Abstract
In Rhodospirillum rubrum, NifA, the transcriptional activator for the nif genes, is posttranslationally activated only by the uridylylated form of GlnB, one of three PII homologs in the organism. We have used the yeast two-hybrid system to detect variants of GlnB that interact better with NifA than does wild-type GlnB. When examined for physiological effects in R. rubrum, these GlnB* variants activated NifA in the presence of NH4+, which normally blocks NifA activation completely, and in the absence of GlnD, whose uridylylation of GlnB is also normally essential for NifA activation. When these variants were tested in the two-hybrid system for their interaction with NtrB, a receptor that should interact with the nonuridylylated form of GlnB, they were uniformly weaker than wild-type GlnB in that interaction. When expressed in R. rubrum either as single-copy integrants or on multiple-copy plasmids, these variants were also dramatically altered in terms of their ability to regulate several other receptors involved in nitrogen metabolism, including GlnE, NtrB/NtrC, and DRAT (dinitrogenase reductase ADP-ribosyl transferase)-DRAG (dinitrogenase reductase-activating glycohydrolase). The consistent pattern throughout is that these GlnB variants partially mimic the uridylylated form of wild-type GlnB, even under nitrogen-excess conditions and in strains lacking GlnD. The results suggest that the role of uridylylation of GlnB is primarily to shift the equilibrium of GlnB from a “nitrogen-sufficient” form to a “nitrogen-deficient” form, each of which interacts with different but overlapping receptor proteins in the cell. These GlnB variants apparently shift that equilibrium through direct structural changes.
The PII family of proteins serves as one of the most broadly distributed regulatory proteins in biology, being found in almost all bacteria, archaea, and many lower eukaryotes and plant organelles (1, 39, 41, 66). The primary function of these proteins is to regulate the function of proteins involved in nitrogen metabolism in response to the carbon-nitrogen balance in the cell. The PII proteins do this by interacting with one set of proteins under nitrogen-excess conditions and with a different but overlapping set of proteins in nitrogen deficiency. In each case, the interaction causes the appropriate effect on the activity of the interacting partners, termed receptor proteins, based on the nomenclature of Ninfa and Atkinson (41).
In enteric bacteria, the primary nitrogen signal molecule affecting PII function is glutamine, which is sensed by GlnD. Under nitrogen deficiency, glutamine levels are relatively low and the bifunctional GlnD serves as a uridylytransferase, modifying each subunit of the PII trimer (23). Under nitrogen-excess conditions, glutamine levels are high and glutamine-bound GlnD has the opposite activity, that of a uridylyl-removing enzyme (UR) to remove the UMP group from the modified PII. In the cyanobacteria, PII is phosphorylated (or not) in response to α-ketoglutarate (αKG) levels, an indicator of nitrogen status (14, 40, 57). Though the kinase remains unknown in these organisms, the phosphatase, PphA, senses and responds to αKG and is unrelated to GlnD (17, 47). In higher organisms, there is no apparent posttranslational modification of the PII homologs (53).
In all cases, however, PII is thought to act as a sensor as well, by directly binding αKG (8, 12, 29, 43). PII can also bind ATP (29), but its physiological role is unknown. In vitro analysis of the well-studied prokaryotic systems has shown that the presence or absence of these small molecules affects the modification of PII noted above (23, 29). αKG also affects the interactions of PII proteins with receptor proteins, at least in vitro (12, 23-25, 29, 33). Finally, αKG can also affect other regulatory proteins such NtcA, a transcriptional regulator in cyanobacteria (56, 58), and NrpR, a repressor for nif expression in Methanococcus maripaludis (31).
PII proteins act as trimers, and several structures have been solved, either with or without ATP, but all have been in the UMP-free form (4, 6, 48, 51, 54, 59-61) (see Fig. 1A, for the E. coli GlnB structure). In some structures, a region termed the T loop extends away from the bulk of the trimer, and it is at the tip of this loop that UMP (or PO4−2 in the case of the cyanobacteria) is attached. ATP binds near the lower portion of the T loop, and indirect evidence in both Escherichia coli and other organisms suggests that αKG binds adjacent to ATP (4, 26, 41). Each PII trimer is capable of binding three molecules each of αKG and ATP (23, 29).
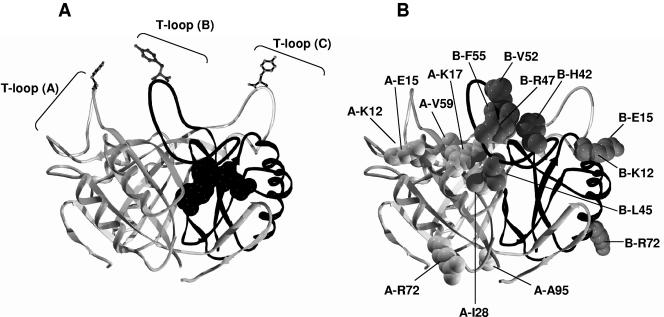
FIG. 1.
Model of the structure of R. rubrum GlnB. (A) Ribbon model of the E. coli GlnB trimer (6). The B monomer is shown with a dark backbone. A single ATP molecule, whose position is based on the structure of ATP-bound GlnK of E. coli (60, 61), is shown at the interface of monomers A and B. The T loops are indicated, and at the tip of each is Tyr51, the site of uridylylation. (B) The same view, but with the R. rubrum wild-type residues altered in the GlnB* variants shown as space filled. Only residues on one face of the trimer are shown, and their respective monomer is indicated by the “A” or “B” preceding the residue name.
The physiological role of PII proteins is somewhat complicated by the presence of multiple homologs in some organisms, and the synthesis of some of these is also regulated by nitrogen status. The homolog that is most commonly found, and that tends to be present under all growth conditions, is termed GlnB. The nitrogen-regulated homologs are most typically termed GlnK (1). Rhodospirillum rubrum, the model organism used in these studies, has a third, termed GlnJ. We will focus on GlnB because studies of this protein in a variety of organisms have shown that it interacts with NtrB and GlnE in its nonuridylylated form and with NifA and GlnE in its uridylylated form. It certainly also interacts with DRAG (dinitrogenase reductase-activating glycohydrolase) and DRAT (dinitrogenase reductase ADP-ribosyl transferase) (see below), but the relevant forms of GlnB for these interactions remain unclear. NtrB is the sensor portion of a two-component regulatory system that has NtrC as the regulated transcriptional activator (42). GlnE (ATase) controls the activity of glutamine synthetase (GS) through posttranslational modification (18, 45, 46, 55). Under nitrogen-excess conditions, GlnE adenylylates GS and thereby lowers its activity. Under nitrogen-limiting conditions, GlnE removes that modification. NifA is a transcriptional activator of the nif genes, which encode nitrogenase and necessary auxiliary functions. DRAT and DRAG are two different enzymes that reversibly control nitrogenase activity posttranslationally. DRAT modifies and inactivates dinitrogenase reductase under conditions of nitrogen excess or energy deficiency, while DRAG removes that modification under conditions appropriate to nitrogen fixation (35).
