Abstract
Pseudomonas aeruginosa secretes two siderophores, pyoverdine and pyochelin, under iron-limiting conditions. These siderophores are recognized at the cell surface by specific outer membrane receptors, also known as TonB-dependent receptors. In addition, this bacterium is also able to incorporate many heterologous siderophores of bacterial or fungal origin, which is reflected by the presence of 32 additional genes encoding putative TonB-dependent receptors. In this work, we have used a proteomic approach to identify the inducing conditions for P. aeruginosa TonB-dependent receptors. In total, 11 of these receptors could be discerned under various conditions. Two of them are only produced in the presence of the hydroxamate siderophores ferrioxamine B and ferrichrome. Regulation of their synthesis is affected by both iron and the presence of a cognate siderophore. Analysis of the P. aeruginosa genome showed that both receptor genes are located next to a regulatory locus encoding an extracytoplasmic function sigma factor and a transmembrane sensor. The involvement of this putative regulatory locus in the specific induction of the ferrioxamine B and ferrichrome receptors has been demonstrated. These results show that P. aeruginosa has evolved multiple specific regulatory systems to allow the regulation of TonB-dependent receptors.
In most environments, the amount of free iron (approximately 10−9 M) is below the concentration required by most microorganisms for growth (49). To fulfill their requirements for iron, bacteria have developed several strategies, including (i) the reduction of ferric to ferrous ions, (ii) the direct acquisition of iron from host proteins, (iii) the secretion of high-affinity iron-chelating compounds, called siderophores, and (iv) the uptake of heterologous siderophores (23). The transport of (heterologous) siderophores and heme/hemophores through the outer membranes of gram-negative bacteria is an energy-dependent process that needs the function of the ExbB-ExbD-TonB system. This system transduces the proton-motive force of the cytoplasmic membrane to the high-affinity outer membrane receptors, which are therefore also known as TonB-dependent receptors (48). The synthesis of siderophores and their transport systems is negatively regulated by the Fur repressor protein (18). In addition, some siderophore transport systems are also positively regulated by the presence of their cognate siderophores (12). This level of regulation often involves extracytoplasmic function (ECF) sigma factors (56). The ECF family of sigma factors constitutes a group of environmentally responsive transcriptional factors of the RpoD (σ70) family that control a wide range of bacterial functions (39). The ECF factors involved in the regulation of iron uptake systems have been classified as the iron starvation class of sigma factors (35). One well-studied example is ferric dicitrate uptake by Escherichia coli (10). Binding of ferric dicitrate to the outer membrane receptor FecA generates a signal that is transmitted by the N terminus of FecA, located in the periplasm, to the C terminus of the cytoplasmic membrane FecR. This protein functions as an anti-sigma factor inhibiting the function of the ECF sigma factor FecI in the absence of ferric dicitrate. After induction, FecI binds to the RNA polymerase core complex and directs it to the promoter region of the genes required for ferric dicitrate transport (fecABCDE). Similar regulatory systems, also called cell surface signaling, have been identified in various bacteria, including Pseudomonas putida (31, 32) and Pseudomonas aeruginosa (6).
P. aeruginosa is a remarkably versatile pathogen. This bacterium, which grows in soil, freshwater, and saltwater habitats, is not only an important opportunistic pathogen for humans but is also able to infect a large range of other species, including plants, insects, and nematodes (Caenorhabditis elegans) (57). Under iron-limiting conditions, P. aeruginosa secretes two siderophores, pyoverdine and pyochelin. Ferri-pyoverdine binding to the outer membrane receptor FpvA induces, via a cell surface signaling pathway consisting of two ECF sigma factors, transcription of the fpvA gene and also that of the genes required for the production of pyoverdine as well as the genes encoding exotoxin A and the protease PrpL (6, 34). Moreover, the presence of a second pyoverdine receptor, FpvB, in P. aeruginosa has recently been reported (21). Besides these two pyoverdine receptors and the characterized receptor for pyochelin (FptA) (2, 27), this bacterium also produces receptors for heterologous iron sources: there are two receptors, PhuR and HasR, involved in the utilization of heme (44), and two, PfeA and PirA, that mediate the uptake of enterobactin (14, 22). However, these receptors are probably just a small sample of the real iron acquisition potential of P. aeruginosa, because in silico analysis of its genome revealed an impressive total of 34 genes encoding putative TonB-dependent outer membrane receptors (11). Here we report that two of these receptors, PA2466 (FoxA) and PA0470 (FiuA), are indeed involved in the transport of the heterologous siderophores ferrioxamine B and ferrichrome, respectively. In addition, we also show the involvement of both receptors in a signaling pathway that regulates their own synthesis in response to the presence of their cognate ligands.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. The P. aeruginosa PAO1 wild-type strain and all the P. aeruginosa transposon insertion mutants used were from the comprehensive P. aeruginosa transposon mutant library at the University of Washington Genome Center (28). The locations of the mutations were confirmed by PCR with primers flanking the insertion sites. The strains' unique identifiers are given in the table, and further information on these mutants can be found at http://www.genome.washington.edu/UWGC/Pseudomonas/index.cfm.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR1 recA1 endA1 gyrA96 thi-1 relA1 | 25 |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 9 |
