Abstract
Induction of competence for natural genetic transformation in Streptococcus pneumoniae depends on pheromone-mediated cell-cell communication and a signaling pathway consisting of the competence-stimulating peptide (CSP), its membrane-embedded histidine kinase receptor ComD, and the cognate response regulator ComE. Extensive screening of pneumococcal isolates has revealed that two major CSP variants, CSP1 and CSP2, are found in members of this species. Even though the primary structures of CSP1 and CSP2 are about 50% identical, they are highly specific for their respective receptors, ComD1 and ComD2. In the present work, we have investigated the structural basis of this specificity by determining the three-dimensional structure of CSP1 from nuclear magnetic resonance data and comparing the agonist activity of a number of CSP1/CSP2 hybrid peptides toward the ComD1 and ComD2 receptors. Our results show that upon exposure to membrane-mimicking environments, the 17-amino-acid CSP1 pheromone adopts an amphiphilic α-helical configuration stretching from residue 6 to residue 12. Furthermore, the pattern of agonist activity displayed by the various hybrid peptides revealed that hydrophobic amino acids, some of which are situated on the nonpolar side of the α-helix, strongly contribute to CSP specificity. Together, these data indicate that the identified α-helix is an important structural feature of CSP1 which is essential for effective receptor recognition under natural conditions.
Infection by the pathogen Streptococcus pneumoniae is a primary source of bacterial pneumonia, otitis media, and meningitis and a common cause of sepsis in patients infected with the human immunodeficiency virus. Disturbingly, the number of reported antibiotic-resistant clinical isolates of this bacterium has increased dramatically over the last 20 years, and multiresistant strains have already emerged (2, 20). The rapid acquisition and spread of antibiotic resistance genes in S. pneumoniae are attributed to the development of a transient phenotype termed competence for natural genetic transformation (5, 10). This phenotype allows cells to actively lyse and take up DNA from noncompetent pneumococci, thus providing competent bacteria with an efficient predatory mechanism for acquisition of genetic material (16, 22, 30, 31). The key event regulating competence development is the interaction between the secreted competence-stimulating pheromone (CSP) and the membrane-associated histidine kinase receptor ComD (5, 12, 13). Two main CSP and ComD variants (CSP1 and CSP2; ComD1 and ComD2) have been identified in S. pneumoniae (16, 25, 26). Both CSP variants, as well as all other CSPs that have been characterized from the mitis and anginosus phylogenetic groups, contain a conserved sequence fingerprint composed of a negatively charged N-terminal residue, an arginine in position 3, and a positively charged C-terminal tail. However, the central region of the peptides displays much less conserved primary structures (16). Extracellular CSP binds to the polytopic transmembrane ComD receptor domain and triggers competence development in the pneumococcal culture when its concentration in the growth medium reaches a critical threshold level. CSP binding presumably brings on a conformational change in the ComD transmembrane domain that ultimately results in activation of ComD kinase activity. Once activated, ComD phosphorylates the transcriptional regulator ComE (5, 32). This enables ComE to activate transcription of the early genes, including comX, which encodes an alternative sigma factor that directs the transcription of a large number of so-called late genes (19). Close to 190 CSP-responsive genes have been identified in S. pneumoniae. However, only 23 of these genes appear to be essential for competence regulation and DNA uptake. This strongly implies that additional processes are regulated through the CSP-ComD signaling mechanism (24). Indeed, there appears to be a tight interplay between competence and virulence. It has previously been demonstrated that comD mutants are attenuated in bacteremia, as well as respiratory tract infection (1, 11, 18), and it was recently shown that competence-mediated cell lysis might be important for release of the cytolytic toxin pneumolysin (8). Furthermore, Oggioni and coworkers have shown that CSP has therapeutic potential. They recently reported that the survival rate of mice injected intravenously with 1.3 μg of CSP and 106 pneumococcal cells was much higher than the corresponding survival rate for mice injected with bacteria alone (23).
