteins (data not shown). Strains were grown in HEPES minimal medium, and supernatants were examined for PlcH by the detection of NPPC activity and Western analysis as previously described. As shown in Fig. 6A, no PlcH activity could be detected in the PAO1Δtat mutant. Complementation of this strain with either the P. aeruginosa tat operon or the E. coli tat operon restored supernatant PlcH activity and PlcH protein to wild-type levels. Complementation with vector alone did not restore extracellular PlcH. These findings indicate that the E. coli Tat secretion system is able to recognize the PlcH signal peptide and transport PlcH as efficiently as the native Tat transport system in a P. aeruginosa host. In addition, these data indicate that expression of the E. coli tat operon in the PAO1 tat mutant strain does not influence PlcH secretion through the outer membrane, as PlcH was able to be further transported by the Xcp machinery and into the extracellular space.
FIG. 6.
(A) PlcH is secreted in a tat mutant of the PAO1 strain complemented with the tat operon of E. coli. PlcH activity is expressed relative to the extracellular activity of the PAO1 strain expressing wild-type PlcH, which was set to 1 (bar 1). Bar 2, PAO1Δtat mutant expressing wild-type PlcH; bar 3, PAO1Δtat mutant expressing wild-type PlcH complemented with vector alone; bar 4, PAO1Δtat mutant expressing wild-type PlcH complemented with the tat operon of E. coli; bar 5, PAO1Δtat mutant expressing wild-type PlcH complemented with the tat operon of P. aeruginosa. Error bars indicate standard errors of the means of three independent experiments. (B) Secretion of pyoverdine in a PAO1 tat mutant complemented with the tat operon of E. coli. Lane 1, PAO1; lane 2, PAO1Δtat mutant; lane 3, PAO1 Δtat mutant complemented with the tat operon of P. aeruginosa; lane 4, PAO1 Δtat mutant complemented with vector alone; lane 5, PAO1 Δtat mutant complemented with E. coli tat operon; lane 6, PAO1 ΔtatC mutant complemented with E. coli tat operon; lane 7, PAO1 ΔtatC mutant.
Our previous studies examining the effect of a tat mutation in P. aeruginosa demonstrated pleiotropic effects on the bacterium, including pyoverdine-mediated iron acquisition (33). Our initial screening of the P. aeruginosa genome for Tat-secreted proteins revealed that several pyoverdine biosynthesis gene products as well as the ferripyoverdine receptor were putative Tat-secreted proteins (33). We tested pyoverdine-mediated iron acquisition in the PAO1 tat mutant as well as the PAO1 tat mutant complemented with vector alone, the P. aeruginosa tat operon, or the E. coli tat operon on Casamino Acid agar. As shown in Fig. 6B, while the tat operon mutant failed to produce the yellow-green fluorescent pigment synonymous with pyoverdine secretion, complementation with either the P. aeruginosa tat operon or the E. coli tat operon restored pyoverdine secretion. Complementation of the PAO1ΔtatC mutant with the E. coli tat operon also restored the pyoverdine phenotype. Complementation with vector alone failed to restore pyoverdine secretion. This indicates that E. coli Tat proteins can recognize and transport an additional Tat substrate in addition to PlcH, but only in the context of a P. aeruginosa host.
DISCUSSION
PlcH of P. aeruginosa is a known virulence factor which has recently been shown to depend on the Tat secretion system for transport across the inner membrane (49, 50). The twin Arg leader sequence of PlcH seems to play a critical role in this transport process. Targeting to the Tat apparatus in chloroplasts demonstrates an absolute dependence on the twin Arg pair within the signal sequence (12, 19). In bacteria, several studies utilizing a variety of Tat-dependent substrates have demonstrated that the leader peptide twin Arg residues are also crucial for the targeting of proteins to the Tat translocon (11, 15, 45). However, while the mutagenesis of the twin Arg motif leads to disruption in the translocation of several Tat substrates, the degree of disruption seems to differ depending upon which Arg within the twin Arg motif is disrupted. The majority of Tat-transported proteins are partially affected in their secretion when the first Arg is disrupted, but more severe abrogation of secretion is observed when the second Arg in the motif is affected (11, 45). In the case of the P. aeruginosa protein PlcH, which contains three Arg residues within the Tat consensus motif (R8, R9, and R10), it appears that Arg-9 and Arg-10 are absolutely required for PlcH translocation by the Tat secretion machinery, while Arg-8 does not seem to play a role in secretion. It is interesting to note that the location of the twin Arg residues (R9 and R10) within the PlcH consensus motif seems to be critical to secretion since Arg-8 cannot substitute for either invariant twin Arg residue. Thus, it appears that the first three residues within the consensus motif for PlcH constitute x-R-R, where the x residue may be an Arg, Lys, Ser, or Ala to obtain efficient PlcH secretion.
