Abstract
Across evolution, type I signal peptidases are responsible for the cleavage of secretory signal peptides from proteins following their translocation across membranes. In Archaea, type I signal peptidases combine domain-specific features with traits found in either their eukaryal or bacterial counterparts. Eukaryal and bacterial type I signal peptidases differ in terms of catalytic mechanism, pharmacological profile, and oligomeric status. In this study, genes encoding Sec11a and Sec11b, two type I signal peptidases of the halophilic archaeon Haloferax volcanii, were cloned. Although both genes are expressed in cells grown in rich medium, gene deletion approaches suggest that Sec11b, but not Sec11a, is essential. For purification purposes, tagged versions of the protein products of both genes were expressed in transformed Haloferax volcanii, with Sec11a and Sec11b being fused to a cellulose-binding domain capable of interaction with cellulose in hypersaline surroundings. By employing an in vitro signal peptidase assay designed for use with high salt concentrations such as those encountered by halophilic archaea such as Haloferax volcanii, the signal peptide-cleaving activities of both isolated membranes and purified Sec11a and Sec11b were addressed. The results show that the two enzymes differentially cleave the assay substrate, raising the possibility that the Sec11a and Sec11b serve distinct physiological functions.
In all three domains of life, i.e., Eukarya, Bacteria, and Archaea, proteins destined to reside beyond the cytoplasm are generally synthesized as preproteins, containing a cleavable N-terminal extension referred to as the signal peptide (SP) that serves to target the preprotein to the appropriate membrane-embedded protein translocation complex (3, 7, 18). Type I signal peptidases (SPases) are integral membrane proteins responsible for the removal of SPs following preprotein translocation across the eukaryal endoplasmic reticulum membrane or the prokaryal plasma membrane (11, 27, 28, 38).
While tending to share little overall resemblance, SPases in Eukarya and Bacteria include five regions of significant sequence homology, termed boxes A to E, with boxes B to E participating in the catalytic cycle of the enzyme (11, 27, 28, 38). Nonetheless, bacterial and eukaryal SPases differ enzymatically and structurally. In the bacterial enzyme, the box B region contains the conserved nucleophilic Ser-90 (Escherichia coli numbering) residue, while the proposed general base Lys-145 is found in box D (11, 27, 28). Ser-90 and Lys-145 are believed to form the catalytic dyad responsible for the proteolytic action of the enzyme (6, 29, 36, 37). By contrast, eukaryal type I SPases have replaced the essential lysine of the bacterial catalytic dyad with a histidine residue (13, 39). Thus, while their catalytic mechanism remains to be elucidated, eukaryal SPases may rely on either a Ser-His dyad or a Ser-His-Asp triad for catalytic activity rather than the Ser-Lys dyad employed by the bacterial enzyme (39). It should be noted, however, that in a limited number of gram-positive bacterial SPases, e.g., Bacillus subtilis SipW, the lysine residue of the catalytic dyad has also been replaced by a histidine, although this histidine could be exchanged for a lysine without hindering enzymatic activity (34, 38). The bacterial and eukaryal enzymes also differ in terms of their oligomeric status. Unlike the bacterial enzyme, which functions independently, i.e., as a single encoded polypeptide, eukaryal SPases function as part of a multisubunit SPase complex (16, 40).
Whereas type I SPases in Bacteria and Eukarya are relatively well described, little is known about signal peptide cleavage in Archaea. While containing the evolutionarily conserved regions of sequence homology, archaeal SPases lack the conserved lysine of the bacterial Ser-Lys catalytic dyad and, like Eukarya, contain a histidine residue at this position (15, 23, 33). Thus, archaeal SPases may rely on a catalytic mechanism similar to that used by the eukaryal enzyme. Indeed, site-directed mutagenesis studies of SPase from the methanoarchaeon Methanococcus voltae have confirmed the essential nature of the Ser-90 and His-145 equivalents (4), as is the case in the eukaryal enzyme (39). However, differences between the modes of action of the eukaryal and archaeal enzymes apparently exist. While the equivalents of the well-conserved Asp-273 and Asp-280 residues (E. coli numbering) are essential for the activity of the yeast enzyme (39), only the latter is essential for M. voltae SPase activity (4). The role assumed by the Asp-280 equivalent in the catalytic mechanism of the archaeal enzyme is unclear, since some Archaea do not contain this residue (4).
Evidence pointing at similarities between archaeal and bacterial SPases also exists. The inability of genomic searches thus far to detect Eukarya-like SPase complex subunits in Archaea suggests that the archaeal enzyme operates independently, as in Bacteria (15). Furthermore, like bacterial SPases, certain archaeal enzymes include a stretch of residues not found in eukaryal SPases that comprises domain II, a structural motif of unknown function (15, 26). Thus, current understanding suggests that archaeal SPase may represent an evolutionary intermediate between present-day eukaryal and bacterial enzymes.
Examination of archaeal SPases could provide insight into signal peptide processing not only in the face of the extreme environments in which Archaea exist but also across evolution, given the apparent hybrid-like nature of the archaeal enzyme. Towards these ends, we have addressed SPase activity in the halophilic archaeon Haloferax volcanii. In the following, we report the cloning of two H. volcanii SPase-encoding genes and consider the expression and essential nature of each. Furthermore, signal peptide removal by isolated membranes and tagged versions of each enzyme purified from transformed H. volcanii cells was characterized by employing an in vitro SPase assay specifically developed for hypersaline conditions.
MATERIALS AND METHODS
Materials.
Cellulose, isopropyl-β-d-thiogalactopyranoside (IPTG), novobiocin, and Triton X-100 were obtained from Sigma (St. Louis, MO). Proteinase K came from Boehringer (Mannheim, Germany). Yeast extract came from Pronadisa (Madrid, Spain), while tryptone came from USB (Cleveland, OH). Molecular weight markers and goat anti-rabbit horseradish peroxidase-conjugated antibodies were from Bio-Rad (Hercules, CA). An ECL enhanced chemiluminescence kit came from Amersham (Buckingham, United Kingdom).
Growth conditions.
H. volcanii was grown in rich medium containing 3.4 M NaCl, 0.15 M MgSO4 · 7H2O, 1 mM MnCl2, 4 mM KCl, 3 mM CaCl2, 0.3% (wt/vol) yeast extract, 0.5% (wt/vol) tryptone, and 50 mM Tris-HCl, pH 7.2, at 40°C (22). In Casamino Acids medium, yeast extract and tryptone were replaced by Casamino Acids (Difco, Detroit, MI) at a final concentration of 0.5% (wt/vol). Escherichia coli was grown in Luria-Bertani medium.
Cloning of the H. volcanii SPase-encoding genes.