From the above, it is evident that an increased understanding of the CSP-ComD interface could be of clinical relevance and lead to the development of CSP analogues with improved therapeutic properties. To gain better insight into the CSP-ComD interaction, this work investigated the three-dimensional structure of CSP1 by means of nuclear magnetic resonance (NMR) spectroscopy. Furthermore, by homologue scanning, CSP1 residues conferring receptor specificity was identified.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S1 and CP1500 strains were derived from S. pneumoniae strain Rx as described previously (31). Construction of S2, the third strain used in this study, is described below. Bacterial strains were grown in casein tryptone (CAT) medium containing (per liter) 167 mmol of K2HPO4, 5 mg of choline chloride, 5 g of tryptone, 10 g of enzymatic casein hydrolysate, 1 g of yeast extract, and 5 g of NaCl (21). After sterilization, glucose was added to a concentration of 0.2%. All incubations were carried out at 37°C, and all density measurements of bacterial cultures were done spectrophotometrically at 550 nm.
Synthetic CSPs.
Synthetic CSPs (Genosphere Biotechnologies) used in this work were purified by reversed-phase high-performance liquid chromatography and characterized by analytical reversed-phase high-performance liquid chromatography and mass spectrometry. The CSP1 peptide used for circular dichroism (CD) and NMR spectroscopy displayed a purity of >95%. All of the peptides used in ComD activation assays displayed a purity of >90%.
CD spectroscopy.
CD spectra were recorded with a Jasco J-810 spectropolarimeter (Jasco International Co., Ltd., Tokyo, Japan) calibrated with ammonium D-camphor-10-sulfonate (Icatayama Chemicals, Tokyo, Japan). All of the measurements were performed with a peptide concentration of 0.15 mg/ml in 0.1% trifluoroacetic acid in the presence of various concentrations of dodecyl phosphocholine (DPC; 0 to 8 mM; CDN Isotopes, Quebec, Canada) or trifluoroethanol (0 to 50% [vol/vol]; Sigma-Aldrich, St. Louis, Mo.). Measurements were performed at 20 to 50°C with a quartz cuvette (Starna, Essex, England) with a path length of 0.1 cm. Samples were scanned five times at 50 nm/min with a bandwidth of 1 nm and a response time of 1 s over a wavelength range 190 to 245 nm. The data were averaged, and the spectrum of a sample-free control was subtracted. The spectra were adjusted according to the volume changes obtained as a result of the volume changes during titration. The α-helical contents of the various peptides were determined by applying the spectral fitting methods in the CDpro package (28, 29).
NMR sample preparation.
For NMR structure elucidation, 3.73 mg of CSP1 was dissolved in 880 μl of 250 mM deuterated DPC (CDN Isotopes) and a water solution with 10% D2O (Cambridge Isotope Laboratories). The final concentration of the sample was 1.9 mM. NMR structures, assignments, and constraints have been posted to the Protein Data Bank (2A1C).
NMR spectroscopy.
The NMR spectra of CSP1 were obtained at 28°C and 38°C on an 800-MHz Varian INOVA 800 NMR spectrometer with four channels, 5 mm 1H {13C, 15N} pfg probe. Total correlation spectroscopy (TOCSY) (3) and nuclear Overhauser effect spectroscopy (NOESY) (15, 35) experiments included in the biopack were performed to assign the molecule. Spectra with mixing times of 32 and 64 ms were acquired for the TOCSY pulse sequence, and spectra with times of 100 and 150 ms were acquired for the NOESY pulse sequence. Watergate water decoupling was applied in the TOCSY and NOESY experiments (27). Two thousand complex data points were obtained in the direct dimension, and 512 were obtained in the indirect dimension. The data were multiplied with a sine bell function and zero filled to double size prior to Fourier transformation. All postprocessing was performed by applying NMR Pipe (7), while spectral assignments and integration were done by applying SPARKY (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
Restraints and structure calculation.