| P. aeruginosa strains | ||
| PAO25 | PAO1 leu arg | 24 |
| PAO1 | Wild type | 28 |
| PA2466 (foxA) | PAO1 with insertion of ISlacZ/hah transposon in nucleotide 266 of the PA2466 (foxA) gene (ID 19564) | 28 |
| PA2467 (foxR) | PAO1 with insertion of ISlacZ/hah transposon in nucleotide 231 of the PA2467 (foxR) gene (ID 9095) | 28 |
| PA0470 (fiuA) | PAO1 with insertion of ISphoA/hah transposon in nucleotide 378 of the PA0470 (fiuA) gene (ID 34018) | 28 |
| PA0471 (fiuR) | PAO1 with insertion of ISphoA/hah transposon in nucleotide 822 of the PA0471 (fiuR) gene (ID 53020) | 28 |
| PA0472 (fiuI) | PAO1 with insertion of ISphoA/hah transposon in nucleotide 39 of the PA0472 (fiuI) gene (ID 52421) | 28 |
| pvdD pchEF | PAO1 with unmarked allelic pvdD pchEF mutations | 21 |
| pvdD pchEF PA0470 | Allelic mutant of PA40470 (fiuA) in pvdD and pchEF mutant background | 21, 22 |
| Plasmids | ||
| pBSL99 | Source of the Km cassette; Apr Kmr | 1 |
| pMMB67EH | IncQ broad-host-range plasmid; lacIq Apr | 20 |
| pMP220 | IncP broad-host-range lacZ fusion vector; Tcr | 53 |
| pRK600 | Helper plasmid with oriColE1, mobRK2, and traRK2; Cmr | 16 |
| pUC18 | Cloning vector with oriColE1, rop mutant, α-lacZ mutant | 42 |
| pUCMA8 | pUC18 carrying at the SmaI site the 2.5-kb PCR fragment containing the P. aeruginosa PAO1 foxI-foxR (PA2468-PA2467) genes; Apr | This study |
| pMUM8 | pMMB67EH carrying the 824-bp SacI-SalI insert from pUCMA8 containing the foxI (PA2468) gene; Apr | This study |
| pMPR8b | foxA (PA2466) promoter fragment cloned upstream of lacZ gene in pMP220; Tcr | This study |
| pMPR8bKm | pMPR8b carrying at the BgIII site, in the opposite direction from the lacZ gene, a 1.2-kb BamHI fragment of pBSL99 containing a Km cassette; Kmr Tcr | This study |
| pMMBFiuI | pMMB67EH carrying the 742-bp XbaI-HindIII PCR fragment containing the fiuI (PA0472) gene; Apr | This study |
| pMPFiuA | fiuA (PA0470) promoter fragment cloned upstream of lacZ gene in pMP220; Tcr | This study |
| pMPFiuAKm | pMPFiuA carrying at the BgIII site, in the opposite direction from the lacZ gene, a 1.2-kb BamHI fragment of pBSL99 containing a Km cassette; Kmr Tcr | This study |
| pMPR2911 | PA2911 promoter fragment cloned upstream of lacZ gene in pMP220; Tcr | This study |
| pMPR2911Km | pMPR2911 carrying at the BgIII site, in the opposite direction from the lacZ gene, a 1.2-kb BamHI fragment of pBSL99 containing a Km cassette; Kmr Tcr | This study |
| pMPR3268 | PA3268 promoter fragment cloned upstream of lacZ gene in pMP220; Tcr | This study |
| pMPR3268Km | pMPR8 carrying at the BgIII site, in the opposite direction from the lacZ gene, a 1.2-kb BamHI fragment of pBSL99 containing a Km cassette; Kmr Tcr | This study |
Apr, Cmr, Kmr, and Tcr, resistance to ampicillin, chloramphenicol, kanamycin, and tetracycline, respectively.
Bacterial strains were routinely grown in liquid Luria-Bertani medium (50) at 37°C on a rotary shaker operated at 200 revolutions per min. For experiments on the effect of the iron concentration on gene expression, P. aeruginosa cells were grown in liquid CAS medium [3.18 mM Ca(NO3)2, 1 mM MgSO4, 50 mM PIPES, pH 6.8, containing 1 mM K2HPO4, 1% (wt/vol) Casamino Acids, and 1% (wt/vol) glycerol] supplemented with either 50 μM FeCl3 (iron-rich conditions) or 400 μM 2,2′-bipyridyl (iron-restricted conditions). The ferrioxamine B and ferrichrome stock solutions (20 mM) were prepared by dissolving desferrioxamine B mesylate (Desferal) and ferrichrome (Sigma) in a Tris-HCl, pH 8.8, solution without (iron-free form) or with 20 mM FeCl3 (iron-loaded form). Pseudobactin PS358 (8) was used at a 20 μM final concentration. When required, antibiotics were used at the following final concentrations (μg ml−1): ampicillin, 100; chloramphenicol, 30; kanamycin, 25 (for E. coli) and 200 (for P. aeruginosa); piperacillin, 25; and tetracycline, 10 (for E. coli) and 20 (for P. aeruginosa).
General molecular biology methods.
Standard molecular biology techniques were used for DNA manipulations (50). DNAs from plasmids pUCMA8 and pMMBFiuI were sequenced by the dideoxy sequence termination method (51) with AmpliTaq DNA polymerase (Big Dye Terminator v3.1 cycle sequencing kit; Applied Biosystems). PCR amplifications were carried out using the Expand high-fidelity system (Roche) containing 0.2 ng μl−1 P. aeruginosa PAO1 chromosomal DNA, a 100 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, and 5% (vol/vol; final concentration) dimethyl sulfoxide. The primers used for amplification of the wild-type PA2468 and PA2467 genes were Puma8F (5′-CTGGCCAACATCGAATTGT-3′) and Puma8R (5′-GAGACGGTCTGGCTGGTTT-3′), and those for the amplification of PA0472 were FiuI5X (5′-TATTCTAGATGATCAGCAAAGGTTCGGC-3′) and FiuI3H (5′-AAAAAGCTTCGATCGTCGGCGCTCACC-3′).
Construction of lacZ transcriptional fusions and β-galactosidase assay.
Transcriptional fusions were constructed by cloning DNA segments with the promoter regions of the PA0470, PA2911, PA2466, and PA3268 genes of P. aeruginosa PAO1, amplified by PCR as EcoRI-XbaI fragments with appropriate primers, into the EcoRI-XbaI sites of pMP220. The fusion constructs were confirmed by DNA sequencing and transferred from E. coli DH5α to P. aeruginosa by triparental mating using the helper plasmid pRK600 as described before (16). β-Galactosidase activity from the fusion plasmid was measured as described previously (38). Each assay was run in duplicate at least three times, and the data given are averages.
Cell envelope preparations.
P. aeruginosa cells were grown until late log phase, and fractions enriched for outer membrane proteins were isolated by centrifugation of ultrasonically disrupted cells (30 min at 10,000 × g) followed by extraction with 1% (wt/vol) Sarkosyl (17). Protein profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in gels containing 7.5% acrylamide as described by Laemmli (33). Samples were incubated for 10 min at 95°C prior to electrophoresis, and proteins were stained with Coomassie blue.
In-gel digestion of proteins and sample preparation for MALDI-mass spectrometry (MALDI-MS) fingerprint mapping.