What then is the molecular role of UMP in affecting the interaction of PII and its receptor proteins? It is broadly believed that the T loop makes direct contact with appropriate regions of at least some receptor proteins (19, 26, 27, 36). We assumed this to be true and supposed that the uridylylated Tyr51 at the tip of the T loop might well also interact with the receptors. In this case, appropriate receptor proteins would or would not have surfaces that directly contact the uridylylated or the unmodified residue. We became somewhat skeptical of this simple model when we converted GlnJ of R. rubrum to perform a GlnB-specific function. The activity of NifA is posttranslationally activated by GlnB-UMP in R. rubrum (65, 68). GlnJ-UMP is unable to activate NifA, and we therefore systematically changed GlnJ residues to those of GlnB to determine the minimal set of residues that defined NifA's specificity for GlnB (68). The critical residues for interaction with NifA mapped to a large part of the GlnB surface, including positions far from the site of uridylylation (68). This seemed to imply that NifA would have to contact a very large GlnB surface, at least if the T loop was extended. Instead we proposed that, at least for this interaction, the UMP-T loop might move closer to the body of GlnB than has been found for the T loop in the solved UMP-free GlnB structures. Consistent with the notion of T-loop flexibility, some structures have failed to resolve this region of PII (4, 48, 59-61).
In the present work, we tried to expand our understanding of the NifA-contacting residues of GlnB by selecting for GlnB variants, termed GlnB*, that interacted better with NifA than did wild-type GlnB in the yeast two-hybrid system. In fact we found these GlnB* variants to be profoundly altered in both nitrogen sensing and interaction with receptor proteins. Based on these results, we propose a model in which the role of uridylylation is to cause a conformational change in GlnB to a form that signals nitrogen deficiency.
MATERIALS AND METHODS
Growth conditions and whole-cell nitrogenase activity assays.
R. rubrum strains were grown in rich (supplemented malate-NH4+ [SMN]) medium or minimal (malate-glutamate ([MG]) medium (11, 30). The whole-cell nitrogenase activity assay has been described previously (62).
Construction of plasmids for two-hybrid analysis.
About 350 bp of R. rubrum wild-type glnB was PCR amplified with two oligonucleotides with EcoRI and SalI sites at both ends, and cloned into pGBDU-C1 (20) to generate an in-frame fusion with the GAL4 DNA-binding domain, yielding pUX679. Similarly, R. rubrum wild-type draT, nifA, ntrB, and glnE were PCR amplified with two oligonucleotides with BamHI and PstI sites at both ends and cloned into pGAD-C1 (20) to generate an in-frame fusion with the GAL4 activation domain, yielding pUX682, pUX686, pUX688 and pUX1959, respectively.
Random PCR mutagenesis and yeast two-hybrid selection of mutants.
Mutations in glnB were generated with pUX679 (GAL4 DNA-binding domain-GlnB fusion) as a template and two oligonucleotides with EcoRI and SalI sites at both ends as primers using the GeneMorph II random mutagenesis kit (Stratagene, La Jolla, California). The mutagenized DNA was digested with EcoRI and SalI, and ligated with pGBUD-C1 (20). After transformation with E. coli DH5α, 5 × 104 independent colonies were pooled and the plasmids were isolated and used to transform Saccharomyces cerevisiae strain pAJ69 harboring pUX686 (GAL4 activation domain-NifA fusion) (UY19) by the lithium acetate method (15, 50). The transformants were selected on SD (a yeast minimal medium) plates lacking leucine, uracil, and histidine and containing 1.5 mM 3-amino-1,2,4-triazol, a competitive inhibitor of His3 protein (9). Plasmids were recovered from fast-growing colonies and then reintroduced into UY19 to exclude false positives. The glnB was sequenced using Big Dye Terminator v.3.1 cycle sequencing kit (Applied Biosystems) to localize the mutation positions.
Construction of R. rubrum mutants.
To construct the glnB glnD mutant, two plasmids pUX1295 (ΔglnD3::accC1) and pUX1299 (ΔglnD4::accC1) (67) were separately conjugated into UR717 (ΔglnB3) (65). Smr Nxr Gmr colonies were selected and then screened for Cms clones resulting from double-crossover events, and the two glnB glnD mutants were designated UR1665 and UR1666. In UR1665, glnD and aacC1 are transcribed in the opposite direction, while in UR1666 they are transcribed in the same direction. An R. rubrum mutant, UR1760 (glnB glnD nifH::lacZ), was obtained by conjugating plasmid pUX1831 (nifH::lacZ) (R. Kerby et al., unpublished results) into R. rubrum UR1665. Smr Nxr Gmr Kmr colonies were selected and replica printed to screen for Cms (Cmr is encoded by the vector sequence) colonies. R. rubrum UR1789 (ΔglnB ΔglnK glnJ::lacZ) was obtained by conjugating pUX2040 (glnJ::lacZ) (D. Wolfe et al., unpublished results) into UR757 (ΔglnB ΔglnK) (65). Smr Nxr Gmr Kmr colonies were selected and then screened for Cms clones resulting from double-crossover recombination events.
Expression of GlnB* variants in R. rubrum.
For integration of glnB alleles in single copy into the chromosome, a 2.5-kb fragment of R. rubrum glnB was cloned into a suicide vector, pUX19 (32), which was designated pUX1906. Six plasmids (from pUX2010 to pUX2015) producing GlnB* variants E15A, K17T, H42Q, L45P, V52D, and F55L, respectively, were constructed with pUX1906 as the template by the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Together with pUX1906, these six plasmids were transformed into E. coli strain S17-7 (52), then conjugated into UR1665 and UR1666 (glnB glnD) separately, and Smr Nxr Gmr Kmr colonies were selected. In the resulting strains (from UR1683 to UR1696), glnB or glnB* is expressed at normal levels from its own promoter. Multicopy plasmids expressing the selected GlnB* variants (pUX1999-2004) were constructed by inserting about 1-kb fragments carrying the glnB promoter and the appropriate glnB allele into the broad-host-range plasmid pRK404 (7). Together with pUX1998 (wild-type glnB), these six plasmids were transferred from E. coli into R. rubrum UR1665/UR1666 (glnB glnD), UR1760 (glnB glnD nifH::lacZ), and UR1789 (ΔglnB ΔglnK glnJ::lacZ) separately by triparental mating, yielding UR1669-1682, UR1761-1767, and UR1834-1840, respectively (16).