Using a partially completed H. volcanii genome sequence (http://zdna2.umbi.umd.edu) as a guide, oligonucleotide primers were designed against regions within contig 2978 (8,221 bp), containing a DNA sequence annotated as SPase (lying between positions 2037 and 2510) as well as upstream and downstream to the proposed open reading frame (ORF). The forward primer Sa1was synthesized to bind to H. volcanii genomic DNA from position 30 of contig 2978, while the reverse primer ASa1 was synthesized to bind to H. volcanii genomic DNA from position 2515 of the contig. Additional forward primers Sa2, synthesized to bind from position 721, and Sa3, synthesized to bind from position 1640, and the reverse primer ASa2, synthesized to bind from position 2086, were also prepared. For cloning of the second H. volcanii SPase gene, oligonucleotide primers were designed against regions within contig 270 of the unfinished version of the H. volcanii genome found at The Institute for Genomic Research (TIGR) website (http://www.tigr.org). The forward primer Sb1 was synthesized to bind to H. volcanii genomic DNA from position 757 of contig 270 (2,912 bp), while the reverse primer ASb1 was synthesized to bind from position 2106 of the contig. Additional forward primers Sb3, synthesized to bind from position 1265, and Sb4, synthesized to bind from position 480, as well as the reverse primers ASb2, synthesized to bind from position 1539, and ASb3, synthesized to bind from position 1385, were also prepared. The various primers were used in a series of PCR amplifications using H. volcanii genomic DNA as the template, prepared as previously described (31). The sequences of the various primers employed are listed in Table 1.
TABLE 1.
Primers used in this study
| Purpose and primer name | Direction | Sequencea |
|---|---|---|
| Sec11a | ||
| Sa1 | Forward | ATGGCCCGATACCACATCGAGACA |
| ASa1 | Reverse | GGGGTTCAAAGAAGCTTCTGG |
| Sa2 | Forward | CGACACGACAAGCTCTACAACTTC |
| ASa2 | Reverse | TCGCCGCGTTCGAGAGTCGGTTG |
| Sa3 | Forward | AGCCGACCCATACCGACTATC |
| Sec11b | ||
| Sb1 | Forward | TTCCCGACGGGTCCGCGGC |
| Sb2 | Forward | GGTCGACGACGGAGAGAACT |
| Sb3 | Forward | ATGGAACCGCACATGCACAAA |
| Sb4 | Forward | TCGTAGCGGTCGCGGAACACCTT |
| ASb1 | Reverse | GTCGTCTCGAAGGGTCGCTA |
| ASb2 | Reverse | GGGCAGTTGGCGAGTTCTC |
| ASb3 | Reverse | CCGGGGCCGCCGAACTTCCGGTA |
| RT-PCR | ||
| Sec11afor | Forward | CGTCGTCGTGACCGCGGAGGTT |
| Sec11arev | Reverse | TCAAAGAAGCTTCTGGACTT |
| Sec11bfor | Forward | ATGGAACCGCACATGCACAAA |
| Sec11brev | Reverse | TCACGCCGCGAGCGCGGCGGTCCGCAT |
| TrpA | ||
| trpAfor | Forward | gggaagcttCGTGGATAAAACCCCTCGTTG |
| trpArev | Reverse | cccgaattcTTATGTGCGTTCCGGATGCG |
| pIDT-Sec11a | ||
| UsSec11a | Forward | gggatcgatCACTCGGAGAAACATCCCTTTCG |
| UsSec11ar | Reverse | cccaagcttCTACCACCTCGTCCCCGAGTTTC |
| DsSec11a | Forward | gggggatccACCCCCAGCGGTCGTCAGCGGT |
| DsSec11ar | Reverse | ccctctagaCGACAACGTGAACTACCTCGTC |
| pIDT-Sec11b | ||
| UsSec11b | Forward | gggatcgatGTGTCCCCTGCGCTTCGGCTG |
| UsSec11br | Reverse | cccaagcttCCTTTTTGCCGCGTGTCGC |
| DsSec11b | Forward | gggggatccCGCGAGTGCACTAAAAGCGTC |
| DsSec11br | Reverse | ccctctagaCTACAAGCGCCTCTGCGAGGAG |
| His6-SP-CBD | ||
| SPCBDf | Forward | cttgtcatgaCAAAGCTCAAA |
| SPCBDr | Reverse | tgatctcgagTACTACACTGCCACCGG |
| CBD-Sec11a | ||
| CBDSec11af | Forward | gggcatATGCCGACGCCGGCAACAAGATTG |
| CBDSec11ar | Reverse | cccggtaccTCAAAGAAGCTTCTGGACTTC CAGC |
| CBD-Sec11b | ||
| CBDSec11bf | Forward | gggcatATGAGTGACGACGATGGCCGC CCGCCCT |
| CBDSec11br | Reverse | cccggtaccTCACGCCGCGAGCGCGGCGGT CCGCAT |
Genomic DNA sequences are shown in uppercase letters.
RT-PCR.
To perform reverse transcriptase PCR (RT-PCR), specific forward and reverse oligonucleotide primers were designed for each version of H. volcanii SPase (Table 1). RNA isolation was carried out according to protocols described in The Halohandbook: Protocols for Halobacterial Genetics, Version 4.9 (http://www.microbiol.unimelb.edu.au/micro/staff/mds/HaloHandbook/). RNA concentrations were determined spectrophotometrically after contaminating DNA had been eliminated with a DNAFree kit (Ambion, Austin, TX). Single-stranded cDNA was prepared for each SPase sequence from the corresponding RNA (1 μg/ml) by using the appropriate reverse primer (2 pmol) in a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). The single-stranded cDNA was then used as a template in a PCR containing the appropriate forward and reverse primer pairs. cDNA amplification was monitored by electrophoresis in 1% agarose gels. The sequences of the PCR products were determined to confirm their identity. In control experiments designed to exclude any contribution from contaminating DNA, PCR amplification was performed on total RNA prior to cDNA preparation.
Deletion of H. volcanii Sec11a- and Sec11b-encoding genes.