Protein bands of interest were excised from Coomassie-stained SDS-PAGE gels. The excised gel bands were washed twice with 400 μl of 50 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile to destain the proteins and dehydrate the gel pieces and then dried completely in a vacuum centrifuge for 30 to 45 min. The dried gel pieces were rehydrated in 50 mM ammonium bicarbonate, pH 8, containing 6.25 ng/μl trypsin (sequencing grade; Promega), and the proteins were digested for 12 to 16 h at 37°C. The peptides were then recovered from the gel particles by performing two extractions. For the first extraction, 50 μl of 1% (vol/vol) trifluoroacetic acid was added, and the samples were vortexed for 15 min. The second extraction was performed with 100 μl of 0.1% (vol/vol) trifluoroacetic acid in 50% (vol/vol) acetonitrile. Peptide extracts were combined and concentrated by reducing the final volumes of the extracts to 20 to 25 μl in a vacuum centrifuge. The tryptic peptides were then desalted and concentrated in reversed-phase C18 Zip Tip pipette tips (Millipore) according to the manufacturer's recommendations. Briefly, Zip Tips were washed with 50% (vol/vol) acetonitrile and equilibrated with 0.1% (vol/vol) trifluoroacetic acid. Peptide extracts were then applied to the Zip Tips, washed with 0.1% (vol/vol) trifluoroacetic acid, and eluted in 3 μl of a saturated matrix solution of 5 μg/μl α-cyano-4-hydroxycinnamic acid in 0.1% (vol/vol) trifluoroacetic acid and 50% (vol/vol) acetonitrile. A 0.5-μl sample of the mixture was immediately spotted onto the matrix-assisted laser desorption ionization (MALDI) target and allowed to dry and crystallize.
MALDI-MS and database searching.
The molecular masses of the tryptic peptides (peptide mass fingerprinting) were determined in a MALDI-time of flight mass spectrometer (4700 Proteomics analyzer; Applied Biosystems), using an acceleration voltage of 20 kV. MS spectra were searched against the NCBI database, using Protein Prospector MS-Fit v4.0.5 software, available at http://prospector.ucsf.edu, to identify the proteins. The protonated trypsin autodigestion products at m/z 842.510 and 2,211.104 were used for internal calibration of the MALDI-MS spectra. The MALDI-MS resolution for the peptides was ∼10,000, and the mass accuracy was within 0.01 to 0.02 Da. For database searching of peptide mass fingerprint data, the mass tolerance was set at 20 ppm.
Computer-assisted analysis.
In silico analysis of the PAO1 genome was performed at http://www.pseudomonas.com. Signal peptides were predicted using the SignalP 3.0 server available at http://www.cbs.dtu.dk/services/SignalP (7).
RESULTS
TonB-dependent receptors under iron-limiting conditions.
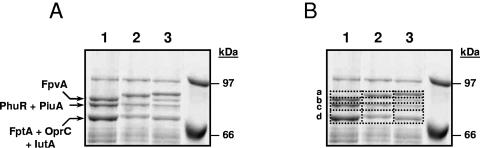
P. aeruginosa PAO1 genome analysis showed that this bacterium has the potential to produce 34 different TonB-dependent receptors (11). We first wanted to establish how many of these receptors are produced under iron-limiting conditions. Therefore, cell envelope preparations, enriched for outer membrane proteins by Sarkosyl extraction, of cells grown in liquid CAS medium supplemented with 400 μM of 2,2′-bipyridyl (iron-limiting conditions) were subjected to SDS-PAGE analysis. At least three high-molecular-weight major outer membrane proteins could be observed in these samples (Fig. 1A, lane 1). In order to unequivocally determine the nature of these proteins, the gel bands shown in Fig. 1B were excised and subjected to tryptic digestion, followed by MALDI-mass spectrometry for peptide mass mapping. The first visible band (sample 1b) contained the pyoverdine receptor FpvA, the second band (sample 1c) contained mainly the heme/hemophore receptor PhuR, and the third band (sample 1d) contained the pyochelin receptor FptA (Table 2). Besides PhuR, sample 1c also contained the putative TonB-dependent receptor PA4514. The amount of this protein was considerably lower than that of PhuR, as judged by the smaller number of tryptic peptides that matched PA4514 (Table 2). Sample 1d not only contained FptA but also had the TonB-dependent receptor PA4675 and the putative TonB-dependent outer membrane protein OprC (PA3790) (Table 2). In addition, in sample 1a, in which no protein band was visible (Fig. 1), another putative TonB-dependent receptor encoded by PA4168 was detected. This protein, which is the second pyoverdine receptor, named FpvB (21), has been visualized previously (26). Although the presence of the FpvA, PhuR, and FptA receptors in the outer membrane of P. aeruginosa has been reported previously, this analysis showed the presence of four other TonB-dependent receptors under iron-limiting conditions, which brings the total to seven. This means that the 27 other putative P. aeruginosa TonB-dependent receptors require additional or other inducing conditions. To substantiate this hypothesis, P. aeruginosa was grown under iron-limiting conditions in the presence of various heterologous siderophores.
FIG. 1.
SDS-PAGE analysis of outer membrane proteins from P. aeruginosa PAO25. P. aeruginosa PAO25 was grown under iron-limiting conditions without siderophores (lane 1), with 20 μM ferrioxamine B (iron-loaded) (lane 2), or with 40 μM ferrichrome (iron-loaded) (lane 3) until late log phase. Outer membrane-enriched preparations were prepared as described in Materials and Methods and separated in a 7.5% acrylamide gel. (A) The positions of the TonB-dependent receptor proteins detected by MALDI-MS in cells grown under iron-limiting conditions are indicated on the left (arrows), and those of the molecular size markers are indicated on the right. (B) The different samples used for MALDI-MS analyses are shown.
TABLE 2.