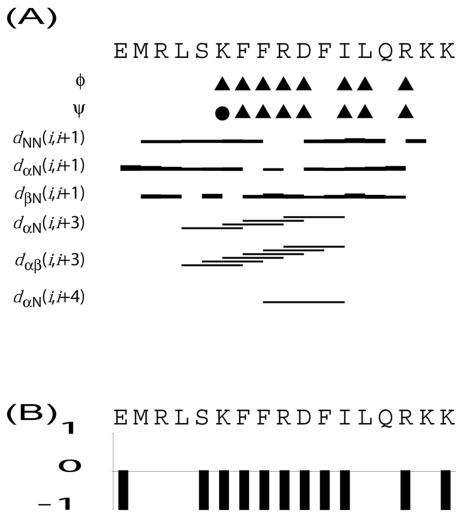
Dihedral angle restraints were obtained by using the TALOS program (6) on the chemical shift values. From this program, we obtained restrictions for the torsion angles of the molecule shown in Fig. 1. The NOESY spectrum of the molecule was manually assigned. A total of 278 NOE distance constrains were obtained, out of which 104 were interresidue and 174 were intraresidue NOE restraints. No long-range NOE restraint distances larger than four residues were observed in either of the spectra. The structure was calculated and annealed with the structure calculation program CYANA (9) and the anneal function. A total of 100 structures were calculated, out of which the 20 best structures were selected for further evaluation. The target function gave an average of 0.0045 ± 0.0032. The final structures were visualized with the MOLMOL program (17).
FIG. 1.
(A) Pattern of interresidue NOEs observed in the NOESY experiment (150-ms mixing time) with CSP1 in the presence of 250 mM DPC. The thickness of the lines is relative to the size of the NOE cross-peak intensities. The triangles indicate where TALOS found dihedral angle restrains that are typically for α-helical structure, while the circle indicates the presence of angles nontypical for α-helices or β-sheets. (B) CSI indexes (33, 34) observed by analysis of the Hα and N chemical shift values of CSP1.
Construction of the S. pneumoniae S2 mutant.
The S2 strain is identical to the S. pneumoniae S1 reporter strain, except that it expresses the ComD2 receptor derived from strain A66 (25). The entire comCDE operon of strain A66 was first amplified by PCR with the primers ArgTF (5′-CGCTGAGAATCTGTGTCAGT-3′) and 2tGlu2 (5′-GGCGGTGTCTTAACCCCTTGACCAACGG-3′). The PCR fragment was subsequently transformed into S. pneumoniae S1 cells by adding 1 μg of DNA and 250 ng of CSP1 to 1 ml of exponentially growing cells at an optical density at 550 nm (OD550) of 0.1. Following incubation for 2.5 h at 37°C, cells were harvested by centrifugation and washed 10 times with CAT medium. The cells were next diluted in 20 ml of CAT medium and grown to an OD550 of 0.05. To select for bacteria in which the comD1 gene had been exchanged with the comD2 gene of strain A66, 1 ml of the culture was subsequently treated with 1 μg of genomic DNA from the novobiocin-resistant S. pneumoniae mutant CP1500 (4) and 150 ng of CSP2. Following incubation for 2.5 h at 37°C, novobiocin-resistant transformants were selected on agar plates containing 5 μg/ml novobiocin. Genomic DNA from one such transformant (S. pneumoniae S2) was subsequently sequenced and found to contain a correct insertion of the comD2 gene.
ComD activation assays.
An overnight culture of exponentially growing S. pneumoniae S1 or S2 cells was diluted to an OD550 of 0.05 in prewarmed (37°C) CAT medium and incubated until an OD550 of 0.2 was reached. At this point, the culture was treated with the desired amount of the appropriate synthetic CSP. The culture was next incubated at 37°C for 30 min and subsequently placed on ice before the final OD550 of the culture was measured. In order to measure the β-galactosidase activity in the culture, the cells were lysed by incubating the culture for 10 min at 37°C with 0.1% Triton X-100. The lysate was kept on ice until assayed. All samples were assayed for β-galactosidase activity as described previously (30).
RESULTS AND DISCUSSION
Structural analysis of CSP1 upon exposure to membrane-mimicking environments.