For overexpression of R. rubrum wild-type GlnB and five GlnB* variants (except E15A) in E. coli, a 0.4-kb fragment of glnB or glnB* was PCR amplified and cloned into pND706 (34) at NdeI and EcoRI sites and transformed into strain BK (ΔglnB Δmdl-glnK::kan) obtained from Alex Ninfa (3). For each strain (UQ2549, and UQ4308-4312), a 10-ml overnight culture was inoculated into 500 ml of 2× LB medium supplemented with 100 μg/ml ampicillin and 25 μg/ml kanamycin. Cultures were incubated at 30°C until the optical density at 600 nm reached 1.2 to 1.7; the temperature was then shifted to 44°C, and growth continued for 4 h. Cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl, pH 7.5, with 1 μg/ml leupeptin, frozen at −20°C overnight, and then lysed by sonication. Following precipitation with 45% (final) ammonium sulfate, the precipitant was resuspended in 50 mM Tris-HCl, pH 7.5, and the GlnB was purified to greater than 90% purity on a Q-Sepharose column. E. coli GlnD was purified from a strain with pDOP1 (28) by a protocol similar to that described for GlnB above, and purification to approximately 50% purity was achieved.
Uridylyltransferase assay.
The uridylyltransferase assay was performed as described previously. Reaction mixtures included 100 mM Tris-HCl, pH 7.5, 100 mM KCl, 25 mM MgCl2, ATP, and αKG as indicated, 1 mM dithiothreitol, 0.3 mg/ml bovine serum albumin, 0.8 μM GlnD, and 8 to 12 μM GlnB (23). Reaction mixtures were preincubated at 30°C for 2 min, started with the addition of 1 mM UTP, and incubated for 1 h at 30°C, before being stopped by the addition of 25 mM EDTA. Uridylylation was detected qualitatively using nondenaturing 12.5% polyacrylamide gel electrophoresis (PAGE) analysis, as described previously (2).
Protein immunoblotting.
A trichloroacetic acid precipitation method was used for rapid protein extraction (63). GlnJ and GS Western blotting was performed as described previously (64, 67).
Assay for β-galactosidase in liquid culture.
R. rubrum cultures were first grown in SMN medium aerobically in the dark for 2 days and then inoculated into MG media at a 65-fold dilution. MG-grown cultures were anaerobically grown under light for 2 days, and NH4+ was added as described. β-Galactosidase activity was measured by the standard protocol (38).
RESULTS AND DISCUSSION
Rationale and working hypothesis.
As stated in the introduction, we wished to probe the surfaces of GlnB that interact with NifA. Yeast two-hybrid analysis (9, 10) has already been used with PII homologs to verify presumed receptor proteins and to find new ones (5, 36, 37, 44, 49). However, we used a version of the yeast system (20) to select for GlnB variants that showed improvement in interaction with NifA as judged by improved growth of the yeast reporter.
We assumed that GlnB would not be uridylylated in yeast, that improved interactions would result from changes in specific contact residues, and that such changes would energetically compensate for the absence of UMP. We expected that these changes would affect PII interaction with receptor proteins other than NifA in a varied fashion, depending on whether or not the changes happened to affect important contacts for those different receptors. In fact, the GlnB variants chosen for improved interaction with NifA were typically reduced in their ability to interact with two other receptors, NtrB and DRAT, in the two-hybrid assay. NtrB is known to interact with the nonuridylylated form of PII in E. coli (22). Although the situation with DRAT remains unknown, it is likely that it too interacts with nonuridylylated form of PII. In contrast to the initial hypothesis, the results described below suggest an admittedly simplistic model for the role of UMP: GlnB exists in two different functional forms, one signaling N sufficiency and the other N deficiency, and each of these forms interacts with one of two different sets of receptor proteins. In this model, uridylylation has the primary effect of shifting the equilibrium toward the N-deficiency form. The substitutions in the GlnB variants are then explained as those changes that shift that equilibrium toward the N-deficiency form, but in the absence of UMP. This in turn explains the uniform reduction in their interaction with NtrB, a receptor that favors the N-sufficiency form. We recognize that αKG and ATP also affect PII interaction with receptors and certainly also affect PII conformation (25, 29, 33); it is likely that multiple structural forms of PII exist, reflecting the nitrogen, carbon, and probably energy status in the cell. However, we only focus on the functional change by UMP here.
The two-hybrid screen with NifA yields variants that no longer require UMP for significant interaction with NifA in R. rubrum.
To screen for GlnB variants that are improved for interaction with NifA, we mutagenized the glnB region in the yeast expression vector by error-prone PCR, producing approximately 1.5 mutations per clone, as determined by direct sequencing. These were then used as bait with an appropriate yeast clone carrying NifA. The wild-type controls showed a low but detectable level of interaction (Table 1 and Fig. 2), and the frequency of mutants with improved growth (and therefore improved interaction) was approximately 0.1%. The plasmids containing the glnB alleles were isolated and returned to an identical yeast strain to verify that they were causal to the improved growth. They were then sequenced, and 15 different variants were detected in a total of 36 mutants (Table 1). These GlnB* variants differed in their specific degrees of interaction in this assay, and most were significantly better than wild-type GlnB, although some, such as E15A and L45M GlnB, were only marginally improved (Table 1 and Fig. 2).
TABLE 1.
GlnB* variants identified for their improved interaction with NifA in the yeast two-hybrid systema
| GlnB* variantb | Growth response |
|---|---|
| Wild type | −/+ |
| K12R | +++ |
| E15Ac | ++ |
| E15D | +++ |
| K17T (2)c | ++++ |
| I28L | +++ |
| H42Q (9)c | ++++ |
| L45P (5)c | +++ |
| L45M | ++ |
| R47P (3) | ++++ |
| V52D (3)c | ++++ |
| V52I | +++ |
| F55L (5)c | ++++ |
| V59A | +++ |
| R72C | +++ |
| A95T | +++ |
The growth response of the yeast strains expressing each variant is shown. Cells were grown in minimal (SD) medium without histidine and in the presence of 3 mM 3-amino-1,2,4-triazol, a competitive inhibitor of His3 protein (9). A sense of the degree of growth that is represented by the plus signs can be gained by the photographs of the selected variants shown in Fig. 2.
The various causative substitutions found in GlnB* variants are shown in the left column, with the number of identical isolates shown in parentheses.
These variants are the selected set used in many of the later experiments.
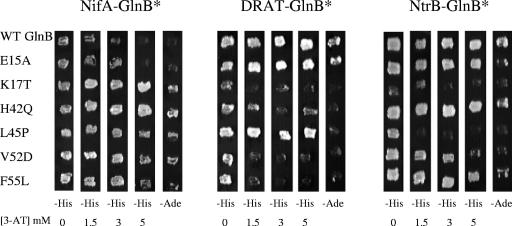
FIG. 2.
Yeast two-hybrid results demonstrate relative interaction between selected GlnB* variants and NifA, DRAT, and NtrB. Strains expressing the proteins indicated were patched onto different plates that are increasingly restrictive for growth and therefore increasingly demanding of GlnB interaction with its receptors, from left to right. In the first four columns, the demand is for growth in SD medium without histidine and in the presence of increasing amounts of 3-amino-1,2,4-triazol (3-AT), a competitive inhibitor of His3 protein (9). The right column demands growth in the absence of adenine (−Ade). WT, wild type. The experiment was performed in quintuplicate, but only a single set of responses is shown.