Our finding that the PlcH R9K and PlcH R10K mutants associate with the inner membrane is somewhat surprising since previous studies examining E. coli Tat substrates with twin Arg substitutions within the consensus motif have found that these mutants localize to the cytosol (10, 18, 41). Some evidence to support membrane integration prior to translocation comes from studies of the thylakoid transport system. Musser and Theg (31) demonstrated that the thylakoid ΔpH/Tat machinery transported pOE17 precursor-avidin constructs bound tightly to thylakoid membranes independently of the translocation machinery pore. A recent model of E. coli Tat translocation put forth by Bruser and Sanders (9), based on their data as well as in vitro studies on HiPIP, the high-potential iron-sulfur protein of Allochromatium vinosum (10), proposes that bacterial Tat translocation occurs in several steps. Our findings fit their hypothesis, suggesting that membrane targeting of PlcH is independent of the translocation event.
Studies with bacterial Tat signal peptides indicate that, while the twin Arg residues are important in targeting to the Tat apparatus, additional conserved residues within the signal peptide also play a role in Tat secretion. A consensus motif around the twin Arg residues, defined as S-R-R-x-F-L-K, has been established, with the frequency of the conserved residues surrounding the twin Arg residues exceeding 50% (1). Previous studies have demonstrated that the residue within the consensus motif directly preceding the twin Arg pair is most frequently a Ser, Thr, asparagine, or aspartate, each being residues that serve to cap NH2 termini of helices (17). Stanley et al. (45) speculated that this residue might serve to stabilize the core of the signal peptide in an α-helical conformation. However, when they tested the importance of this residue in the consensus motif of the E. coli SufI Tat protein by mutating the conserved serine to an alanine, they found no effect on the export kinetics of the SufI protein.
Unlike the SufI protein of E. coli or the majority of other Tat-secreted proteins, PlcH contains an Arg at the position just prior to the twin Arg residues within the consensus motif. Mutagenesis of this Arg to a Lys, a Ser, or an Ala, however, had no effect on overall PlcH secretion, mirroring the findings of Stanley et al. (45). That is, the first residue within the Tat consensus motif of PlcH may be an Arg, Lys, Ser, or Ala to obtain efficient PlcH secretion. However, the twin Arg residues within the consensus motif are absolutely required for secretion.
In addition to the residues within the consensus sequence of the leader peptide, basic residues at the C terminus of the leader sequence have been shown to play a role in the secretion of Tat substrates both in thylakoids and bacteria (4, 5, 15, 26). These residues have been found to function, not in targeting to the Tat translocon, but in preventing the functional interaction of a Tat substrate with the Sec apparatus (5, 3). In this study, we found that substitution of the C-terminal Arg for Ala within the leader sequence of PlcH did not have any apparent effect on secretion of this phospholipase through the Tat system. Further, inactivation of the Tat secretion system in these mutants prevented PlcH secretion, indicating that the secretion was not redirected to the Sec pathway after removal of the C-terminal Arg. These results are in corroboration with the findings of Stanley et al. (45) but differ from those of Blaudeck et al. (3), who showed that a spontaneous mutant of the TorA-MalE hybrid precursor that had lost the positively charged residues in the C terminus gained the ability to use the Sec system. The PlcH protein, on the other hand, requires the association of two cofactor proteins, PlcR1 and PlcR2, for efficient secretion (14 and unpublished data). Thus, PlcH is secreted through the Tat system as part of a complex of folded proteins and may therefore be Sec-incompatible irrespective of the presence or absence of the “Sec avoidance” motif. It is curious that PlcH would have not just one, but two positively charged residues at the C terminus of the leader sequence if they are dispensable for PlcH Tat transport.
Although the mutagenesis of the consensus motif Arg prior to the twin Arg sequence as well as the two Arg residues prior to the cleavage site did not affect PlcH secretion, we did observe an effect on the overall level of PlcH detected in these mutants, and this effect (decrease) corresponded to the severity of the mutation. It is possible that mutating these Arg residues has an effect on the secondary structure of the plcH message, which would affect the levels of PlcH translation. Indeed, Punginelli et al. (36) showed that the signal sequence coding region of the E. coli formate dehydrogenase N, a Tat-secreted substrate, could fold into a stable secondary structure and that this stem-loop mediated translational control of formate dehydrogenase N synthesis. Alternatively, the Arg residues within the signal sequence may play a role in PlcH interaction with the PlcR2 chaperon that was shown to be important for PlcH secretion through the inner membrane (14). It is also clear that signal peptides and chaperones both play a role in the folding and export of Tat substrates (2).