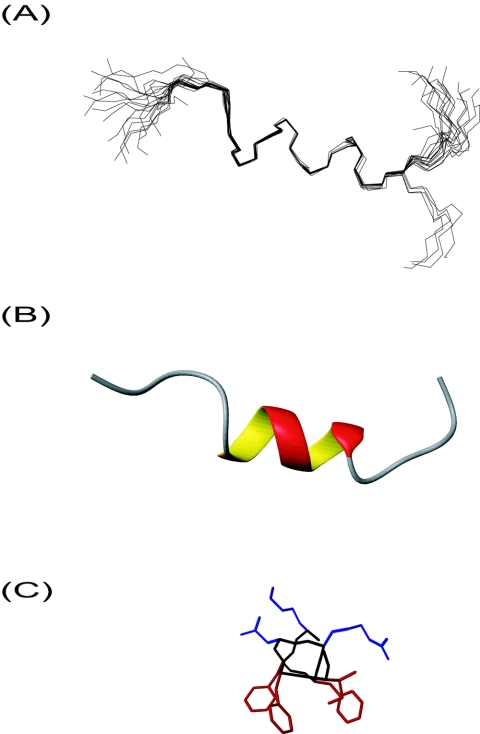
Initial CD analysis of CSP1 or CSP2 in water revealed little or no structuring of the pheromones. In contrast, both peptides became structured upon exposure to membrane-mimicking environments such as trifluoroethanol and micelles, indicating that structuring of the pheromones is initiated upon interaction with the membrane of target bacteria. Under such conditions, CSP1 displayed an α-helical content between 40 and 46%, suggesting that seven or eight residues of the pheromone were contained within the structured part. Based on these results, we next performed NMR spectroscopy analysis of CSP1 in the presence of DPC. The TOCSY and NOESY NMR spectra were analyzed and assigned according to standard methods. The intraresidue NOEs and dihedral angles obtained with the TALOS program are shown in Fig. 1A. The existence of αH-HN (i,i+3), αH-HN (i,i+4), and αH-HN (i,i+5) NOEs indicated that there is a possible α-helix stretching from residue 4 to residue 12 in CSP1. The torsion angles obtained by TALOS analysis of the chemical shift values clearly indicated the formation of an α-helix between residues 6 and 12. The latter prediction was also supported by the chemical shift index (Fig. 1B). The final 20 best structures obtained after annealing of the structure are shown in Fig. 2A. Superimposing the structures from residue 6 to residue 12 revealed the presence of a well-defined α-helix in this region, clearly reflected in the low backbone and heavy atom root mean square distances (0.13 ± 0.08 and 1.00 ± 0.19, respectively). Figure 2A shows that the residues located outside this helical region are relatively unstructured. Although NOE connections between Leu-4 and Phe-7 were observed and chemical shift indexing indicated that residue 5 might be contained within the α-helical part of the peptide (Fig. 1), it was not possible to find NMR data indicating restrictions that in DYANA calculations extended the α-helix to residue 4 or 5. The length of the observed α-helical stretch in the three-dimensional model presented in Fig. 2B is also in good agreement with the data obtained from the CD analysis. Interestingly, the α-helical region of CSP1 is amphiphilic, with the nonpolar residues Phe-7, Phe-8, Phe-11, and Ile-12 facing one side of the helix and Lys-6, Arg-9, and Asp-10 facing the opposite side (Fig. 2C). As the primary structures of CSP1 and CSP2 diverge considerably in this region, we considered it unlikely that the observed amphiphilicity is accidental. We therefore speculated that the hydrophobic patch formed by Phe-7, Phe-8, Phe-11, and Ile-12 determines the specificity of the CSP1-ComD1 interaction and exploited the closely related CSP2-ComD2 system to test this hypothesis.
FIG. 2.
Structure of CSP1. (A) Backbone superposition of the 20 best structures of CSP1 derived by NMR analysis. The structures are superimposed over the backbone atoms of residues 6 to12. (B) Cartoon of CSP1 depicting the α-helix. (C) Helical region residues 6 to 12 viewed from the N-terminal side. Polar side chains (Lys-6, Arg-9, and Asp-10) are blue, and hydrophobic side chains (Phe-7, Phe-8, Phe-11, and Ile-12) are red.