TonB-dependent receptors of P. aeruginosa identified by MALDI-MS analysisa
| Sample | Rank | Protein name | PA numberb | No. of peptides matching protein | % Protein coverage | Protein scorec |
|---|---|---|---|---|---|---|
| 1a | 1 | FpvA | PA2398 | 12 | 20 | 1.6e+08 |
| 2 | FpvB | PA4168 | 11 | 17 | 0.61e+04 | |
| 2a | 1 | Probable TonB-dependent receptor | PA2466 | 29 | 44 | 1.2e+10 |
| 3a | 1 | Probable hydroxamate-type ferrisiderophore receptor | PA0470 | 42 | 55 | 4.1e+17 |
| 1b | 1 | FpvA | PA2398 | 49 | 58 | 1.4e+25 |
| 2b | 1 | FpvA | PA2398 | 23 | 30 | 3.2e+08 |
| 2 | Probable TonB-dependent receptor | PA3268 | 16 | 22 | 1.6e+05 | |
| 3b | 1 | FpvA | PA2398 | 23 | 25 | 1.8e+08 |
| 2 | Probable TonB-dependent receptor | PA3268 | 17 | 25 | 7e+05 | |
| 1c | 1 | PhuR | PA4710 | 47 | 68 | 5.8e+22 |
| 2 | Hydroxamate-type ferrisiderophore receptor (PiuA) | PA4514 | 16 | 28 | 2.3e+06 | |
| 2c | 1 | Hydroxamate-type ferrisiderophore receptor | PA4514 | 36 | 53 | 1.9e+14 |
| 2 | PhuR | PA4710 | 32 | 42 | 3.4e+13 | |
| 3 | Probable TonB-dependent receptor | PA2911 | 19 | 28 | 1.3e+09 | |
| 3c | 1 | Probable TonB-dependent receptor | PA2911 | 18 | 30 | 7.9e+08 |
| 2 | Hydroxamate-type ferrisiderophore receptor | PA4514 | 12 | 18 | 1.1e+05 | |
| 1d | 1 | FptA | PA4221 | 48 | 70 | 9e+21 |
| 2 | OprC | PA3790 | 28 | 41 | 6.6e+10 | |
| 3 | Probable TonB-dependent receptor (IutA) | PA4675 | 20 | 29 | 1.3e+07 | |
| 2d | 1 | Probable TonB-dependent receptor | PA4675 | 33 | 43 | 2.1e+11 |
| 2 | OprC | PA3790 | 22 | 34 | 9.6e+07 | |
| 3d | 1 | OprC | PA3790 | 52 | 67 | 6.1e+20 |
| 2 | Probable TonB-dependent receptor | PA4675 | 26 | 33 | 1e+10 |
For the mass peptide fingerprint search, only mass deviations of <20 ppm were allowed (high-stringency search).
PA number in the P. aeruginosa genome annotation project (http://www.pseudomonas.com).
Scores are −10 log(P) values, where P is the probability that the observed match is a random event (http://prospector.ucsf.edu). Protein scores given are significant (P < 0.05).
Induction of P. aeruginosa outer membrane proteins by ferrioxamine B and ferrichrome.
In order to determine the inducing conditions for other TonB-dependent receptors, P. aeruginosa was grown in the presence of different iron sources, such as the iron-loaded form of the heterologous siderophores ferrioxamine B, ferrichrome, and pseudobactin PS358. In the presence of pseudobactin PS358, P. aeruginosa did not express any other readily detectable outer membrane proteins compared with those induced under normal iron-restricted conditions (data not shown). However, new outer membrane proteins with different apparent molecular weights were expressed when P. aeruginosa was cultured in the presence of ferrioxamine B or ferrichrome (Fig. 1, lanes 2 and 3, respectively). The expression of these inducible proteins was completely suppressed upon growth under iron-rich conditions, even if ferrioxamine B or ferrichrome was present (not shown). Apparently, both the absence of iron and the presence of the heterologous siderophores are needed for induction.
To determine the nature of the induced proteins, the gel bands shown in Fig. 1B (lanes 2 and 3) were excised and analyzed by MALDI-MS. A protein database search revealed that the ferrioxamine B-induced protein (Fig. 1B, lane 2, sample 2a) was the putative TonB-dependent receptor PA2466 (Table 2), whereas the ferrichrome-induced protein (Fig. 1B, lane 3, sample 3a) showed high identity with the putative TonB-dependent receptor PA0470 (FiuA) (Table 2). These two identified proteins (PA2466 and PA0470) could not be detected in the corresponding gel fragment in lane 1 (sample 1a). To determine whether any other receptors were induced by ferrioxamine B or ferrichrome, the gel fragments corresponding to regions b, c, and d were also analyzed by MALDI-MS analysis. This analysis showed that the proteins in region b (lanes 2 and 3) contained, in addition to FpvA, the TonB-dependent receptor PA3268, whereas in region c (lanes 2 and 3), the previously identified PA4514 protein (PiuA) and the novel putative TonB-dependent receptor PA2911 were detected (Table 2). Proteins in sector d (lanes 2 and 3) were shown to be the previously identified OprC and PA4675 proteins (Table 2). These results indicate that the addition of ferrioxamine B and ferrichrome not only resulted in the induction of a specific receptor but that both siderophores possibly also induced PA2911 and PA3268 synthesis, since these proteins were not detected in the absence of these siderophores (samples 1b and 1c, respectively).
Characteristics of ferrioxamine B- and ferrichrome-induced proteins.
The ferrioxamine B-induced protein PA2466 is an 820-amino-acid protein that contains a predicted signal peptide of 47 amino acids and has a predicted molecular mass of 85,273 Da (after cleavage of the signal peptide). The PA2466 gene is annotated in the Pseudomonas Genome Project database (www.pseudomonas.com) as a probable TonB-dependent receptor gene named optS (outer protein S). A BLAST search of the P. aeruginosa PA2466 protein sequence showed 43 to 47% identities with the characterized TonB-dependent ferrioxamine B FoxA receptors of Salmonella enterica serovar Typhimurium (29), Yersinia enterocolitica (5), and Erwinia amylovora (15). By analogy with these proteins, we propose renaming the P. aeruginosa PA2466 gene foxA. The foxA gene contains a strong putative Fur box in its promoter region, suggesting its regulation by the Fur repressor protein. This is consistent with the observation that the synthesis of FoxA is repressed under iron-rich conditions, even in the presence of ferrioxamine B (not shown). The mature FoxA protein contains an N-terminal periplasmic extension (Fig. 2A), which is a typical feature of TonB-dependent receptors involved in cell surface signaling pathways (30). Moreover, the two open reading frames adjacent to the foxA gene are annotated as a probable transmembrane sensor gene (PA2467) and a probable σ70 gene of the ECF subfamily (PA2468), respectively. The PA2467 (designated foxR in this work) and PA2468 (designated foxI) genes are clustered in an operon that also contains a putative Fur box in its promoter region. In fact, Ochsner and coworkers (46) have shown that expression of the foxI-foxR operon considerably increases under iron-limiting conditions.
FIG. 2.
N-terminal extension of mature P. aeruginosa FoxA and FiuA proteins. (A) N-terminal extension of mature P. aeruginosa FoxA compared to mature Y. enterocolitica, E. amylovora, and S. enterica serovar Typhimurium FoxA proteins. (B) N-terminal extension of mature P. aeruginosa FiuA compared to mature E. coli FhuA. The proposed TonB box sequences are underlined. Positions at which identical or similar residues are present in at least three of the four sequences are shown in bold. The numbering of amino acid residues is indicated.