To test the essential natures of sec11a and sec11b, each gene was deleted as previously described (1). Briefly, the SPase-encoding genes were replaced by the H. volcanii trpA gene by using plasmid pIDT-Sec11a or pIDT-Sec11b. To construct pIDT-Sec11a and pIDT-Sec11b, 400-bp fragments lying upstream from the first codons of ORFs containing the H. volcanii SPase genes were PCR amplified using forward primers UsSec11a and UsSec11b and reverse primers UsSec11ar and UsSec11br, respectively, which were designed to introduce ClaI and HindIII restriction sites at the 5′ and 3′ ends of the fragments, respectively. Four-hundred-base-pair fragments encoding the downstream flanking regions of the ORFs containing each SPase-encoding gene were PCR amplified using forward primers DsSec11a and DsSec11b and reverse primers DsSec11ar and DsSec11br, respectively, which were designed to introduce XbaI and BamHI restriction sites at the 5′ and 3′ ends of the fragments, respectively. The amplified fragments were digested with ClaI and HindIII (upstream fragments) or XbaI and BamHI (downstream fragments) and purified by electrophoresis in 1% agarose gels with a Nucleospin Extract kit (Macherey-Nagel, Duren, Germany). The appropriate fragments were then ligated into plasmid pTA131 (1), which was predigested with the appropriate restriction enzymes. To generate plasmids pIDT-Sec11a and pIDT-Sec11b, the H. volcanii trpA gene was cloned by PCR from plasmid pTA132 (1) using forward and reverse plasmids trpAfor and trpArev, respectively, and inserted between the EcoRI and HindIII sites of plasmid pTA131 containing the Sec11a or Sec11b flanking regions. H. volcanii WR536 (also referred to as strain H53 [1]) was subsequently transformed with plasmid pIDT-Sec11a or pIDT-Sec11b and plated onto Casamino Acids medium. Transformants were screened for integration of the plasmid-derived genes at the corresponding SPase locus by PCR analysis. Excision of plasmids pIDT-Sec11a and pIDT-Sec11b was then performed by propagating the selected colonies in rich medium and plating on agar in Casamino Acids medium supplemented with 10 μg/ml uracil and 50 μg/ml 5-fluoroorotic acid (5-FOA).
Assay of H. volcanii SPase activity.
H. volcanii membranes were prepared as follows. Subcellular fractionation was achieved by sonication (2 s on and 1 s off for 30 s, 35% output, Misonix XL2020 ultrasonicator) followed by centrifugation (8,000 × g, 20 min) to clear unbroken cells and ultracentrifugation (Sorvall Discovery M120 ultracentrifuge, S120AT2 rotor, 190,000 × g, 10 min, 4°C) to pellet the membrane fraction. To confirm the efficiency of the fractionation, immunoblotting was performed using antibodies raised against H. volcanii dihydrofolate reductase-1 (a gift from Moshe Mevarech, Tel Aviv University) or against the H. volcanii S-layer glycoprotein (14), markers of the soluble and membrane fractions of the cell, respectively. Cleavage by H. volcanii SPase was assayed by incubating aliquots of the membrane pellet (30 μg) resuspended 3 M NaCl, 50 mM Tris-HCl, pH 7.2, with substrate (15 μg) in the presence of 0.7% Triton X-100 at 40°C. The substrate employed was a polyhistidine-tagged version of the previously described chimera formed between the 34-amino-acid-residue SP of the H. volcanii S-layer glycoprotein (32) and a Clostridium thermocellum cellulose-binding domain (CBD) (19). To assess SPase-catalyzed cleavage of the SP-CBD substrate, aliquots from reaction mixtures containing H. volcanii membranes and SP-CBD were removed at increasing intervals, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotted using anti-CBD antibodies (a gift form Yuval Shoham, Technion Israel Institute of Technology). When the assay was performed using purified CBD-Sec11a or CBD-Sec11b (see below) instead of membrane fragments, detection was via Coomassie blue staining.
A C-terminally polyhistidine-tagged version of SP-CBD was prepared by PCR amplifying the SP-CBD fragment from plasmid pWL-SP-CBD, which was previously used to introduce SP-CBD into H. volcanii cells (19), and cloning theamplified fragment into plasmid pET-24d (Novogen, Nottingham, United Kingdom) to generate plasmid pET-24d-SP-CBD. In preparing plasmid pET-24d-SP-CBD, forward (SPCBDf) and reverse (SPCBDr) oligonucleotide primers designed to introduce BspHI and XhoI restriction sites at the 5′ and 3′ ends of the SP-CBD-encoding fragment, respectively, as well as to delete the native stop codon of the CBD-encoding sequence were employed. Insertion into the pET-24d vector at the NcoI and XhoI restriction sites introduced a polyhistidine tag at the C terminus of the chimera. Plasmid pET-24d-SP-CBD was then used to transform Escherichia coli BL21(DE3) cells. The transformed cells were grown to an optical density at 600 nm of 0.4 and induced with 0.5 mM IPTG for 3 h. Solubilization of isolated inclusion bodies in a series of urea solubilization steps (incubation in 0.5 M urea, 0.1 M Tris-HCl, pH 8.5, followed by centrifugation) followed by Ni-nitrilotriacetic acid chromatography and elution with 0.5 M imidazole was then performed to isolate the SP-CBD chimera. N-terminal amino acid sequencing (Biological Services, Weizmann Institute of Science, Rehovot, Israel) confirmed the purified protein as SP-CBD.
Purification of CBD-Sec11a and CBD-Sec11b from transformed H. volcanii cells.
To generate a plasmid encoding CBD-Sec11a, forward (CBDSec11af) and reverse (CBDSec11ar) oligonucleotide primers were employed. Since the N-terminal amino acid residue of Sec11a has yet to be experimentally determined, forward primer CBDSec11af was designed to introduce an NdeI restriction site followed by a methionine-encoding codon 11 residues (35 bp) upstream of the transmembrane domain of the protein. The reverse primer CBD-Sec11ar was designed to introduce a KpnI restriction site at the 3′ end of the Sec11a-encoding gene. The PCR-amplified fragment was digested with NdeI and KpnI, purified by electrophoresis in 1% agarose gels with a Nucleospin Extract kit (Macherey-Nagel), and then ligated to the NdeI and KpnI sites of plasmid pWL-CBD (18) to yield plasmid pCBD-Sec11a.
To generate a plasmid encoding CBD-Sec11b, forward (CBDSec11bf) and reverse (CBDSec11br) oligonucleotide primers designed to introduce NdeI and KpnI restriction sites at the 5′ and 3′ ends of the Sec11b-encoding gene, respectively, were generated. Since the N-terminal residue of Sec11b has yet to be experimentally determined, forward primer CBDSec11bf was designed to introduce an NdeI restriction site immediately upstream from the methionine-encoding codon found 40 residues (119 bp) upstream of the transmembrane domain of the protein, arbitrarily chosen as the N terminus of the protein. The PCR-amplified fragment was digested with NdeI and KpnI, purified by electrophoresis in 1% agarose gels with a Nucleospin Extract kit (Macherey-Nagel), and then ligated to the NdeI and KpnI sites of plasmid pWL-CBD (19) to yield plasmid pCBD-Sec11b.
H. volcanii WR341 cells were transformed with these plasmids as described previously (10). Following isolation and solubilization of the membrane fraction of the transformed cells, the CBD-containing chimeras were purified on cellulose beads as described previously (20).