CSP1 and CSP2 receptor activation and specificity.
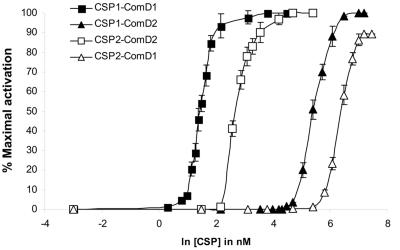
Lacking a direct pheromone-receptor binding assay, we utilized the S1 reporter strain and its derivative S2 for further studies of CSP agonist function. The S1 strain contains a chromosomally located lacZ reporter gene under control of the promoter of the late genes orf62 and orf51 (31), whose products were recently named CibA and CibB, respectively (8). Hence, expression of the β-galactosidase protein is under control of the competence-specific sigma factor ComX, which is synthesized in direct response to CSP1-mediated activation of ComD1. This strain also contains a deletion of the comA gene, a feature which prevents secretion of endogenously produced pheromone and consequently autoinduction of competence. As shown in Fig. 3, dose-dependent activation of the ComD1 receptor was observed upon exogenous addition of synthetic CSP1 to S1 cells, with half-maximal activation being reached at approximately 4.5 nM (Fig. 3). In contrast, the ComD1 receptor displayed very poor sensitivity toward CSP2, which appeared to be around 200 times less potent than CSP1 as an activator. Nevertheless, high concentrations of CSP2 could activate ComD1 at levels comparable to the maximal activation observed upon addition of CSP1. To be able to study the ComD2 receptor in the same genetic background as ComD1, the comCDE operon of strain S1 (CSP1, ComD1) was exchanged with the corresponding operon from strain A66 (CSP2, ComD2), giving rise to strain S2 (see Materials and Methods for details). As expected, ComD2 displayed dose-dependent activation when subjected to increasing concentrations of synthetic CSP2 (Fig. 3). The ComD2 receptor could also be activated by high concentrations of CSP1, with half-maximal activation being reached at around 220 nM. We next asked whether the poor agonist function of CSP2 toward ComD1 was due to a low affinity for the receptor or a poor ability to activate the receptor following binding of the peptide. If the poor agonist activity of CSP2 toward ComD1 was simply due to a lack of activator potential and not due to a lack of binding affinity, it would be reasonable to assume that CSP2 could antagonize the CSP1-ComD1 interaction by competing for the receptor binding site. However, when S1 cells activated with subsaturating concentrations of CSP1 were challenged with 200 nM CSP2, no decrease in receptor activation could be observed, suggesting that CSP2 is a poor binder of the ComD1 receptor (data not shown).
FIG. 3.
Dose-response assays of ComD1 and ComD2. Exponentially growing S1 (ComD1) or S2 (ComD2) cells were incubated in the presence of increasing concentrations of CSP1 or CSP2 and assayed for β-galactosidase activity. At least five independent experiments were performed for each CSP concentration indicated. The data are presented as percent maximal activation plotted against the natural logarithm of [CSP] in nanomolar. Half-maximal activation of ComD1 was reached at 4.4 nM CSP1 or 600 nM CSP2, while half-maximal activation of ComD2 was reached at 14 nM CSP2 or 220 nM CSP1.
CSP receptor recognition depends on hydrophobic interactions.
To characterize the amino acids in CSP1 that govern the specificity of receptor recognition, homologue scanning mutagenesis of CSP2 was performed (Table 1). Previous alanine scanning mutagenesis of CSP1 indicated that certain hydrophobic residues, including Phe-7, Phe-8, and Phe-11, are essential for pheromone bioactivity (14). Interestingly, our results show that these three phenylalanine residues, together with Ile-12, form a hydrophobic patch along one side of the amphiphilic α-helix in CSP1. We therefore concentrated our efforts on characterizing the role of these hydrophobic positions with respect to receptor binding and activation. As shown in Table 1, replacing a single hydrophobic residue in CSP2 with the corresponding residue from CSP1 in most cases resulted in a modest but significant increase in activity toward ComD1 (CSP2.1 to CSP2.7). However, the most active hybrid peptides turned out to be those containing a leucine in position 4 together with a phenylalanine in positions 7 and/or 8 (CSP2.9, CSP2.10, and CSP2.15). Together, the data in Table 1 show that amino acids situated in the hydrophobic patch of CSP1 strongly contribute to specificity, although Ile-12 appears to be considerably less important than the two phenylalanines. Replacement of Arg-6 or Leu-9 in CSP2 with the corresponding amino acid in CSP1 (Lys-6 or Arg-9) only resulted in an incremental increase in agonist activity toward ComD1 (data not shown), a result that further emphasizes the importance of hydrophobic residues in CSP receptor specificity.