The ferrichrome-induced protein encoded by the PA0470 gene is a protein of 802 amino acids that contains a predicted signal peptide of 26 amino acids and has a predicted molecular mass of 85,489 Da (after cleavage of the signal peptide). This gene is annotated in the P. aeruginosa genome as a probable hydroxamate-type ferri-siderophore receptor gene named fiuA. The FiuA protein shows 31.6% identity with the ferrichrome receptor FhuA of E. coli (19), but in contrast to FhuA, the mature FiuA protein contains an N-terminal extension (Fig. 2B). The fiuA gene contains a putative Fur box in its promoter region and is preceded by two genes, named fiuR (PA0471) and fiuI (also called fiuS) (PA0472), encoding a probable transmembrane sensor and an ECF sigma factor, respectively. The expression of the fiuI-fiuR gene cluster has been shown to be under the control of the iron-responsive Fur repressor protein (45).
Involvement of the Fox system and ferrioxamine B in the production of FoxA.
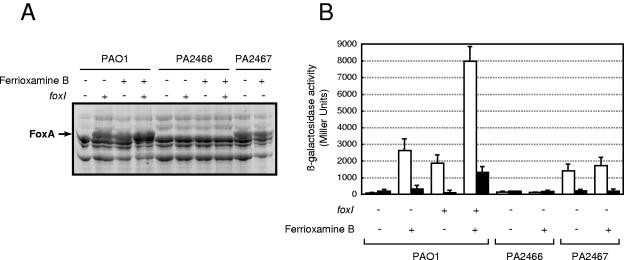
To analyze the function of the putative ECF sigma factor FoxI (PA2468), the foxI gene was amplified by PCR, cloned into the pMMB67EH plasmid (yielding pMUM8) (Table 1), and introduced in P. aeruginosa. In pMUM8, foxI is constitutively expressed from the Ptac promoter, and this plasmid also contains the lacIq gene, which allows regulated expression of the sigma factor. The introduction of pMUM8 into P. aeruginosa PAO1 resulted in the specific induction of an outer membrane protein in both noninduced cultures (Fig. 3) and cultures induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (not shown). The induction by FoxI was independent of the presence or absence of ferrioxamine B in the culture medium, and the apparent molecular weight of the FoxI-induced protein was similar to that of the ferrioxamine B-induced receptor FoxA (Fig. 3A). Subsequently, the pMUM8 plasmid was transferred to the PA2466 mutant strain, which contains a transposon insertion in the foxA gene (Table 1). Both the FoxI- and ferrioxamine B-induced outer membrane proteins were absent in the foxA mutant (Fig. 3A), indicating that the protein is in fact FoxA.
FIG. 3.
Analysis of Fox signaling system. P. aeruginosa PAO1 (wild type), PA2466 (foxA mutant), and PA2467 (foxR mutant) bearing the plasmid pMMB67EH (foxI negative) or pMUM8 (foxI positive) were grown under iron-restricted conditions without (−) or with (+) 20 μM ferrioxamine B (iron-free form) until late exponential growth phase. (A) Cell envelope preparations enriched for outer membrane proteins by Sarkosyl extraction were separated in a 7.5% acrylamide gel. The position of the ferrioxamine B receptor FoxA (PA2466) is indicated on the left. (B) P. aeruginosa strains containing the plasmid pMPR8bKm (PfoxA::lacZ transcriptional fusion) and the plasmid pMMB67EH (foxI negative) or pMUM8 (foxI positive) were grown under iron-restricted conditions (white bars) or iron-rich conditions (black bars) in either the presence (+) or absence (−) of 20 μM ferrioxamine B (iron-free form).
The involvement of ferrioxamine B and FoxI in regulating the production of FoxA was tested further using the plasmid pMPR8bKm, which contains the putative promoter region of the foxA gene fused to a promoterless lacZ reporter gene. As shown in Fig. 3B, the addition of ferrioxamine B to the iron-restricted culture medium of the wild-type PAO1 strain led to a 33-fold increase in the activity of the foxA promoter. Overexpression of foxI has a similar effect and resulted in a 25-fold increase in foxA expression in the absence of ferrioxamine B and a 100-fold increase in the presence of the siderophore, both under iron-restricted conditions. This effect of ferrioxamine B and FoxI on foxA expression was severely reduced under iron-rich conditions (Fig. 3B). The requirement of FoxI for foxA expression is consistent with FoxI being a sigma factor that directs the expression of foxA in the presence of ferrioxamine B.
By analogy with other cell surface signaling systems, the activity of FoxI would be regulated by the transmembrane sensor FoxR (PA2467), which would act as an anti-sigma factor, and by the FoxA protein itself. To test this, first the production of FoxA in the PA2467 foxR mutant was examined. As expected, the FoxR mutant showed induction of FoxA independent of ferrioxamine B (Fig. 3A). However, the ISlacZ/hah transposon used to generate the PA2467 gene mutation could have polar effects on the expression of the downstream gene PA2466 encoding FoxA (28). Therefore, the involvement of FoxR in foxA expression was also examined using the pMPR8bKm plasmid. Also, in this situation, the expression of foxA in the foxR mutant occurred in the absence of ferrioxamine B (Fig. 3B) and was higher under iron-restricted than iron-rich conditions. These data are consistent with FoxR being an anti-sigma factor for FoxI, which affects FoxA production in the absence of ferrioxamine B through its inhibitory effect on FoxI activity. In fact, the specific binding of FoxR to FoxI has been previously reported (36). In addition, the level of foxA promoter activity in the PA2466 foxA mutant strain was also examined. foxA promoter activity in the presence of ferrioxamine B was completely abolished in this mutant (Fig. 3B), which indicates that the ferrioxamine B signal was not properly transmitted.
All of these data are consistent with a model in which binding of ferrioxamine B to FoxA results in signal transmission to FoxR and, consequently, increased activity of FoxI, enabling foxA expression.
Involvement of the Fiu system and ferrichrome in the production of FiuA.