Other methods.
Protein concentration was determined using Bradford reagent (Bio-Rad), with bovine serum albumin as a standard. Antibody binding was detected using goat anti-rabbit horseradish peroxidase-conjugated antibodies and enhanced chemiluminescence.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in GenBank and assigned accession numbers AY940668 (sec11a) and AY940669 (sec11b).
RESULTS
Cleavage of a signal peptide by membrane-associated H. volcanii SPase activity.
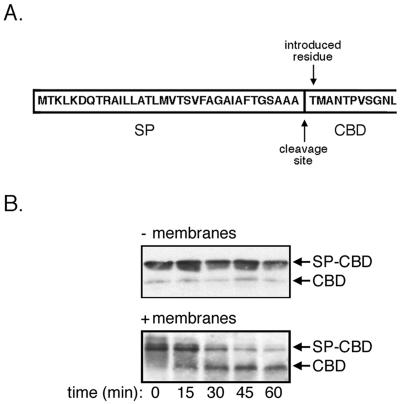
As a first step in characterizing H. volcanii SPase activity, the in vitro cleavage of the signal peptide from a reporter substrate by isolated membranes was assessed. The choice of SP-CBD as a reporter was based on earlier in vivo studies showing that the same chimera could be secreted by transformed H. volcanii cells and that the secreted protein underwent cleavage at the junction between the SP and CBD moieties (Fig. 1A) (19), consistent with the actions of type I SPase. In the present study, H. volcanii membranes were incubated with the chimera for increasing intervals, after which time the samples were separated by SDS-PAGE and immunoblotted using anti-CBD antibodies (Fig. 1B). At time zero, only a single protein band, corresponding to SP-CBD, was detected. With time, a faster-migrating antibody-stained band, corresponding in size to CBD, began to accumulate as the level of SP-CBD decreased (Fig. 1B, lower panel). When the reaction was repeated in the absence of membranes, no such faster-migrating antibody-stained band accumulated above the background level of the CBD-sized band that copurified with SP-CBD, visible from time zero (Fig. 1B, upper panel).
FIG. 1.
In vitro assay of H. volcanii SPase activity. A. Amino acid sequence of SP-CBD up to the 11th amino acid residue of the CBD moiety. B. Assay of H. volcanii SPase activity. Purified SP-CBD (15 μg) was incubated in the presence or absence of H. volcanii membranes (30 μg) for 0 to 60 min in 3 M NaCl, 0.7% Triton X-100, 50 mM Tris-HCl, pH 7.2, at 40°C. Following separation by SDS-PAGE, the reaction products were probed by immunoblotting using anti-CBD antibodies. The positions of SP-CBD and CBD are indicated.
To confirm that the faster-migrating anti-CBD antibody-stained species generated in the presence of H. volcanii membranes indeed corresponded to CBD lacking its engineered signal peptide, the antibody-labeled protein was purified by exploiting the salt-insensitive interaction between C. thermocellum CBD and cellulose (19, 25). N-terminal amino acid sequencing of the purified faster-migrating band yielded the sequence TMANT, corresponding to the first four residues of mature CBD preceded by the threonine residue introduced during cloning (Fig. 1A) (19). Thus, the H. volcanii membrane preparation contains SPase activity that is able to effectively remove the signal peptide from a preprotein substrate.
The salt dependence of the cleavage reaction was next considered by combining H. volcanii membranes with SP-CBD in buffer containing from 0.5 to 3 M NaCl. Optimal cleavage was achieved in the presence of 3 M NaCl (not shown). Peptidase activity could not be tested at higher salt levels, since in the presence of over 3 M NaCl a precipitate began to appear in the reaction mixtures, likely due to the presence of detergent in the assay mixture. Similar results were obtained when KCl was used in place of NaCl (not shown). The salt preference of the haloarchaeal reaction is in striking contrast to E. coli SPase activity, which is inhibited by salt concentrations of above 160 mM (41), yet is fitting with the behavior of haloarchaeal enzymes, which are designed to fold and function optimally in the molar salt concentrations encountered in the halophilic archaeal cytoplasm (21).
H. volcanii contains two type I SPase-encoding genes.
Having shown that H. volcanii membranes possess SPase activity, attention was next focused on identifying the responsible protein(s). Accordingly, the original version of the partially completed H. volcanii genome sequence (http://zdna2.umbi.umd.edu) was consulted (October 2004) to determine whether an SPase-encoding gene had been identified in that effort. Such analysis revealed a predicted ORF in contig 2978 containing four of the five sequence motifs present in SPases across evolution and annotated as encoding a type I signal peptidase (11, 27, 28, 38). Surprisingly, this H. volcanii ORF failed to predict the existence of a membrane-spanning domain, in contrast to the case for other type I signal peptidases (11, 27, 28, 38). PCR amplification of H. volcanii genomic DNA using oligonucleotide forward primers directed at various positions upstream of the predicted start site of the putative H. volcanii SPase gene together with reverse primers directed at various positions within the SPase-coding region revealed that an extra cytosine at position 2036 of the contig had been erroneously included in the partially completed genome sequence. The corrected gene sequence, annotated as sec11a (GenBank accession number AY940668), now predicts the existence of a single membrane-spanning domain located near the N terminus of the protein.
Given the existence of two putative SPases in the Haloarcula marismortui genome sequence (Sec11a [GenBank entry AAV47480] and Sec11b [GenBank entry AAV47481]) (2), the partially completed TIGR version of the H. volcanii genome was probed for the presence of a second SPase gene. By employing the Haloarcula marismortui Sec11b sequence as a probe in a BLAST scan of this partially completed H. volcanii genome, an additional SPase-encoding gene was identified (contig 270, nucleotides 1786531 to 1787442). The computational prediction was confirmed when PCR amplification of H. volcanii genomic DNA, using the appropriate oligonucleotide primers, yielded a DNA fragment of the predicted length and sequence, which was subsequently annotated as encoding Sec11b (GenBank accession number AY940669).