Because we were concerned that the yeast system might yield GlnB variants that improved interaction in yeast, but were physiologically irrelevant to the normal R. rubrum situation, we reconstructed six alleles encoding GlnB* variants with a range of responses in the two-hybrid assay into a low-copy plasmid capable of replication in R. rubrum. To avoid complications from the wild-type GlnB, we used a glnB glnD recipient and expressed the GlnB* variants from the normal promoter to decrease complications due to inappropriate protein accumulation. We have previously shown that there is no detectable GlnB uridylylation in a glnD mutant, and this mutant failed to activate NifA and NtrC under nitrogen-limiting conditions (67). We then tested the ability of these variants to support NifA activation under different conditions. We first examined their ability to activate NifA with a nifH::lacZ reporter fusion, which requires active NifA for function. In the absence of NH4+ there was little expression in the control strain expressing wild-type GlnB, because the glnD mutation eliminates GlnB uridylylation, which is normally essential for NifA activation (65, 67) (Table 2). In contrast, the strains with the tested GlnB* variants displayed substantial NifA activity, indicating that the GlnB* variants can activate NifA even without uridylylation. The experiment was also performed in the identical strains but in the presence of 17.8 mM NH4+ (Table 2). NifA activity was low in the strain with wild-type GlnB. Most but not all GlnB* variants showed decreased nifH::lacZ expression, though they remained substantially above that of the wild-type GlnB. This response in strains lacking the normal pathway for sensing NH4+ by the glutamine-binding GlnD suggests that these variants sense NH4+ by another means, perhaps through αΚG binding. We suppose that this GlnD-independent nitrogen regulation of GlnB can occur in a wild-type background as well, but it is less striking because the regulation is overshadowed by the uridylylation event. The relatively high level of nifH::lacZ expression remaining in these GlnB* mutants indicates that they still partially mimic the uridylylated form even under NH4+-excess conditions. H42Q GlnB seems to be particularly resistant to this NH4+ addition, so perhaps its interaction with NifA happens not to be perturbed by αKG binding because of the H42Q substitution. We do not know the reason for the increase in activity in the presence of NH4+ in the strain with E15A GlnB.
TABLE 2.
NifA activation by GlnB* variants under several conditionsa
| Wild type or glnB mutant | β-Galactosidase activity (Miller units) of:
|
Nitrogenase activity (−NH4+) ofc:
|
||
|---|---|---|---|---|
|
glnBD mutant nifH::lacZ multicopy glnB*
|
glnBD mutant multicopy glnB* | glnBD mutant single-copy glnB*b | ||
| −NH4+ | +NH4+ | |||
| Wild-typeGlnBd | 150 ± 8 | 90 ± 7 | <10 | <10 |
| E15A | 540 ± 34 | 1020 ± 124 | <10 | <10 |
| K17T | 4280 ± 55 | 480 ± 10 | 580 ± 67 | 20 ± 4 |
| H42Q | 6150 ± 80 | 6120 ± 100 | 510 ± 40 | 520 ± 91 |
| L45P | 5130 ± 22 | 1160 ± 16 | 570 ± 29 | 470 ± 60 |
| V52D | 5630 ± 152 | 1510 ± 40 | 530 ± 98 | 680 ± 32 |
| F55L | 3880 ± 40 | 1370 ± 82 | 680 ± 43 | 570 ± 73 |
| UR2e | ND | ND | 830 ± 52 | 840 ± 85 |
The data in columns 2, 3, and 4 were from a glnB glnD strain background with the GlnB* variants or the wild-type GlnB expressed from a low-copy plasmid.
In this column, the strain background is glnB glnD with the GlnB* variants or wild-type GlnB expressed in single copy.
Nitrogenase activity is expressed in nmol of ethylene produced h−1 ml of culture−1 at on optical density at 600 nm of 1.
Wild-type GlnB was expressed identically to the various GlnB* variants.
UR2 is wild-type R. rubrum; because it lacks the nifH::lacZ fusion, β-galactosidase activities were not determined (ND).
We also monitored NifA activation by measuring nitrogenase activity in vivo to verify that the above results were not an artifact of the reporter fusion. In this case, activity requires the successful activation of several nif operons, as well as the maturation and function of nitrogenase. In this assay, all GlnB* variants (except E15A) supported sufficient NifA activation to provide substantial nitrogenase activity (Table 2). Finally, the same set of mutations was moved in single copy into the R. rubrum chromosome, using a suicide vector to verify that the results were not an artifact of overexpression. The results were quite similar to those seen with the multicopy situation for all variants except for K17T GlnB. The lower nitrogenase activity in the strain this variant expressed from single copy suggests that there is a weak activation of NifA by K17T GlnB in R. rubrum that is masked by high GlnB expression. Because K17T GlnB is generally effective with other receptors (see below), we assume that the K17T substitution happens to affect NifA interaction specifically. Consistent with this, Lys17 is one of the GlnB residues found to be critical for its activation of NifA activity in our previous study (68). Overall, the results show that GlnB* variants, chosen for better interaction with NifA in yeast, can productively interact with NifA even without uridylylation. Most of these variants continue to sense NH4+ addition by an unknown mechanism independent of GlnD uridylylation.
Two-hybrid analyses of the GlnB* variants show that interactions with other PII targets are also altered, and in a pattern consistent with the GlnB* variants mimicking GlnB-UMP.
The original hypothesis was that the GlnB* variants would be specifically improved for NifA interaction, but we could not predict the effect of the substitutions on interactions with other receptor proteins. We had already identified two other R. rubrum receptor proteins that interacted well with wild-type GlnB in the yeast system, NtrB and DRAT (Fig. 2), and these receptors had also been detected in the yeast system by others with homologous proteins from other organisms (36, 44, 49). NtrB is thought to interact exclusively with the nonuridylylated form of GlnB (21, 23). Based on the present data, we assume that the nonuridylylated form of GlnB is also critical for interaction with DRAT. We then tested the ability of the selected set of GlnB* variants to interact with these receptors in the yeast system. Each of these GlnB* variants interacted differently with NtrB and DRAT, but the striking result was that all but E15A interacted less well with each of these two receptors than did wild-type GlnB (Fig. 2). The exception, E15A, remained similar to the wild type for NtrB interaction, but it was also the variant with the least improvement in NifA interaction, so this correlation is not surprising. We note that the specific interactions of each GlnB* variant with DRAT and NtrB are not identical, but they are uniformly weaker than those of the wild type. We interpret the different patterns of interaction for each GlnB* to mean that the substitution in each GlnB* variant does cause some receptor-specific perturbations, presumably by either direct or indirect effects on GlnB structure. However, the pattern of uniformly decreased interactions (compared to those of the wild type) with both receptors that typically bind the non-UMP form requires a rather different explanation, such as the equilibrium shift model described above.