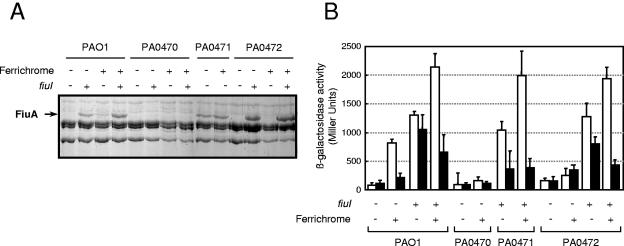
The role of the Fiu system in the ferrichrome-mediated production of FiuA was analyzed in a similar way. The fiuI gene was constitutively expressed in P. aeruginosa from the plasmid pMMBFiuI. Introduction of this plasmid into the P. aeruginosa wild-type PAO1 strain resulted in the specific induction of an outer membrane protein (Fig. 4A) with an apparent molecular weight similar to that of the ferrichrome-induced receptor FiuA (Fig. 4A). Both the FiuI- and ferrichrome-induced proteins were absent in the PA0470 (fiuA) mutant (Fig. 4A), confirming that in both cases the induced protein was the FiuA receptor. The role of FiuI was analyzed using a fiuI (PA0472) mutant strain. As shown in Fig. 4A, this strain did not produce FiuA after growth in ferrichrome. Complementation of the fiuI mutation with the pMMBFiuI plasmid resulted in normal amounts of FiuA in both the absence and presence of ferrichrome (Fig. 4A), indicating that FiuI is required for FiuA production. Subsequently, the reporter construct pMPFiuAKm, in which the fiuA promoter region was cloned in front of a lacZ gene (Table 1), was used to show that FiuI is a sigma factor that causes expression from the fiuA promoter. The presence of ferrichrome under iron-restricted conditions led to a sevenfold increase in the activity of the fiuA promoter (Fig. 4B). This induction, however, was lacking in the PA0472 fiuI mutant strain (Fig. 4B). Constitutive expression of fiuI from the pMMBFiuI plasmid in both the wild-type PAO1 and fiuI mutant strains resulted in 13.5- and 20-fold increases in fiuA promoter activity in the absence and presence of ferrichrome, respectively (Fig. 4B).
FIG. 4.
Analysis of Fiu signaling system. P. aeruginosa PAO1 (wild type), PA0470 (fiuA mutant), PA0471 (fiuR mutant), and PA0472 (fiuI mutant) bearing the plasmid pMMB67EH (fiuI negative) or pMMBFiuI (fiuI positive) were grown under iron-restricted conditions without (−) or with (+) 40 μM ferrichrome (iron-free form) until late exponential growth phase. (A) Cell envelope preparations enriched for outer membrane proteins by Sarkosyl extraction were separated in a 7.5% acrylamide gel. The position of the ferrichrome receptor FiuA (PA0470) is indicated on the left. (B) P. aeruginosa strains containing the plasmid pMPFiuAKm (PfiuA::lacZ transcriptional fusion) and the plasmid pMMB67EH (fiuI negative) or pMMBFiuI (fiuI positive) were grown under iron-restricted conditions (white bars) or iron-rich conditions (black bars) in either the presence (+) or absence (−) of 20 μM ferrichrome (iron-free form).
The role of FiuR in the ferrichrome-mediated signaling pathway was analyzed using a fiuR mutant (PA0471 strain) (Table 1). As expected, the production of FiuA in the fiuR mutant was independent of the presence or absence of ferrichrome (Fig. 4A) due to the constitutive expression of fiuA in this mutant (Fig. 4B). This is consistent with a role for FiuR as an anti-sigma factor for FiuI. The constitutive expression of fiuA in PA0471 was higher under iron-restricted than iron-rich conditions (Fig. 4B). In contrast, fiuA promoter activity was completely abolished in the PA0470 fiuA mutant, even in the presence of ferrichrome (Fig. 4B). This confirmed the role of the FiuA receptor in sensing and transmitting the signal that induces its own synthesis.
These data are in agreement with a model in which the FiuA receptor transmits a signal to FiuR after sensing the presence of ferrichrome, which increases FiuI activity and, in turn, fiuA expression.
Role of FoxA and FiuA in the uptake of ferrioxamine B and ferrichrome.
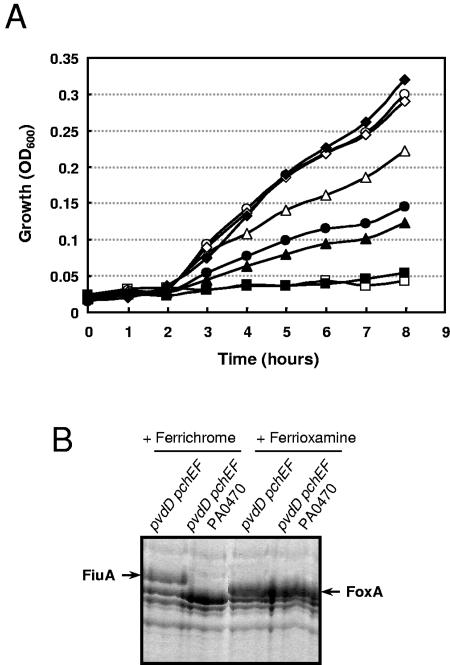
FoxA and FiuA are part of a signaling pathway, induced by ferrioxamine B and ferrichrome, respectively, that regulates their own synthesis. To determine whether these receptors are, in addition to their role in signal transduction, also involved in the uptake of their corresponding ligands, the growth of the PA2466 (foxA) and PA0470 (fiuA) mutants under iron-restricted conditions supplemented with either ferrioxamine B or ferrichrome was assayed. P. aeruginosa cells were grown in CAS medium containing 800 μM 2,2′-bipyridyl (iron-restricted conditions) supplemented with different concentrations of the iron-free forms of the siderophores. At higher siderophore concentrations (10 and 100 μM), no growth differences between the mutants and the wild-type strain were observed (data not shown). But at lower siderophore concentrations (0.5 and 1 μM), a very slight defect in growth rate was detected in the PA2466 mutant growing in ferrioxamine B and in the PA0470 mutant growing in ferrichrome (not shown). In order to rule out any effect caused by the endogenous siderophores pyoverdine and pyochelin, a P. aeruginosa PA0470 mutant in a siderophore-free background (pvdD pchEF mutant) was used. As reported before (21, 22), the pvdD pchEF mutant could not grow in the presence of 100 μM of the iron chelator 2,2′-bipyridyl unless an exogenous source of iron, i.e., ferrioxamine B or ferrichrome, was present in the medium (Fig. 5A). The mutation of PA0470 (fiuA) in this genetic background significantly delayed growth in the presence of ferrichrome, whereas this mutation had no effect on growth in the presence of ferrioxamine B (Fig. 5A). In agreement with that observation, the ferrichrome receptor FiuA was not detectable in this mutant, whereas the ferrioxamine B receptor FoxA could be detected (Fig. 5B). These results show that FiuA is directly involved in the uptake of ferrichrome but that P. aeruginosa is also able to take up this siderophore via another receptor(s).