TABLE 1.
Homologue scanning of CSP2
| Pheromone | Amino acid sequencea | ComD1b activity (% of maximal activation ± SE) at pheromone concn (nM) of:
|
||
|---|---|---|---|---|
| 44 | 110 | 220 | ||
| CSP1 | EMRLSKFFRDFILQRKK | 100 | 100 | 100 |
| CSP2 | EMRISRIILDFLFLRKK | —b | — | — |
| CSP2.1 | EMRLSRIILDFLFLRKK | — | 5 ± 1 | 27 ± 4.6 |
| CSP2.3 | EMRISRFILDFLFLRKK | — | 1 ± 0.1 | 7 ± 1 |
| CSP2.4 | EMRISRIFLDFLFLRKK | — | — | 2 ± 0.2 |
| CSP2.6 | EMRISRIILDFIFLRKK | — | — | 1 ± 0.1 |
| CSP2.7 | EMRISRIILDFLLLRKK | — | — | — |
| CSP2.9 | EMRLSRFILDFLFLRKK | 1 ± 0.1 | 69 ± 4.1 | 95 ± 1 |
| CSP2.10 | EMRLSRIFLDFLFLRKK | 1 ± 0.2 | 52 ± 2.5 | 94 ± 1.4 |
| CSP2.11 | EMRLSRIILDFIFLRKK | — | 10 ± 2 | 52 ± 3 |
| CSP2.12 | EMRISRFFLDFLFLRKK | — | 6 ± 1 | 35 ± 5.4 |
| CSP2.13 | EMRISRFILDFIFLRKK | — | — | 4 ± 0.3 |
| CSP2.14 | EMRISRIFLDFIFLRKK | — | — | — |
| CSP2.15 | EMRLSRFFLDFLFLRKK | 15 ± 3.9 | 82 ± 6 | 99 ± 0.4 |
| CSP2.16 | EMRLSRFILDFIFLRKK | 6 ± 1 | 82 ± 7.9 | 97 ± 1 |
| CSP2.17 | EMRLSRIFLDFIFLRKK | — | 28 ± 4.1 | 85 ± 2.7 |
Amino acids in bold are residues derived from CSP1.
—, no activation detected.
The results presented in Table 1 demonstrate that certain positions in CSP1, including Leu-4, Phe-7, and Phe-8, are essential for specificity, i.e., for the recognition of this pheromone's cognate receptor. To determine whether the same hydrophobic positions in CSP2 are important for this pheromone's specific interaction with ComD2, hybrid pheromones were designed by successively exchanging Leu-4, Phe-7, and Phe-8 from CSP1 with the corresponding amino acids from CSP2 (CSP1.30 to CSP1.33). As shown in Table 2, an increased number of swapped positions shifted the hybrid pheromones' specificity toward ComD2, while the activity toward ComD1 was gradually lost. These results confirm that the same key hydrophobic amino acid positions are essential for receptor recognition in CSP1 and CSP2.
TABLE 2.