The GlnB* variants continued to interact with GlnE (data not shown), but this result is complicated by the apparent fact that both GlnB and GlnB-UMP are thought to interact with GlnE, albeit at different sites (18). As a consequence, an equilibrium shift in GlnB might change the site of GlnE interaction, but not necessarily its magnitude. Further analysis using fragments of GlnE should prove informative. No interaction of wild-type GlnB or the GlnB* variants was detected with DRAG or AmtB1, but such negative results are not interpretable and might be due to a disoriented fusion protein (data not shown).
The important substitutions in the GlnB* variants are found predominantly, but not exclusively, near the T loop.
Figure 1A shows a model of R. rubrum GlnB based on the known structure of E. coli GlnB (6, 60), and the three T loops and an ATP-binding site are indicated. The wild-type residues altered in the GlnB* variants are shown as space filled in same structure in Fig. 1B. For clarity, only GlnB* residues on one face of the trimer are shown, although residues Lys12, Glu15, and Arg72 from two monomers are repeated on both the left and right sides. Several points are evident. (i) While a number of changes are found in and near the T loop, many GlnB* residues identified in the two-hybrid analysis lie elsewhere. (ii) There is not an obvious clustering of substitutions near the known ATP-binding site, although indirect effects on this region cannot be ruled out. Though we propose that these substitutions might each have the effect of shifting the GlnB equilibrium to a new structure, it is premature to speculate on that structure. We recognize that several substitutions, such as T29M, A49P, and T83N, have been found in E. coli GlnB that altered its interaction with other receptors (26). All of these wild-type residues are conserved in all three R. rubrum PII homologs, but substitutions at these positions were not detected in our selection. It is possible that the phenotypes of these variants simply do not satisfy the initial selection that we imposed.
When expressed at normal levels, the GlnB* variants are altered in their regulation of NtrB and GlnE.
We examined the ability of these GlnB* variants to regulate the two receptors that are most critical for cell growth, NtrB and GlnE. First, we examined the effects of GlnB* variants on GlnE activity by Western analysis of GS from cells grown under nitrogen-limiting conditions and then after NH4+ addition. As shown in Fig. 3, GS in the UR2 (wild-type and glnD+) strain (lanes 15 and 16) was substantially nonadenylylated before NH4+ addition and adenylylated afterwards. In the glnD mutant background, the wild-type GlnB (UR1683) lacks uridylylation and therefore signals nitrogen sufficiency, so that GS is adenylylated under both conditions (lanes 1 and 2), similar to previous results (67). In a number of the GlnB* variants, except for E15A, whose relatively weak phenotype was noted above, a distinct nonadenylylated band is detectable before NH4+ addition, consistent with these variants mimicking the uridylylated form even in the absence of GlnD. In some GlnB* variants, especially in H42Q (UR1689), L45P (UQ1691), and F55L (UQ1695), the deadenylylated band predominates, indicating that the regulation of GlnE activity by these GlnB* variants is poised toward demodification of GS even in this GlnD− background. More interestingly, all GS became adenylylated after NH4+ addition, again indicating that these GlnB* variants are still able to respond to a nitrogen signal in the absence of GlnD. It is important to note that the pattern of effectiveness of GlnB* variants for regulating GlnE does not completely match the pattern for NifA described above, nor does it match that for NtrB to be described below. This might either mean that the specific substitutions in the GlnB* variants have also altered contact surfaces for different receptor proteins or that these substitutions alter either αKG or ATP binding or their effect on receptor interaction.
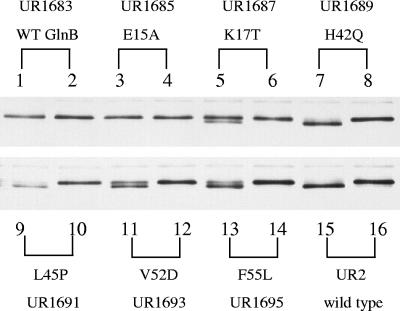
FIG. 3.
Perturbed modification of GS in strains expressing GlnB* variants demonstrates altered regulation of GlnE activity. The figure shows a Western blot of a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel developed with antibody to GS, where adenylylated GS appears as the upper of the two bands. All strains are glnB glnD but express wild-type (WT) GlnB (UR1683) or GlnB* variants (UR1685, -1687, -1689, -1691, -1693, and -1695) in single copy from an integrated plasmid. For each pair of lanes, the first shows GS from the cells grown in MG medium (nitrogen deficient, since glutamate serves as sole nitrogen source), and the second shows the GS 60 min after the addition of 10 mM NH4Cl. Lanes 15 and 16 contain similar preparations from UR2 (wild-type R. rubrum).
To analyze GlnB* effects on NtrB function, we used two methods. First, because glnJ is expressed under the control of NtrC (67), we analyzed the effects of the GlnB* variants on the expression of a glnJ::lacZ fusion in strains that lacked all other PII homologs, to avoid any possible complications caused by their presence. Because of complications in strain construction, the GlnB alleles were expressed from the low-copy plasmid noted above in a glnB glnJ::lacZ glnK background. The β-galactosidase activity was measured in cultures grown in MG medium with or without NH4+. All of these variants and wild-type GlnB supported high levels of lacZ expression from this reporter in MG medium without NH4+ (Table 3). In the presence of NH4+, GlnD causes the deuridylylation of GlnB, resulting in poor NtrC activation, as seen in the strain expressing wild-type GlnB (Table 3). Two strains with GlnB* variants (E15A and H42Q) had slightly higher β-galactosidase activity than that seen with wild-type GlnB, but the other four GlnB* variants showed high activity. The consistency of these results with those of the GlnB*-NtrB interaction detected in the yeast is surprisingly good (Fig. 2), given that there are a number of differences between the two assays. These results suggest that conformations of these four GlnB* variants decrease their interaction with NtrB, which results in activation of NtrC even in the presence of NH4+. We lack a good explanation for the fact that β-galactosidase activity is higher in the presence of NH4+ than in its absence in the case of two GlnB* variants, K17T and L45P, but the result was reproducible.
TABLE 3.
NtrC activation by GlnB* variants
| R. rubrum strain | Mutation in glnB on plasmid | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|---|
| −NH4+b | +NH4+c | ||
| UR1834 | Wild type | 510 ± 88 | 28 ± 3 |
| UR1835 | E15A | 1040 ± 52 | 35 ± 4 |
| UR1836 | K17T | 500 ± 100 | 930 ± 127 |
| UR1837 | H42Q | 670 ± 40 | 41 ± 3 |
| UR1838 | L45P | 560 ± 77 | 950 ± 70 |
| UR1839 | V52D | 400 ± 48 | 350 ± 87 |
| UR1840 | F55L | 420 ± 43 | 130 ± 18 |
All strains are glnB glnJ::lacZ glnK (where the glnJ mutation eliminates GlnJ function), and they express wild-type GlnB or the identified GlnB* variants from the normal glnB promoter on a low-copy plasmid. All values are the average of three independent analyses.
Cultures were grown in MG (malate-glutamate) medium.
Cultures were grown in MG medium plus 17.8 mM NH4Cl.