FIG. 5.
Growth phenotype of P. aeruginosa PA0470 (FiuA) mutant. (A) P. aeruginosa PAO1 pvdD pchEF (white symbols) and pvdD pchEF PA0470 (black symbols) mutant strains were grown in CAS medium containing 100 μM 2,2′-bipyridyl and no siderophores (squares), 0.5 μM ferrichrome (triangles), 2 μM ferrichrome (circles), or 0.5 μM ferrioxamine B (diamonds) (iron-free forms of the siderophores were used). Growth is expressed as an increase in the optical density measured at 600 nm. A representative growth curve from three separate experiments is shown. (B) SDS-PAGE analysis of outer membrane proteins from P. aeruginosa PAO1 pvdD pchEF mutant and its isogenic fiuA mutant (pvdD pchEF PA0470) after overnight growth in CAS medium containing 100 μM 2,2′-bipyridyl and 50 μM ferrichrome or ferrioxamine B, respectively. The position of the ferrichrome receptor FiuA (PA0470) is indicated on the left, and that of the ferrioxamine B receptor FoxA (PA2466) is indicated on the right.
Analysis of PA2911 and PA3268 gene expression.
Proteomic analysis showed that ferrioxamine B and ferrichrome not only induce the synthesis of the specific receptors FoxA and FiuA, respectively, but that they both also seem to induce the production of two other TonB-dependent receptors, i.e., PA2911 and PA3268 (Table 2). In order to confirm this result, the reporter constructs pMPR2911Km and pMPR3268Km were generated, in which the promoter regions of the PA2911 and PA3268 genes, respectively, were fused to a promoterless lacZ gene. Subsequently, these constructs were introduced into the P. aeruginosa wild-type PAO1 strain. In contrast to what was expected, the addition of ferrioxamine B or ferrichrome to the iron-restricted medium did not result in the induction of PA2911 and PA3268 expression (not shown). Also, overexpression of the sigma factors foxI and fiuI from pMUM8 and pMMBFiuI, respectively, in the strains containing the reporter constructs did not result in increased expression of the PA2911 and PA3268 genes (not shown). These results show that neither these siderophores nor the signaling pathway activated by them is directly involved in the induction of PA2911 and PA3268 synthesis and that the presence of these proteins in siderophore-containing cultures must have another reason.
DISCUSSION
P. aeruginosa contains an impressive number of genes (34) coding for TonB-dependent receptors. Using a proteomic approach, we showed that at least seven of these TonB-dependent outer membrane receptors are produced in significant amounts under iron-limiting conditions (Fig. 1 and Table 2). The expression of some of these receptors, but not all of them (i.e., IutA and OprC), has been shown previously by microarray analysis to be induced under similar conditions (46, 47). This could be due to the fact that growth conditions (different carbon sources or iron chelators) can influence the expression of TonB receptor genes (21), which means that the receptors expressed under one condition could not be expressed under another one. In addition, we have identified four TonB-dependent receptors that are induced by the presence of heterologous siderophores. The receptors PA2911 and PA3268 were detected in cells cultured in the presence of either ferrioxamine B or ferrichrome (Table 2), whereas two other receptor genes, PA0470 (fiuA) and PA2466 (called foxA in this work), were specifically induced under iron-restricted conditions in the presence of the heterologous siderophores ferrichrome and ferrioxamine B, respectively.
Most siderophores fall into two chemical groups, namely, catecholates and hydroxamates. Ferrichrome and ferrioxamines both belong to the latter group. Ferrichrome is produced by a number of fungi, whereas ferrioxamines are produced by different bacterial species. In addition to these ferrioxamine-producing species, many other bacteria have also been shown to utilize ferrioxamines. In E. coli, a low level of ferrioxamine B transport is mediated by the outer membrane FhuE receptor (52), and the existence of a second ferrioxamine B receptor has been proposed (40, 41). Y. enterocolitica, S. enterica serovar Typhimurium, and Serratia marcescens take up ferrioxamine B efficiently through an outer membrane protein encoded by a Fur-repressible gene (5), although it has not been determined whether the synthesis of these receptors is induced by ferrioxamine. In Vibrio vulnificus, a ferrioxamine B receptor has been recently identified whose synthesis is induced by the presence of the siderophore under iron-limiting conditions via an AraC-like regulator (3, 54). This work shows that in P. aeruginosa, ferrioxamine B and ferrichrome regulate the production of their own outer membrane receptors (FoxA and FiuA, respectively) through a cell surface signaling pathway composed of the receptor itself, an ECF sigma factor, and an anti-sigma factor. These signaling systems show mechanistically high similarities to the E. coli FecIRA and P. putida PupIRB systems (10, 32). However, whereas the anti-sigma factors FecR and PupR are required for maximal activities of the corresponding sigma factors (32, 43), our data show that FoxR and FiuR are not required for maximal activities of FoxI and FiuI, respectively.
It has been stated previously that, based on unpublished data, the FiuA receptor is involved in the uptake of ferrioxamine B, and that the FiuR and FiuI proteins are responsible for the induction of FiuA in the presence of this compound (55). However, our analysis shows that the FiuA (PA0470) receptor is induced by ferrichrome, whereas the FoxA protein (PA2466) is induced by ferrioxamine B. This result was also confirmed by analyzing a P. aeruginosa fiuA mutant strain. In this mutant, the ferrioxamine-induced protein could be still detected, whereas the ferrichrome-induced protein was not detectable (Fig. 4A and 5B). In addition, the fiuA promoter activity was not increased in the presence of ferrioxamine B (not shown), and the growth of the fiuA mutant was compromised when ferrichrome was used as a source of iron, but not when ferrioxamine B was used (Fig. 5A). However, very recent data suggest that not only FoxA but also FiuA facilitates ferrioxamine B uptake (4). These two proteins share high sequence identity (40%). This probably means that FiuA can functionally replace FoxA for the uptake of ferrioxamine B in the absence of this main ferrioxamine B receptor and vice versa, although its synthesis is not induced by the siderophore. This is consistent with the fact that growth of the foxA mutant in ferrioxamine B and of the fiuA mutant in ferrichrome was not completely abolished (not shown and Fig. 5A), suggesting the involvement of other receptors in the uptake of the siderophores. In silico analysis of the P. aeruginosa PAO1 genome has already suggested a certain degree of redundancy among the putative receptor genes in such a way that different receptors would facilitate the uptake of a single siderophore (11), as is the case for the siderophores pyoverdine and enterobactin (21, 22).