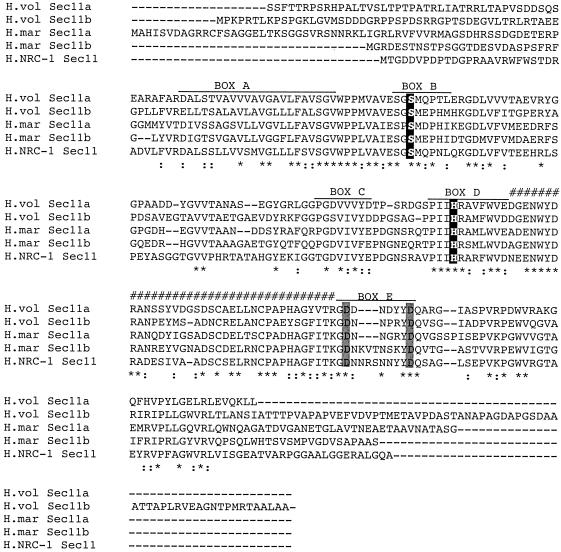
Alignment of five predicted haloarchaeal SPases amino acid sequences currently available (i.e., H. volcanii and Haloarcula marismortui Sec11a and Sec11b and Halobacterium sp. strain NRC-1 SPase) revealed that the haloarchaeal enzymes all include those regions of sequence homology termed boxes B to E as well as the membrane-spanning box A domain (Fig. 2) (11, 27, 28, 38). Like other archaeal and eukaryal SPases (12, 15, 23, 33, 39), the haloarchaeal enzymes have replaced the lysine residue of the serine-lysine pair responsible for catalytic activity in bacterial SPases with a histidine residue in box D. Box E of the different haloarchaeal sequences also includes the pair of conserved aspartic acid residues recently addressed in Methanococcus voltae SPase (4). The essential requirement of only one of those residues for signal peptide cleavage by the methanoarchaeal SPase remains, however, to be confirmed for the haloarchaeal enzymes. Finally, analysis of the haloarchaeal SPase sequences, including those from H. volcanii, revealed the presence of the domain II sequence region (22). Domain II, found in bacterial but not eukaryal SPase sequences, was previously observed in other selected archaeal SPase sequences (15).
FIG. 2.
Alignment of haloarchaeal SPases. Haloarchaeal SPase amino acid sequences were aligned using ClustalW (http://clustalw.genome.ad.jp). In each sequence, the positions of boxes B to E are indicated, as are identical residues (asterisks) and conserved residues (colons). The SPase sequences shown are from Haloferax volcanii (H.vol Sec11a and Sec11b; GenBank accession numbers AY940668 and AY940669, respectively), Halobacterium sp. strain NRC-1 (H.NRC Sec11; GenBank accession number NP_281023), and Haloarcula marismortui (H.mar Sec11a and Sec11b; GenBank accession numbers AAV47480 and AAV47481, respectively). The conserved box B lysine and box D histidine residues are shown against a black background, the equivalents of Asp-273 and Asp-280 are shown against a gray background, and domain II is indicated by hatch marks.
Both H. volcanii Sec11a and Sec11b are expressed, but only Sec11b is essential.
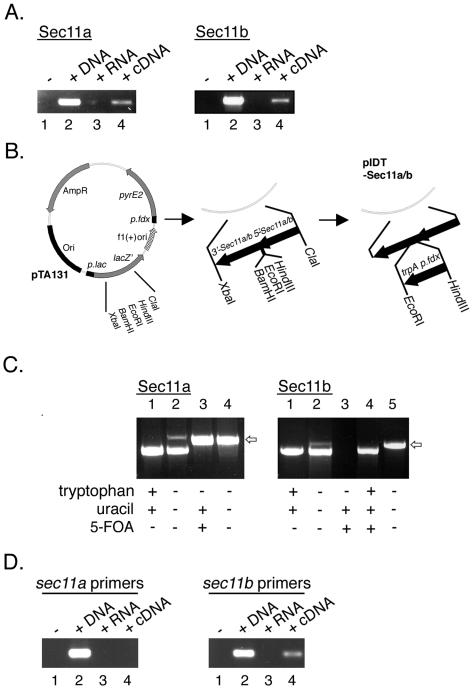
To determine whether both H. volcanii SPase genes are expressed, PCR was performed using cDNA prepared from total RNA isolated from cells grown to mid-exponential phase and primers specific for each SPase gene sequence. In each case, as shown in Fig. 3A, PCR amplification led to the appearance of the same single band when either genomic DNA (lanes 2) or cDNA (lanes 4) served as the template. Sequencing confirmed the bands as encoding either Sec11a (left panel) or Sec11b (right panel). No such bands appeared in control experiments when RNA served as the template (lanes 3) or when no template was included (lanes 1).
FIG. 3.
Both Sec11a and Sec11b are expressed in H. volcanii, but only Sec11b is essential. A. RNA was extracted from H. volcanii cells grown to exponential phase and used to direct the synthesis of cDNA. PCR was then performed in the absence of nucleic acids (−) (lanes 1) or in the presence of genomic DNA (+DNA) (lanes 2), total RNA (+RNA) (lanes 3), or cDNA (+cDNA) (lanes 4), using primers against either sec11a (left panel) or sec11b (right panel). B. Schematic diagram of the plasmids used to delete the Sec11a- and Sec11b-encoding genes. C. PCR amplification of H. volcanii cells transformed with plasmid pIDT-Sec11a or pIDT-Sec11b, designed to delete either sec11a or sec11b, respectively. The arrows show the positions of the PCR amplification product encoding the Sec11a or Sec11b flanking regions separated by the H. volcanii trpA gene, presented in the last lane of each panel. Also shown in each panel are PCR products obtained using genomic DNA (lanes 1), genomic DNA including the integrated pIDT-Sec11a or pIDT-Sec11b plasmids (lanes 2), and the same DNA taken from cells grown in tryptophan-free, 5-FOA- and uracil-containing medium, i.e., following expulsion of the plasmid and native Sec11a- or Sec11b-encoding gene (lanes 3) as template. In the right panel, lane 4 shows the PCR product of the cells considered in lane 3 but returned to tryptophan-containing medium such that DNA encoding the sec11a or sec11b flanking regions separated by the H. volcanii trpA gene is expelled from the genome with the plasmid. D. RNA was extracted from H. volcanii cells from which the Sec11a-encoding gene had been deleted and used to direct the synthesis of cDNA. PCR was then performed in the absence of nucleic acids (−) (lane 1) or in the presence of genomic DNA taken from untreated cells (+DNA) (lane 2) or total RNA (+RNA) (lane 3) or cDNA (+cDNA) (lane 4) from the deletion strain, using primers against either sec11a (left panel) or sec11b (right panel).
The essential natures of Sec11a and Sec11b were next considered by deleting either gene by using a “pop-in/pop-out” protocol developed for use with H. volcanii (1, 5). Briefly, sec11a or sec11b was replaced with DNA containing the upstream and downstream flanking 400 nucleotides of either gene separated by the H. volcanii tryptophan synthase-encoding trpA gene. The replacement sequences were inserted into the pyrE-containing plasmid pTA131 (1) (Fig. 3B), which was then integrated into the genome of H. volcanii WR536 (auxotrophic for uracil and tryptophan, also referred to as strain H53) (1) by plating onto Casamino Acids medium lacking uracil and tryptophan. To replace the native Sec11a- or Sec11b-encoding genes, the transformed cells are grown in the absence of tryptophan but in the presence of uracil and 5-FOA, a toxic uracil analogue. Cells become resistant to 5-FOA in the absence of pyrE. In this manner, the integrated pyrE-containing plasmid and the native gene are expelled, while the sequences encoding the sec11a or sec11b flanking regions, separated by the trpA gene, remain integrated in the genome at the sec11a or sec11b locus (i.e., pop-out).