Several studies have raised the possibility that the Tat leader sequence plays a role not only in the translocation of the Tat substrate through the Tat system but also in the overall stability of the Tat protein (for review, see reference 2). Sambasivarao et al. (38) showed that the disruption of the Tat leader sequence of the dimethyl sulfoxide reductase of E. coli led to a loss of reductase activity. In this study, we found that the removal of the PlcH Tat leader sequence essentially abrogated PlcH expression. This loss was due to severely decreased plcH transcript levels, although whether it was through the loss of expression or transcript instability is not clear at this time. However, since transcript levels did increase over time after induction with IPTG for the plcHΔss mutant, it seems likely that there is an issue relating to message stability. Further studies will have to be conducted to support this contention, but these results do suggest that the sequence encoding the Tat leader sequence plays a key role in the transcription of plcH.
Previous reports (4) have suggested that there exists a degree of species specificity in the recognition between the signal peptide of a Tat-secreted substrate and the Tat translocon. Blaudeck et al. (4), for example, demonstrated that the Zymomonas mobilis pre-GFOR, a Tat-secreted protein, is not exported by the E. coli Tat machinery despite being secreted efficiently in its natural host. On the other hand, sequence signal recognition by a heterologous host has also been reported (32). Our results in this regard indicate that while the expression of PlcH in an E. coli host did not allow PlcH secretion, the expression of PlcH in a P. aeruginosa Tat mutant host complemented with the E. coli Tat transport machinery did restore PlcH secretion as well as secretion of an additional Tat-secreted protein pyoverdine. A possible explanation for the failure of PlcH to be exported in E. coli is the lack of accessory proteins necessary for Tat transport, which are present in the natural P. aeruginosa host, but are not expressed in E. coli. Such accessory proteins could be specific to PlcH (e.g., PlcR) or may be broader in their function as Tat chaperones.
Acknowledgments
We thank Timothy Yahr for serum against the E. coli Tat proteins and Tracy Palmer for her kind advice and assistance with regard to the E. coli Tat system.
This work was supported by NIH grants AI15940 and HL06208 and by a research grant (VASIL03GO) from the Cystic Fibrosis Foundation to Michael Vasil.
REFERENCES
- 1.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 3.Blaudeck, N., P. Kreutzenbeck, R. Freudl, and G. A. Sprenger. 2003. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J. Bacteriol. 185:2811-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaudeck, N., G. A. Sprenger, R. Freudl, and T. Wiegert. 2001. Specificity of signal peptide recognition in tat-dependent bacterial protein translocation. J. Bacteriol. 183:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogsch, E., S. Brink, and C. Robinson. 1997. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 16:3851-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 7.Bolhuis, A. 2002. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology 148:3335-3346. [DOI] [PubMed] [Google Scholar]
- 8.Brunschwig, E., and A. Darzins. 1992. A two-component T7 system for the overexpression of genes in Pseudomonas aeruginosa. Gene 111:35-41. [DOI] [PubMed] [Google Scholar]
- 9.Bruser, T., and C. Sanders. 2003. An alternative model of the twin arginine translocation system. Microbiol. Res. 158:7-17. [DOI] [PubMed] [Google Scholar]
- 10.Bruser, T., T. Yano, D. C. Brune, and F. Daldal. 2003. Membrane targeting of a folded and cofactor-containing protein. Eur. J. Biochem. 270:1211-1221. [DOI] [PubMed] [Google Scholar]
- 11.Buchanan, G., F. Sargent, B. C. Berks, and T. Palmer. 2001. A genetic screen for suppressors of Escherichia coli Tat signal peptide mutations establishes a critical role for the second arginine within the twin-arginine motif. Arch. Microbiol. 177:107-112. [DOI] [PubMed] [Google Scholar]
- 12.Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klosgen, and C. Robinson. 1995. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 14:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, K. J., J. M. Ingram, and J. W. Costerton. 1970. Release of alkaline phosphatase from cells of Pseudomonas aeruginosa by manipulation of cation concentration and of pH. J. Bacteriol. 104:748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cota-Gomez, A., A. I. Vasil, J. Kadurugamuwa, T. J. Beveridge, H. P. Schweizer, and M. L. Vasil. 1997. PlcR1 and PlcR2 are putative calcium-binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect. Immun. 65:2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristobal, S., J. W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doig, A. J., M. W. MacArthur, B. J. Stapley, and J. M. Thornton. 1997. Structures of N-termini of helices in proteins. Protein Sci. 6:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreusch, A., D. M. Burgisser, C. W. Heizmann, and W. G. Zumft. 1997. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim. Biophys. Acta 1319:311-318. [DOI] [PubMed] [Google Scholar]