Second, we monitored NtrC function by the accumulation of GlnJ using Western analysis. In this case, we used glnB glnD strains with the GlnB* variants expressed from a single copy in the chromosome under nitrogen-poor growth conditions (MG medium). Wild-type R. rubrum (UR2; glnB+ glnD+) accumulated higher levels of GlnJ than did the variants (Fig. 4). However, a strain expressing wild-type GlnB in the glnB glnD background accumulated negligible GlnJ because the nonuridylylated form of GlnB activates the phosphatase activity of NtrB, which prevents NtrC activation. This is similar to our previous results demonstrating that GlnD mutants have very low levels of GlnJ (67). Strains with E15A and K17T GlnB had only a modest effect on GlnJ accumulation, while the other GlnB* alleles allowed dramatic GlnJ accumulation (Fig. 4), consistent with GlnD-independent activation of NtrC in these variants. There are certainly some differences in NtrC activation seen in these two different assays (Fig. 2 and Table 3), especially in K17T and H42Q GlnB. However, there are a number of possible explanations for these, including differences in GlnB accumulation because of copy number, complications due to heterotrimers formed between GlnB* and GlnJ, and secondary physiological effects because of the different genetic backgrounds.
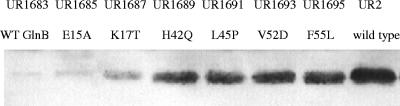
FIG. 4.
GlnJ accumulates in a glnB glnD background when GlnB* variants are expressed in single copy from an integrated plasmid. Cells were grown in MG medium, and proteins from crude extracts were separated by SDS-PAGE and immunoblotted with antibody against R. rubrum GlnJ. UR2 is the R. rubrum wild type (WT). UR1683 contains wild-type GlnB, while UR1685, -1687, -1689, -1691, -1693, and -1695 contain the GlnB* variants.
Taken together, these results show that the GlnB* variants are profoundly perturbed in their interactions with a variety of different proteins. Although it is impossible to quantitatively compare these differences in R. rubrum with differences in interaction in the yeast system, the broad conclusion is clear: these GlnB* variants behave as if they are at least partially shifted toward a form that mimics the UMP-bound (N deficient) form of wild-type GlnB, and this affects all of their interactions with different receptors. The fact that some perturbations are more profound than others presumably reflects the extent of the equilibrium shift, the degree to which a specific substitution affects interaction with a specific receptor, and the degree of interaction necessary to cause a discernible effect on the cell's phenotype.
The GlnB* variants are perturbed in their requirements for uridylylation in vitro.
Because of the substantially altered behavior of the GlnB* variants in vivo, we presumed that their in vitro behavior would also be different and informative. We therefore purified and analyzed the modification requirements of a subset of GlnB* variants, using GlnD purified from E. coli.
Under our assay conditions, wild-type GlnB is substantially uridylylated only in the presence of high levels of αKG and ATP (Fig. 5). This is somewhat in contrast to the case of E. coli GlnB, using the same GlnD, where rather lower levels (33 μM) of αKG are sufficient for full modification (23).
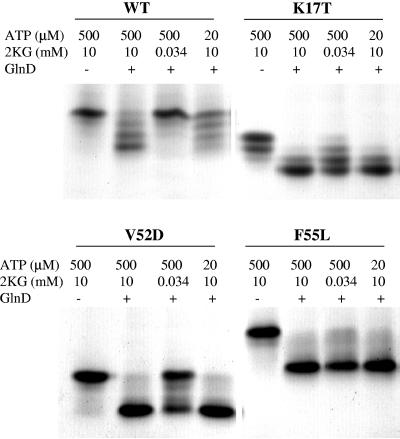
FIG. 5.
In vitro uridylyltransferase activity assays on wild-type (WT) GlnB and GlnB* variants show that many of these variants are dramatically altered in their properties as GlnD substrates. Wild-type GlnB and four GlnB* proteins were overexpressed and purified from E. coli (UQ2549, UQ4308, UQ4310, UQ4311, and UQ4312) and then used as substrates of E. coli GlnD as described in Materials and Methods. The specific GlnB protein used in each portion of the gel is shown at the top, and below that are the specific conditions for the samples in each lane. These are native gels, so that the protein trimer runs at as many as four positions, depending on the number of UMP groups per trimer, with the fully uridylylated form moving the fastest.
In our native gels, K17T and V52D GlnB migrate faster than does wild-type GlnB because of the charge change of the substitutions. With this in mind, Fig. 5 shows that K17T GlnB is substantially modified at both low and high αKG and ATP levels, demonstrating a higher propensity for uridylylation than wild-type GlnB. (The slight modification of K17T GlnB before treatment apparently reflects a slight difference in culture conditions from which this variant was harvested.) V52D GlnB is almost completely modified at high αKG with either low or high ATP levels, while F55L GlnB is almost completely modified at both low αKG and low ATP levels. L45P and H42Q GlnB were both rather similar to wild-type GlnB in their uridylylation properties under these conditions (data not shown), and E15A GlnB was not analyzed.
We cannot directly interpret these differences in terms of biological function. Nevertheless, the results do show that many of these variants are more readily uridylylated under ATP- and αKG-limiting conditions than is wild-type GlnB. The simplest explanation might be that these variants mimic the form of the protein that is stabilized by that modification and are therefore better substrates for uridylylation. While we do not know why all tested variants do not behave more similarly, it is possible that these substitutions happen to have a secondary effect on GlnB binding by GlnD. The different uridylylation requirements of these GlnB* variants need to be further investigated.
Working hypothesis.
The notion that uridylylation has a profound effect on PII function and that it shifts PII from the N-sufficiency form to the N-deficiency form is well known. Our results suggest a model in which a primary role of uridylylation is to stimulate a global conformational change, which is substantially mimicked by these GlnB* variants. We will discuss the current paradigms of PII regulation by covalent modification and then put our results in a larger context.
Work from Ninfa's laboratory has elegantly shown that αKG and ATP bind to PII synergistically and that they have the potential to affect uridylylation and deuridylylation (23). Similar results have been seen in cyanobacteria, although PII is regulated by phosphorylation (13). These analyses have led to a multistate model of PII, where the major effect is the result of uridylylation, but where αKG has additional conformational effects that affect certain PII-receptor interactions. The situation in the cyanobacteria has differences noted in the introduction, but similarities as well. Here again PII binds αKG and ATP, and the presence of these is thought to be necessary for phosphorylation in vivo (12).
We recognize the complications inherent in the synergistic binding of ATP and αKG and the anti-cooperative binding of αKG (23, 29), but suggest that the UMP effect might be viewed more simply. That is, because GlnB* variants chosen for improved interaction with NifA in yeast are also uniformly poorer than wild-type GlnB for interaction with NtrB and DRAT, a two-state model for these differences is the simplest hypothesis. Moreover, because these GlnB* variants mimic the uridylylated form of GlnB by several analyses, it is our hypothesis that such a two-state model can account for uridylylation effects as well. We assume these two states represent a global conformational change, perhaps involving in the T loop, but the nature of that change is completely unclear at present.