A series of PCR amplifications were next performed with genomic DNA taken from cells transformed with plasmid pIDT-Sec11a or pIDT-Sec11b as the template. As shown in Fig. 3C, lanes 2, the transformed cells were able to integrate both plasmid pIDT-Sec11a and plasmid pIDT-Sec11b into the genome. When the plasmid pID19T-Sec11a-incorporating cells were grown in medium to which uracil and 5-FOA were added, the native Sec11a-encoding gene was exchanged for plasmid-derived genetic material in which the SPase-encoding sequence had been replaced by the H. volcanii trpA gene (Fig. 3C, left panel, lane 3). By contrast, no growth of plasmid pIDT-Sec11b-incorporating cells was observed upon transfer to tryptophan-free, uracil- and 5-FOA-containing growth medium, conditions under which the plasmid and native sec11b gene are expelled (right panel, lane 3). If, however, the plasmid pIDT-Sec11b-integrating cells were returned to medium containing tryptophan, the integrated plasmid was expelled from the genome, leaving only the unperturbed, native sec11b locus (right panel, lane 4).
To confirm that the native Sec11a-encoding gene was indeed deleted in the plasmid pIDT-Sec11a-challenged cells, RT-PCR was performed (Fig. 3D). In control experiments where PCR amplification was performed using genomic DNA taken from untreated cells and oligonucleotide primers directed against the sec11a gene (left panel), a single band, shown by sequencing to correspond to sec11a, was detected (lane 2). No such bands appeared when the same reaction was performed using cDNA prepared from RNA obtained from the sec11a deletion strain as the template (Fig. 3D, left panel, lane 4). Similarly, no such bands appeared in control experiments when RNA from the deletion strain served as the template (lane 3) or when no template was included in the reaction (lane 1). When the experiment was repeated, however, using primers directed against sec11b (Fig. 3D, right panel), a single band of the expected size and sequence was detected in reactions containing either genomic DNA from untreated cells (lane 2) or cDNA prepared from RNA obtained from the sec11a deletion strain (lane 4) as the template.
Thus, studies performed at both the DNA and RNA levels suggest that Sec11b, but not Sec11a, is an essential protein in H. volcanii under the growth conditions tested.
Signal peptide cleavage by purified H. volcanii Sec11a and Sec11b.
To confirm that H. volcanii Sec11a and Sec11b indeed correspond to type I SPases, H. volcanii cells were transformed to express chimeras comprising the C. thermocellum CBD fused to either Sec11a or Sec11b (Fig. 4). In each case, the CBD moiety was attached to the SPase upstream of the predicted transmembrane domain lying close to the N-terminal region, so as to avoid any potential problems associated with translocating the CBD entity across the plasma membrane, as would be required were the CBD moiety positioned at the C terminus of Sec11a or Sec11b. In constructing CBD-Sec11a, the CBD moiety was attached to an engineered N-terminal methionine residue arbitrarily introduced at 11 amino acid positions from the start of the transmembrane domain, beginning at nucleotide 127 of the Sec11a-encoding gene, since no methionine residues are predicted to lie upstream of the transmembrane domain of the enzyme. Similarly, since the N-terminal residue of H. volcanii Sec11b has yet to be experimentally determined, the position for attachment of the CBD moiety was arbitrarily selected as the first predicted methionine upstream from the start of the membrane-spanning domain that begins at nucleotide 364 of the ORF containing the Sec11b-encoding gene.
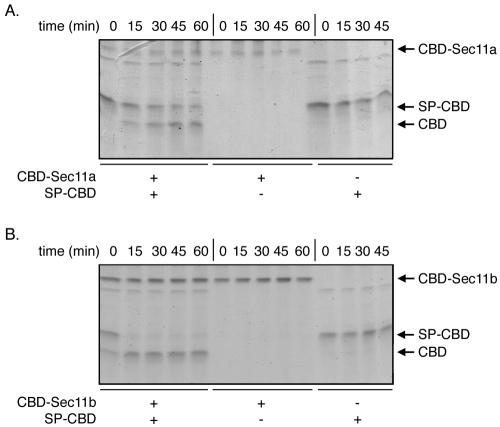
FIG. 4.
Purified CBD-Sec11a and CBD-Sec11b are active. CBD-Sec11a (A) and CBD-Sec11b (B) were purified as described in Materials and Methods and tested for their ability to release the SP domain from the reporter substrate SP-CBD. Incubations were conducted in the presence of purified enzyme and substrate, enzyme alone, or substrate alone, from 0 to 60 min. The collected aliquots were analyzed by SDS-PAGE and Coomassie blue stained.
The chimeras were expressed in the high-salt environment of H. volcanii cells to ensure proper folding of the haloarchaeal SPases, purified on cellulose beads, and tested for signal peptidase activity. As shown in Fig. 4, incubation of the cellulose-bound purified enzymes with SP-CBD resulted in the cleavage of the substrate, although Sec11a and Sec11b displayed strikingly distinct cleaving efficiencies. Whereas Sec11b (Fig. 4B) readily removed the SP from the SP-CBD substrate, Sec11a (Fig. 4A) was only able to partially cleave the reporter. In each case, incubation of SP-CBD with cellulose alone under the same assay conditions did not result in SP cleavage. Finally, to confirm that the CBD that appeared upon incubation of the substrate with the cellulose-bound enzyme did not originate from breakdown of CBD-Sec11a or CBD-Sec11b, the stabilities of the cellulose-bound fusion proteins containing the enzyme were considered. Over the course of the signal peptidase assay, both CBD-Sec11a and CBD-Sec11b remained intact.
Given earlier results showing that soluble truncated versions of bacterial SPases, i.e., versions lacking their membrane-spanning sections, were functional (9, 35), H. volcanii cells were transformed to express either CBD-ΔSec11a or CBD-ΔSec11b. In the former construct, the CBD moiety was linked to the first methionine residue (nucleotide 220 of the Sec11a-encoding ORF) downstream of the transmembrane domain. Similarly, the CBD entity was fused to the first methionine residue downstream of the transmembrane domain, corresponding to nucleotide 556 of the Sec11b-encoding ORF, in the latter. Although CBD-ΔSec11a could be expressed by the transformed cells, the purified protein was inactive (not shown). By contrast, CBD-ΔSec11b could not be expressed (not shown).