Agonist activity of CSP1 mutants
| Pheromone | Pheromone varianta | ComD1 activity (% of maximal activation ± SE) at pheromone concn (nM) of:
|
ComD2 activity (% of maximal activation ± SE) at pheromone concn (nM) of:
|
||||
|---|---|---|---|---|---|---|---|
| 4.4 | 22 | 44 | 26 | 44 | 110 | ||
| CSP1 | EMRLSKFFRDFILQRKK | 52 ± 2.5 | 100 | 100 | —b | — | 3 ± 0.1 |
| CSP2 | EMRISRIILDFLFLRKK | — | — | — | 86 ± 5.4 | 93 ± 3.2 | 100 |
| CSP1.30 | EMRISKFFRDFILQRKK | — | 85 ± 2 | 100 | — | — | — |
| CSP1.31 | EMRLSKIFRDFILQRKK | — | 82 ± 1 | 100 | — | — | 32 ± 1 |
| CSP1.32 | EMRISKIFRDFILQRKK | — | — | 8 ± 0.3 | — | — | 37 ± 1.5 |
| CSP1.33 | EMRISKIIRDFILQRKK | — | — | — | — | 21 ± 3 | 61 ± 6 |
Amino acids in bold are residues derived from CSP1.
—, no activation detected.
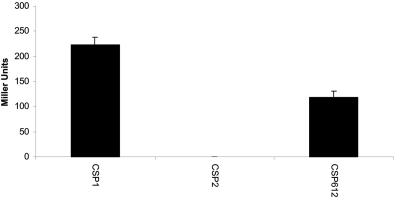
Our laboratory has previously determined the amino acid sequences of a large number of CSPs from S. mitis and S. oralis, species that are closely related to S. pneumoniae (16). To further examine the contribution of positions 7 and 8 in the CSP-receptor interaction, a CSP variant produced by S. mitis strain SK612 (N-ESRLSRLLRDFIFQIKQ-C) (16) was synthesized. In the region determined to have an α-helical configuration in CSP-1, CSP612, and CSP1 differ in three positions. CSP612 contains arginine in position 6 instead of lysine and leucines in positions 7 and 8 instead of phenylalanines. In addition, CSP612 and CSP1 differ in positions 2, 13, 15, and 17. As depicted in Fig. 4, the CSP612 pheromone displays relatively high agonist activity toward ComD1, with half-maximal saturation of the receptor being reached at approximately 40 nM. Hence, the ComD1 receptor is able to interact with peptides containing either Phe or Leu residues in positions 7 and 8 but interacts very poorly with pheromones containing Ile in these positions. This result clearly demonstrates that Phe-7 and Phe-8 in the CSP1 pheromone do not control receptor specificity through sequence-specific hydrophobic contacts with ComD1.
FIG. 4.
CSP612-mediated activation of ComD1. Exponentially growing S1 cells were incubated with CSP1, CSP2, or CSP612 at a final concentration of 44 nM. The respective CSPs are indicated on the x axis, while the y axis indicates the amount of β-galactosidase activity produced. Whereas no activity could be detected with CSP2, approximately half-maximal activation of ComD1 was observed with CSP612.
Concluding remarks.
Based on the results presented in this paper, we propose the following steps for the early events in CSP1-mediated activation of ComD1. Structuring of the secreted CSP1 is initiated upon interaction with membrane environments, resulting in the formation of an amphiphilic α-helix in the middle part of the pheromone. Together with Leu-4, a hydrophobic patch in this α-helix allows CSP1 to specifically recognize ComD1. However, the ability of the CSP612 pheromone to efficiently cross-induce ComD1 (Fig. 4) strongly indicates that receptor recognition does not depend on the formation of highly specific hydrophobic contacts between the α-helical part of CSP1 and ComD1. The exact mechanism by which the hydrophobic patch facilitates receptor recognition and activation thus remains elusive. Interestingly, the positively charged residue in position 3 (Arg-3), which has previously been demonstrated to be absolutely required for pheromone bioactivity (14), is conserved in all CSPs characterized from the mitis and anginosus phylogenetic groups (16). Besides, a negatively charged N-terminal amino acid (Glu-1 or Asp-1) is found in most CSPs. These charged amino acids are essential for pheromone activity but do not seem to be involved in specificity, as they are highly conserved in most streptococcal competence pheromones. We therefore hypothesize that the conserved charged residues in the N terminus are crucial for receptor activation and that the hydrophobic patch in CSP1 functions to correctly position the N terminus into a conserved pocket in the ComD1 receptor. Whether this positioning relies exclusively on CSP1-ComD1 protein-protein interactions or also includes interactions between the amphiphilic region of CSP1 and the bacterial membrane remains to be shown.