The fact that the GlnB* variants are altered in their interaction with so many receptor proteins is most easily explained by such a global conformational change. However, the fact that different GlnB* variants show somewhat different responses to specific receptors is consistent with the idea that some of these substitutions also directly or indirectly alter residues that are more or less critical for interaction with specific receptors.
The results presented here have revealed a phenotypically striking class of GlnB variants in which single substitutions profoundly alter central nitrogen regulation. The results imply a model in which the role of uridylylation is to promote a conformational change in the protein.
Acknowledgments
This work was supported by the College of Agricultural and Life Sciences, University of Wisconsin—Madison, and NIGMS grant GM65891 to G.P.R.
We thank Alex Ninfa for generously providing a number of strains critical for GlnB and GlnD overexpression, Betty Craig and Philip James for the yeast strains and plasmids for the yeast two-hybrid system, and David Ollis for expression vectors. We thank Jose Serate for purifying GlnD and GlnB proteins, Edward Pohlmann and Jonathan Jacobs for their help with some experiments, Robert Kerby and David Wolfe for providing unpublished plasmids, and Hwan Youn for generating structure pictures. We also thank David Wolfe and Hwan Youn for helpful criticisms of the work and the manuscript.
REFERENCES
- 1.Arcondéguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., E. S. Kamberov, R. L. Weiss, and A. J. Ninfa. 1994. Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 269:28288-28293. [PubMed] [Google Scholar]
- 3.Atkinson, M. R., and A. J. Ninfa. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431-447. [DOI] [PubMed] [Google Scholar]
- 4.Benelli, E. M., M. Buck, I. Polikarpov, E. M. Souza, L. M. Cruz, and F. O. Pedrosa. 2002. Herbaspirillum seropedicae signal transduction protein PII is structurally similar to the enteric GlnK. Eur. J. Biochem. 269:3296-3303. [DOI] [PubMed] [Google Scholar]
- 5.Burillo, S., I. Luque, I. Fuentes, and A. Contreras. 2004. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J. Bacteriol. 186:3346-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheah, E., P. D. Carr, P. M. Suffolk, S. G. Vasudevan, N. E. Dixon, and D. L. Ollis. 1994. Structure of the Escherichia coli signal transducing protein PII. Structure 2:981-990. [DOI] [PubMed] [Google Scholar]
- 7.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X.-W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 8.Dodsworth, J. A., N. C. Cady, and J. A. Leigh. 2005. 2-Oxoglutarate and the PII homologues NifI1 and NifI2 regulate nitrogenase activity in cell extracts of Methanococcus maripaludis. Mol. Microbiol. 56:1527-1538. [DOI] [PubMed] [Google Scholar]
- 9.Fields, S. 1993. The two-hybrid system to detect protein-protein interactions. Methods (Orlando) 5:116-124. [Google Scholar]
- 10.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 11.Fitzmaurice, W. P., L. L. Saari, R. G. Lowery, P. W. Ludden, and G. P. Roberts. 1989. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol. Gen. Genet. 218:340-347. [DOI] [PubMed] [Google Scholar]
- 12.Forchhammer, K. 2004. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28:319-333. [DOI] [PubMed] [Google Scholar]
- 13.Forchhammer, K., and A. Hedler. 1997. Phosphoprotein PII from cyanobacteria: analysis of functional conservation with the PII signal-transduction protein from Escherichia coli. Eur. J. Biochem. 244:869-875. [DOI] [PubMed] [Google Scholar]
- 14.Forchhammer, K., and N. Tandeau de Marsac. 1994. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 176:84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 16.Grunwald, S. K., D. P. Lies, G. P. Roberts, and P. W. Ludden. 1995. Posttranslational regulation of nitrogenase in Rhodospirillum rubrum strains overexpressing the regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase activating glycohydrolase. J. Bacteriol. 177:628-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irmler, A., and K. Forchhammer. 2001. A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803. Proc. Natl. Acad. Sci. USA 98:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaggi, R., W. C. van Heeswijk, H. V. Westerhoff, D. L. Ollis, and S. G. Vasudevan. 1997. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J. 16:5562-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaggi, R., W. Ybarlucea, E. Cheah, P. D. Carr, K. J. Edwards, D. L. Ollis, and S. G. Vasudevan. 1996. The role of the T-loop of the signal transducing protein PII from Escherichia coli. FEBS Lett. 391:223-228. [DOI] [PubMed] [Google Scholar]
- 20.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, P., M. R. Atkinson, C. Srisawat, Q. Sun, and A. J. Ninfa. 2000. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry 39:13433-13449. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, P., and A. J. Ninfa. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37:12782-12794. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of Ntr gene transcription in Escherichia coli. Biochemistry 37:12795-12801. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37:12802-12810. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, P., P. Zucker, M. R. Atkinson, E. S. Kamberov, W. Tirasophon, P. Chandran, B. R. Schefke, and A. J. Ninfa. 1997. Structure/function analysis of the PII signal transduction protein of Escherichia coli: genetic separation of interactions with protein receptors. J. Bacteriol. 179:4342-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, P., P. Zucker, and A. J. Ninfa. 1997. Probing interactions of the homotrimeric PII signal transduction protein with its receptors by use of PII heterotrimers formed in vitro from wild-type and mutant subunits. J. Bacteriol. 179:4354-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamberov, E. S., M. R. Atkinson, J. Feng, P. Chandran, and A. J. Ninfa. 1994. Sensory components controlling bacterial nitrogen assimilation. Cell. Mol. Biol. Res. 40:175-191. [PubMed] [Google Scholar]
- 29.Kamberov, E. S., M. R. Atkinson, and A. J. Ninfa. 1995. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 270:17797-17807. [DOI] [PubMed] [Google Scholar]
- 30.Lehman, L. J., and G. P. Roberts. 1991. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J. Bacteriol. 173:5705-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lie, T. J., G. E. Wood, and J. A. Leigh. 2005. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280:5236-5241. [DOI] [PubMed] [Google Scholar]
- 32.Lies, D. P. 1994. Ph.D. thesis. University of Wisconsin—Madison, Madison, Wis.