DISCUSSION
Type I signal peptidases act to release the signal peptide, i.e., the N-terminal extension that serves to target a protein to the translocation machinery, from proteins translocated across the eukaryal endoplasmic reticulum membrane or the prokaryotic plasma membrane. In the present study, two genes encoding type I signal peptidases from the halophilic archaeon H. volcanii, identified by the presence of five regions of sequence homology found in SPases across evolution (11, 27, 28, 38), were cloned, expressed, and purified. Despite substantial sequence homology (55% identity over the region spanning immediately upstream of box B and downstream of box E), differences between H. volcanii Sec11a and Sec11b exist. For instance, Sec11b is predicted to include a C-terminal extension not found in Sec11a or, indeed, in other haloarchaeal SPases. The functional significance of this extension is not known. The N-terminal regions upstream from the putative membrane-spanning domains of the H. volcanii enzymes are also specific to each protein, although the actual N-terminal residues have not been identified in either case. Nonetheless, both H. volcanii SPases contain the equivalents of Ser-90, His-145, Asp-273, and Asp-280 (E. coli numbering), residues implicated in SPase action in other organisms (6, 11, 12, 27-29, 34, 36-39). While the roles played by these residues in the H. volcanii enzymes' activity have yet to be addressed, earlier site-directed mutagenesis studies have shown that in the M. voltae enzyme, which is the only other archaeal SPase studied at the molecular level, the Ser-90, His-145, and Asp-280 equivalents are critical for activity (4).
Given their overall similarity, one can ask why H. volcanii encodes two type I signal peptidases. Whereas the presence of several signal peptidase-encoding genes in a single genome is not unusual (11, 27, 28, 38), the justification for expressing multiple versions of the same enzyme in the same organism is not evident in most instances. In the case of H. volcanii, one possibility is that Sec11a and Sec11b are differentially expressed as a function of growth stage or in response to environmental conditions, with each enzyme assuming a predominate role at different times or in different situations. Alternatively, H. volcanii Sec11a and Sec11b may differ in their substrate preferences. In B. subtilis, where five different signal peptidases are expressed, evidence for the substrate preference of at least one enzyme isoform has been presented (33, 34, 38). The differences in the degree to which SP-CBD was processed by purified CBD-Sec11a and CBD-Sec11b could reflect such substrate preferences. While it could be imagined that the presence of two SPases in H. volcanii reflects the reported prevalent use of the Tat export pathway in halophilic archaea relative to other microorganisms (8, 30), whereby one SPase preferentially cleaves Tat pathway substrates while the second preferentially removes signal peptides recognized by the Sec pathway, no experimental evidence for this proposal has been provided. Indeed, examination of those halophilic archaea for which complete genome sequences are available, i.e., Haloarcula marismortui (2), Halobacterium sp. strain NRC-1 (24), and Natronomonas pharaonis (17), reveals that while Haloarcula marismortui encodes two SPases (like H. volcanii), the latter two organisms seemingly contain only a single SPase-encoding gene. Furthermore, in other genomes, such as those of Pyrococcus strains, the presence of more than one SPase is predicted despite that fact that components of the Tat pathway are apparently absent (13). Yet another explanation for the presence of two SPases in H. volcanii is that one protein (i.e., Sec11b) serves as the main processing enzyme, while the second enzyme (i.e., Sec11a) fulfils a backup role or comes into play in those instances when the former is incapable of coping with an increased amount of signal peptide-bearing substrate.
As noted with other archaeal SPases (4, 15, 23, 33), the H. volcanii enzymes present a mosaic of eukaryal, bacterial, and archaeal properties. Like other archaeal and eukaryal SPases (12, 15, 23, 33, 39), the H. volcanii enzymes have replaced the conserved lysine of the serine-lysine catalytic dyad found in bacterial SPases with a histidine. Thus, at a first approximation, the H. volcanii enzymes appear to rely on a catalytic mechanism similar to that employed by their eukaryal counterparts. On the other hand, CBD-Sec11a and CBD-Sec11b were purified as single polypeptides able to cleave the signal peptide from a reporter protein, suggesting that the H. volcanii enzymes function independently, as do their bacterial counterparts. Eukaryal SPases exist as part of a multisubunit complex, in which polypeptides other than Sec11 are essential for activity (16, 40). The absence of SPase complex components apart from Sec11a and Sec11b in H. volcanii would thus further argue against a shared mode of action between the archaeal and eukaryal enzymes. Still, the possibility remains that H. volcanii SPases operate optimally only when in complex with additional subunits not captured under the conditions considered in the present study or identified in previous genome searches. Indeed, one can speculate that the inactivity of purified CBD-ΔSec11a and the inability of transformed H. volcanii cells to express CBD-ΔSec11b, both corresponding to truncated versions of the H. volcanii enzymes lacking their transmembrane domains, reflects the need for stabilizing partner proteins that interact through the absent membrane-spanning regions. By contrast, bacterial SPases lacking their transmembrane domains are active (9, 35). Of course, the inactivity of purified CBD-ΔSec11a and the inability of transformed H. volcanii cells to express CBD-ΔSec11b may simply be due to the fact that both chimeras are generated by recombinant DNA technology, with the point of connection between the CBD and Sec11 moieties chosen arbitrarily.
Given that archaeal SPases such as H. volcanii Sec11a and Sec11b clearly differ from both their eukaryal counterparts and the majority of bacterial SPases, including the well-studied E. coli enzyme (26, 27), it is possible that archaeal SPases are closer to SipW-like SPases. Like the archaeal enzyme, the bacterial SPase family exemplified by B. subtilis SipW and thus far only detected in sporulating gram-positive Bacteria has also replaced the lysine of the E. coli-type enzyme's catalytic dyad with a histidine residue (38). Site-directed mutagenesis studies have, moreover, confirmed the involvement of this histidine residue in the cleavage reaction catalyzed by SipW (34). Nonetheless, differences between the archaeal enzyme and SipW are evident. Whereas the His-145 equivalent of the archaeal enzyme is essential for the activity of the archaeal enzyme (4), the same residue in SipW can be replaced with a lysine without a loss of enzymatic function (34). Furthermore, in contrast to its archaeal counterpart, as exemplified by the M. voltae enzyme, SipW requires the Asp-273, but not the Asp-280, equivalent for optimal activity (34).