- 19.Henry, R., M. Carrigan, M. McCaffrey, X. Ma, and K. Cline. 1997. Targeting determinants and proposed evolutionary basis for the Sec and the Delta pH protein transport systems in chloroplast thylakoid membranes. J. Cell Biol. 136:823-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinsley, A. P., N. R. Stanley, T. Palmer, and B. C. Berks. 2001. A naturally occurring bacterial Tat signal peptide lacking one of the ‘invariant’ arginine residues of the consensus targeting motif. FEBS Lett. 497:45-49. [DOI] [PubMed] [Google Scholar]
- 21.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 22.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 23.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 25.Hutcheon, G. W., and A. Bolhuis. 2003. The archaeal twin-arginine translocation pathway. Biochem. Soc. Trans. 31:686-689. [DOI] [PubMed] [Google Scholar]
- 26.Ize, B., F. Gerard, M. Zhang, A. Chanal, R. Voulhoux, T. Palmer, A. Filloux, and L. F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 27.Kurioka, S., and M. Matsuda. 1976. Phospholipase C assay using p-nitrophenylphosphoryl-choline together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281-289. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno, T., and M. Kageyama. 1978. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J. Biochem. (Tokyo) 84:179-191. [DOI] [PubMed] [Google Scholar]
- 30.Muller, M. 2005. Twin-arginine-specific protein export in Escherichia coli. Res. Microbiol. 156:131-136. [DOI] [PubMed] [Google Scholar]
- 31.Musser, S. M., and S. M. Theg. 2000. Characterization of the early steps of OE17 precursor transport by the thylakoid ΔpH/Tat machinery. Eur. J. Biochem. 267:2588-2598. [DOI] [PubMed] [Google Scholar]
- 32.Niviere, V., S. L. Wong, and G. Voordouw. 1992. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a beta-lactamase fusion protein. J. Gen. Microbiol. 138:2173-2183. [DOI] [PubMed] [Google Scholar]
- 33.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostroff, R. M., B. Wretlind, and M. L. Vasil. 1989. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 57:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pop, O., U. Martin, C. Abel, and J. P. Muller. 2002. The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 277:3268-3273. [DOI] [PubMed] [Google Scholar]
- 36.Punginelli, C., B. Ize, N. R. Stanley, V. Stewart, G. Sawers, B. C. Berks, and T. Palmer. 2004. mRNA secondary structure modulates translation of Tat-dependent formate dehydrogenase N. J. Bacteriol. 186:6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 38.Sambasivarao, D., R. J. Turner, J. L. Simala-Grant, G. Shaw, J. Hu, and J. H. Weiner. 2000. Multiple roles for the twin arginine leader sequence of dimethyl sulfoxide reductase of Escherichia coli. J. Biol. Chem. 275:22526-22531. [DOI] [PubMed] [Google Scholar]
- 39.Santini, C. L., B. Ize, A. Chanal, M. Muller, G. Giordano, and L. F. Wu. 1998. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sargent, F., N. R. Stanley, B. C. Berks, and T. Palmer. 1999. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073-36082. [DOI] [PubMed] [Google Scholar]
- 42.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 43.Settles, A. M., A. Yonetani, A. Baron, D. R. Bush, K. Cline, and R. Martienssen. 1997. Sec-independent protein translocation by the maize Hcf106 protein. Science 278:1467-1470. [DOI] [PubMed] [Google Scholar]
- 44.Shortridge, V. D., A. Lazdunski, and M. L. Vasil. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol. 6:863-871. [DOI] [PubMed] [Google Scholar]
- 45.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 46.Stonehouse, M. J., A. Cota-Gomez, S. K. Parker, W. E. Martin, J. A. Hankin, R. C. Murphy, W. Chen, K. B. Lim, M. Hackett, A. I. Vasil, and M. L. Vasil. 2002. A novel class of microbial phosphocholine-specific phospholipases C. Mol. Microbiol. 46:661-676. [DOI] [PubMed] [Google Scholar]
- 47.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 48.Terada, L. S., K. A. Johansen, S. Nowbar, A. I. Vasil, and M. L. Vasil. 1999. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect. Immun. 67:2371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Titball, R. W. 1993. Bacterial phospholipases C. Microbiol. Rev. 57:347-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. A. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 52.Wu, L. F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]
- 53.Yahr, T. L., and W. T. Wickner. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20:2472-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]