- 33.Little, R., F. Reyes-Ramirez, Y. Zhang, W. C. van Heeswijk, and R. Dixon. 2000. Signal transduction to the Azotobacter vinelandii NIFL-NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 19:6041-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love, C. A., P. E. Lilley, and N. E. Dixon. 1996. Stable high-copy-number bacteriophage lambda promoter vectors for overproduction of proteins in Escherichia coli. Gene 176:49-53. [DOI] [PubMed] [Google Scholar]
- 35.Ludden, P. W., and G. P. Roberts. 1989. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr. Top. Cell. Regul. 30:23-56. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Argudo, I., and A. Contreras. 2002. PII T-loop mutations affecting signal transduction to NtrB also abolish yeast two-hybrid interactions. J. Bacteriol. 184:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Argudo, I., P. Salinas, R. Maldonado, and A. Contreras. 2002. Domain interactions on the ntr signal transduction pathway: two-hybrid analysis of mutant and truncated derivatives of histidine kinase NtrB. J. Bacteriol. 184:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Moorhead, G. B., and C. S. Smith. 2003. Interpreting the plastid carbon, nitrogen, and energy status. A role for PII? Plant Physiol. 133:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276:38320-38328. [DOI] [PubMed] [Google Scholar]
- 41.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 42.Ninfa, A. J., M. R. Atkinson, E. S. Kamberov, J. Feng, and E. G. Ninfa. 1995. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria, p. 67-88. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 43.Ninfa, A. J., and P. Jiang. 2005. PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8:168-173. [DOI] [PubMed] [Google Scholar]
- 44.Pawlowski, A., K.-U. Riedel, W. Klipp, P. Dreiskemper, S. Groß, H. Bierhoff, T. Drepper, and B. Masepohl. 2003. Yeast two-hybrid studies on interaction of proteins involved in regulation of nitrogen fixation in the phototrophic bacterium Rhodobacter capsulatus. J. Bacteriol. 185:5240-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee, S. G., P. B. Chock, and E. R. Stadtman. 1985. Glutamine synthetase from Escherichia coli. Methods Enzymol. 113:213-241. [DOI] [PubMed] [Google Scholar]
- 46.Rhee, S. G., S. C. Park, and J. H. Koo. 1985. The role of adenylyltransferase and uridylyltransferase in the regulation of glutamine synthetase in Escherichia coli. Curr. Top. Cell. Regul. 27:221-232. [DOI] [PubMed] [Google Scholar]
- 47.Ruppert, U., A. Irmler, N. Kloft, and K. Forchhammer. 2002. The novel protein phosphatase PphA from Synechocystis PCC 6803 controls dephosphorylation of the signalling protein PII. Mol. Microbiol. 44:855-864. [DOI] [PubMed] [Google Scholar]
- 48.Sakai, H., H. Wang, C. Takemoto-Hori, T. Kaminishi, H. Yamaguchi, Y. Kamewari, T. Terada, S. Kuramitsu, M. Shirouzu, and S. Yokoyama. 2005. Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8. J. Struct. Biol. 149:99-110. [DOI] [PubMed] [Google Scholar]
- 49.Salinas, P., and A. Contreras. 2003. Identification and analysis of Escherichia coli proteins that interact with the histidine kinase NtrB in a yeast two-hybrid system. Mol. Genet. Genomics 269:574-581. [DOI] [PubMed] [Google Scholar]
- 50.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 51.Schwarzenbacher, R., F. von Delft, P. Abdubek, E. Ambing, T. Biorac, L. S. Brinen, J. M. Canaves, J. Cambell, H. J. Chiu, X. Dai, A. M. Deacon, M. DiDonato, M. A. Elsliger, S. Eshagi, R. Floyd, A. Godzik, C. Grittini, S. K. Grzechnik, E. Hampton, L. Jaroszewski, C. Karlak, H. E. Klock, E. Koesema, J. S. Kovarik, A. Kreusch, P. Kuhn, S. A. Lesley, I. Levin, D. McMullan, T. M. McPhillips, M. D. Miller, A. Morse, K. Moy, J. Ouyang, R. Page, K. Quijano, A. Robb, G. Spraggon, R. C. Stevens, H. van den Bedem, J. Velasquez, J. Vincent, X. Wang, B. West, G. Wolf, Q. Xu, K. O. Hodgson, J. Wooley, and I. A. Wilson. 2004. Crystal structure of a putative PII-like signaling protein (TM0021) from Thermotoga maritima at 2.5 Å resolution. Proteins 54:810-813. [DOI] [PubMed] [Google Scholar]
- 52.Simon, R., U. B. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 53.Smith, C. S., N. A. Morrice, and G. B. Moorhead. 2004. Lack of evidence for phosphorylation of Arabidopsis thaliana PII: implications for plastid carbon and nitrogen signaling. Biochim. Biophys. Acta 1699:145-154. [DOI] [PubMed] [Google Scholar]
- 54.Smith, C. S., A. M. Weljie, and G. B. Moorhead. 2003. Molecular properties of the putative nitrogen sensor PII from Arabidopsis thaliana. Plant J. 33:353-360. [DOI] [PubMed] [Google Scholar]
- 55.Stadtman, E. R. 2001. The story of glutamine synthetase regulation. J. Biol. Chem. 276:44357-44364. [DOI] [PubMed] [Google Scholar]
- 56.Tanigawa, R., M. Shirokane, S. Maeda Si, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsinoremas, N. F., A. M. Castets, M. A. Harrison, J. F. Allen, and N. Tandeau de Marsac. 1991. Photosynthetic electron transport controls nitrogen assimilation in cyanobacteria by means of posttranslational modification of the glnB gene product. Proc. Natl. Acad. Sci. USA 88:4565-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71-74. [DOI] [PubMed] [Google Scholar]
- 59.Xu, Y., P. D. Carr, P. Clancy, M. Garcia-Dominguez, K. Forchhammer, F. Florencio, S. G. Vasudevan, N. Tandeau de Marsac, and D. L. Ollis. 2003. The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803. Acta Crystallogr. Sect. D 59:2183-2190. [DOI] [PubMed] [Google Scholar]
- 60.Xu, Y., P. D. Carr, T. Huber, S. G. Vasudevan, and D. L. Ollis. 2001. The structure of the PII-ATP complex. Eur. J. Biochem. 268:2028-2037. [DOI] [PubMed] [Google Scholar]
- 61.Xu, Y., E. Cheah, P. D. Carr, W. C. van Heeswijk, H. V. Westerhoff, S. G. Vasudevan, and D. L. Ollis. 1998. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol. 282:149-165. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1995. Comparison studies of dinitrogenase reductase ADP-ribosyl transferase/dinitrogenase reductase activating glycohydrolase regulatory systems in Rhodospirillum rubrum and Azospirillum brasilense. J. Bacteriol. 177:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1993. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J. Bacteriol. 175:6781-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., A. D. Cummings, R. H. Burris, P. W. Ludden, and G. P. Roberts. 1995. Effect of an ntrBC mutation on the posttranslational regulation of nitrogenase activity in Rhodospirillum rubrum. J. Bacteriol. 177:5322-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2000. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J. Bacteriol. 182:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2003. Regulation of nitrogen fixation by multiple PII homologs in the photosynthetic bacterium Rhodospirillum rubrum. Symbiosis 35:85-100. [Google Scholar]
- 67.Zhang, Y., E. L. Pohlmann, and G. P. Roberts. 2005. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J. Bacteriol. 187:1254-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, Y., E. L. Pohlmann, and G. P. Roberts. 2004. Identification of critical residues in GlnB for its activation of NifA activity in the photosynthetic bacterium Rhodospirillum rubrum. Proc. Natl. Acad. Sci. USA 101:2782-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]