With the sequences of H. volcanii Sec11a and Sec11b now known, purification of each protein in its native high-salt surroundings possible, and an in vitro system for enzyme characterization developed, questions related to the catalytic mechanism as well as other aspects of archaeal SPase biology can be addressed. Such studies will likely be complemented by studies relying on the in vitro assay developed for M. voltae SPase (23). These systems can also be used to address questions related to those features of the signal peptide recognized by the archaeal enzyme. For instance, the ability of isolated H. volcanii Sec11and Sec11b to cleave a native signal peptide fused to a nonnative protein suggests that the archaeal enzyme may tolerate variety at positions downstream from the actual cleavage site. Continued manipulation of the amino acid composition of the signal peptide and the N-terminal region of the mature domain of SPase substrates will thus provide further insight into the behavior of the archaeal SPase and possibly highlight differences between the two H. volcanii enzymes.
Acknowledgments
We thank Vered Caspi for her assistance with bioinformatics-based studies; Moshe Mevarch (Tel Aviv University) and Thorsten Allers (University of Nottingham) for strains, guidance, and encouragement in performing gene replacement experiments; and Mevarech and Yuval Shoham (Technion Israel Institute of Technology) for their gifts of antibodies.
This work was supported by the Israel Science Foundation (grant 433/03).
REFERENCES
- 1.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chiu, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14:2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides—a comparative survey at the genome level. Protein Sci. 12:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardy, S. L., S. Y. Ng, D. S. Carnegie, and K. F. Jarrell. 2005. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal Methanococcus voltae. J. Bacteriol. 187:1188-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, M. T. 1993. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J. Bacteriol. 175:4957-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blobel, G., and B. Dobberstein. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67:835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolhuis, A. 2002. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology 148:3335-3346. [DOI] [PubMed] [Google Scholar]
- 9.Carlos, J. L., M. Paetzel, G. Brubaker, A. Karla, C. M. Ashwell, M. O. Lively, G. Cao, P. Bullinger, and R. E. Dalbey. 2000. The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J. Biol. Chem. 275:38813-38822. [DOI] [PubMed] [Google Scholar]
- 10.Cline, S. W., L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171:4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalbey, R. E., M. O. Lively, S. Bron, and J. M. van Dijl. 1997. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalbey, R. E., and G. von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci. 17:474-478. [DOI] [PubMed] [Google Scholar]
- 13.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichler, J. 2000. Novel glycoproteins of the halophilic archaeon Haloferax volcanii. Arch. Microbiol. 173:445-448. [DOI] [PubMed] [Google Scholar]
- 15.Eichler, J. 2002. Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. Evol. 54:411-415. [DOI] [PubMed] [Google Scholar]
- 16.Evans, E. A., R. Gilmore, and G. Blobel. 1986. Purification of microsomal signal peptidase as a complex. Proc. Natl. Acad. Sci. USA 83:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15:1336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fekkes, P., and A. J. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irihimovitch, V., and J. Eichler. 2003. Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278:12881-12887. [DOI] [PubMed] [Google Scholar]
- 20.Irihimovitch, V., G. Ring, T. Elkayam, Z. Konrad, and J. Eichler. 2003. Isolation of fusion proteins containing SecY and SecE components of the protein translocation complex from the halophilic archaeon Haloferax volcanii. Extremophiles. 7:71-77. [DOI] [PubMed] [Google Scholar]
- 21.Madern, D., C. Ebel, and G. Zaccai. 2000. Halophilic adaptation of enzymes. Extremophiles 4:91-98. [DOI] [PubMed] [Google Scholar]
- 22.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162:461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng, S. Y., and K. F. Jarrell. 2003. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae. J. Bacteriol. 185:5936-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortenberg, R., and M. Mevarech. 2000. Evidence for post-translational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J. Biol. Chem. 275:22839-22846. [DOI] [PubMed] [Google Scholar]
- 26.Paetzel, M., R. E. Dalbey, and N. C. J. Strynadka. 1998. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature 396:186-190. [DOI] [PubMed] [Google Scholar]
- 27.Paetzel, M., R. E. Dalbey, and N. C. J. Strynadka. 2000. The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target. Pharmacol. Therapeut. 87:27-49. [DOI] [PubMed] [Google Scholar]
- 28.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 29.Paetzel, M., N. C. J. Strynadka, W. R. Tschantz, R. Casareno, P. R. Bullinger, and R. E. Dalbey. 1997. Use of site-directed chemical modification to study an essential lysine in Escherichia coli leader peptidase. J. Biol. Chem. 272:9994-10003. [DOI] [PubMed] [Google Scholar]
- 30.Rose, R. W., T. Bruser, J. C. Kissinger, and M. Pohlschroder. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45:943-950. [DOI] [PubMed] [Google Scholar]
- 31.Rosenshine, I., T. Zusman, R. Werczberger, and M. Mevarech. 1987. Amplification of specific DNA sequences correlates with resistance of the archaebacterium Halobacterium volcanii to the dihydrofolate reductase inhibitors trimethoprim and methotrexate. Mol. Gen. Genet. 208:518-522. [Google Scholar]
- 32.Sumper, M., E. Berg, R. Mengele, and I. Strobel. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tjalsma, H., A. Bolhuis, M. L. van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. J. Quax, G. Venema, S. Bron, and J. M. van Dijl. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12:2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tjalsma, H., A. G. Stover, A. Driks, G. Venema, S. Bron, and J. M. van Dijl. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102-25108. [DOI] [PubMed] [Google Scholar]
- 35.Tschantz, W. R., M. Paetzel, G. Cao, D. Suciu, M. Inouye, and R. E. Dalbey. 1995. Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34:3935-3941. [DOI] [PubMed] [Google Scholar]
- 36.Tschantz, W. R., M. Sung, V. M. Delgado-Partin, and R. E. Dalbey. 1993. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J. Biol. Chem. 268:27349-27354. [PubMed] [Google Scholar]
- 37.van Dijl, J. M., A. de Jong, G. Venema, and S. Bron. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. Structural and functional similarities with LexA-like proteases. J. Biol. Chem. 270:3611-3618. [DOI] [PubMed] [Google Scholar]
- 38.van Roosmalen, M. L., N. Geukens, J. D. Jongbloed, H. Tjalsma, J. Y. Dubois, S. Bron, J. M. van Dijl, and J. Anne. 2004. Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta 1694:279-297. [DOI] [PubMed] [Google Scholar]
- 39.van Valkenburgh, C., X. Chen, C. Mullins, H. Fang, and N. Green. 1999. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J. Biol. Chem. 274:11519-11525. [DOI] [PubMed] [Google Scholar]
- 40.YaDeau, J. T., C. Klein, and G. Blobel. 1991. Yeast signal peptidase contains a glycoprotein and the Sec11 gene product. Prot. Natl. Acad. Sci. USA 88:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwizinski, C., and W. Wickner. 1980. Purification and characterization of leader (signal) peptidase from Escherichia coli. J. Biol. Chem. 255:7973-7977. [PubMed] [Google Scholar]