Acknowledgments
We thank Gøran Karlson at the Swedish NMR Center in Gothenburg for technical assistance.
This work was supported by grants from the Research Council of Norway.
REFERENCES
- 1.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed] [Google Scholar]
- 2.Bogaert, D., P. W. M. Hermans, P. V. Adrian, H. C. Rümke, and R. de Groot. 2004. Pneumococcal vaccines: an update on current strategies. Vaccine 22:2209-2220. [DOI] [PubMed] [Google Scholar]
- 3.Braunschweiler, L., and R. R. Ernst. 1983. Coherence transfer by isotropic mixing—application to proton correlation spectroscopy. J. Magn. Reson. 53:521-528. [Google Scholar]
- 4.Cato, A., and W. R. Guild. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J. Mol. Biol. 37:157-178. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., and L. S. Håvarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7d:1798-1814. [DOI] [PubMed] [Google Scholar]
- 6.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 7.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 8.Guiral, S., T. J. Mitchell, B. Martin, and J. P. Claverys. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. USA 102:8710-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guntert, P., C. Mumenthaler, and K. Wuthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 10.Hakenbeck, R. 2000. Transformation in Streptococcus pneumoniae: mosaic genes and the regulation of competence. Res. Microbiol. 151:453-456. [DOI] [PubMed] [Google Scholar]
- 11.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 12.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 14.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum-sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-26. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signalling in bacteria. ASM Press, Washington, D.C.
- 15.Jeener, J., B. H. Meier, P. Bachmann, and R. R. Ernst. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71:4546-4553. [Google Scholar]
- 16.Johnsborg, O., T. Blomqvist, M. Kilian, and L. S. Håvarstein. Biologically active peptides in streptococci. In R. Hakenbeck and S. Chhatwal (ed.), The molecular biology of streptococci, in press. Horizon Scientific Press, Norwich, United Kingdom.
- 17.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14:51-55. [DOI] [PubMed] [Google Scholar]
- 18.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 19.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 20.Mera, R. M., L. A. Miller, J. J. Daniels, J. G. Weil, and A. R. White. 2005. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States over a 10-year period: Alexander project. Diagn. Microbiol. Infect. Dis. 51:195-200. [DOI] [PubMed] [Google Scholar]
- 21.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moscoso, M., and J. P. Claverys. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 54:783-794. [DOI] [PubMed] [Google Scholar]
- 23.Oggioni, M. R., F. Ianelli, S. Ricci, D. Chiavolini, R. Parigi, C. Trappetti, J. P. Claverys, and G. Pozzi. 2004. Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:4725-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 25.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Håvarstein, L. Piccoli, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez, M., D. A. Morrison, and A. Tomasz. 1997. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb. Drug Resist. 3:39-52. [DOI] [PubMed] [Google Scholar]
- 27.Sklenar, V., M. Piotto, R. Leppik, and V. Saudek. 1993. Gradient-tailored water suppression for tH-1 N-HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 102:241-245. [Google Scholar]
- 28.Sreerama, N., and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252-260. [DOI] [PubMed] [Google Scholar]
- 29.Sreerama, N., and R. W. Woody. 2004. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 13:100-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinmoen, H., E. Knutsen, and L. S. Håvarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinmoen, H., A. Teigen, and L. S. Håvarstein. 2003. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J. Bacteriol. 185:7176-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]
- 33.Wishart, D. S., B. D. Sykes, and F. M. Richards. 1991. Simple techniques for the quantification of protein secondary structure by 1H NMR spectroscopy. FEBS Lett. 293:72-80. [DOI] [PubMed] [Google Scholar]
- 34.Wishart, D. S., B. D. Sykes, and F. M. Richards. 1991. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222:311-333. [DOI] [PubMed] [Google Scholar]
- 35.Wuthrich, K. 1986. NMR of proteins and nucleic acids. Wiley Interscience, New York, N.Y.