The described FoxIRA and FiuIRA surface signaling systems are not the first examples of siderophore-mediated regulation in P. aeruginosa. The ferri-enterobactin receptor gene (pfeA) is not only repressed by Fur in the presence of iron but is also subject to positive control by the enterobactin-responsive PfeR-PfeS two-component regulatory system (13). The involvement of alternative sigma factors in the regulation of pyoverdine uptake has also been reported (6, 10, 32). In fact, analysis of the P. aeruginosa genome sequence revealed the presence of 10 other ECF sigma factors associated with an anti-sigma-like factor (56). The involvement of these regulatory systems in the expression of siderophore receptors provides an additional level of regulation, enabling P. aeruginosa to respond to the presence of siderophores in the environment as well as to levels of intracellular iron. This seems to be an important issue for environmental bacteria, as other species of Pseudomonas also contain large numbers of ECF sigma factors (37, 56). In agreement with that, a genome-wide analysis of TonB-dependent receptors involved in regulatory pathways has indicated that these signaling systems are commonly found in several environmental bacteria but are only seldom present in dedicated human and animal pathogens (30).
Acknowledgments
M. Llamas acknowledges financial support from the EU through a Marie Curie postdoctoral fellowship (contract no. MCFI-2002-01109).
We thank M. A. Jacobs, C. Manoil, and M. Olson for providing us with the P. aeruginosa mutants from the University of Washington Genome Center P. aeruginosa PAO1 transposon mutant library and P. Cornelis for the P. aeruginosa PAO1 pvdD pchEF and pvdD pchEF PA0470 mutant strains.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63-67. [DOI] [PubMed] [Google Scholar]
- 2.Ankenbauer, R. G. 1992. Cloning of the outer membrane high-affinity Fe(III)-pyochelin receptor of Pseudomonas aeruginosa. J. Bacteriol. 174:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aso, H., S. Miyoshi, H. Nakao, K. Okamoto, and S. Yamamoto. 2002. Induction of an outer membrane protein of 78 kDa in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. FEMS Microbiol. Lett. 212:65-70. [DOI] [PubMed] [Google Scholar]
- 4.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 102:11076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler, A. J., and K. Hantke. 1992. Ferrioxamine uptake in Yersinia enterocolitica: characterization of the receptor protein FoxA. Mol. Microbiol. 6:1309-1321. [DOI] [PubMed] [Google Scholar]
- 6.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 7.Bendtsen, J., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bitter, W., J. D. Marugg, L. A. de Weger, J. Tommassen, and P. J. Weisbeek. 1991. The ferric-pseudobactin receptor PupA of Pseudomonas putida WCS358: homology to TonB-dependent Escherichia coli receptors and specificity of the protein. Mol. Microbiol. 5:647-655. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V. 1997. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch. Microbiol. 167:325-331. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 12.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, C. R., S. Neshat, and K. Poole. 1996. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, C. R., and K. Poole. 1993. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellagi, A., D. Reis, B. Vian, and D. Expert. 1999. Expression of the ferrioxamine receptor gene of Erwinia amylovora CFBP 1430 during pathogenesis. Mol. Plant-Microbe Interact. 12:463-466. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 17.de Weger, L. A., R. van Boxtel, B. van der Burg, R. A. Gruters, F. P. Geels, B. Schippers, and B. Lugtenberg. 1986. Siderophores and outer membrane proteins of antagonistic, plant-growth-stimulating, root-colonizing Pseudomonas spp. J. Bacteriol. 165:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fecker, L., and V. Braun. 1983. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J. Bacteriol. 156:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fürste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 21.Ghysels, B., B. T. Dieu, S. A. Beatson, J. P. Pirnay, U. A. Ochsner, M. L. Vasil, and P. Cornelis. 2004. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150:1671-1680. [DOI] [PubMed] [Google Scholar]
- 22.Ghysels, B., U. Ochsner, U. Mollman, L. Heinisch, M. Vasil, P. Cornelis, and S. Matthijs. 2005. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett. 246:167-174. [DOI] [PubMed] [Google Scholar]
- 23.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 24.Haas, D., and B. W. Holloway. 1976. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol. Gen. Genet. 144:243-251. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 26.Heim, S., M. Ferrer, H. Heuer, D. Regenhardt, M. Nimtz, and K. N. Timmis. 2003. Proteome reference map of Pseudomonas putida strain KT2440 for genome expression profiling: distinct responses of KT2440 and Pseudomonas aeruginosa strain PAO1 to iron deprivation and a new form of superoxide dismutase. Environ. Microbiol. 5:1257-1269. [DOI] [PubMed] [Google Scholar]
- 27.Heinrichs, D. E., L. Young, and K. Poole. 1991. Pyochelin-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. Infect. Immun. 59:3680-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kingsley, R. A., R. Reissbrodt, W. Rabsch, J. M. Ketley, R. M. Tsolis, P. Everest, G. Dougan, A. J. Baumler, M. Roberts, and P. H. Williams. 1999. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol. 65:1610-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 31.Koster, M., J. van de Vossenberg, J. Leong, and P. J. Weisbeek. 1993. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol. Microbiol. 8:591-601. [DOI] [PubMed] [Google Scholar]
- 32.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leoni, L., N. Orsi, V. de Lorenzo, and P. Visca. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182:1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahren, S., S. Enz, and V. Braun. 2002. Functional interaction of region 4 of the extracytoplasmic function sigma factor FecI with the cytoplasmic portion of the FecR transmembrane protein of the Escherichia coli ferric citrate transport system. J. Bacteriol. 184:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Bueno, M. A., R. Tobes, M. Rey, and J. L. Ramos. 2002. Detection of multiple extracytoplasmic function (ECF) sigma factors in the genome of Pseudomonas putida KT2440 and their counterparts in Pseudomonas aeruginosa PA01. Environ. Microbiol. 4:842-855. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, M., C. J. Carrano, and P. J. Szaniszlo. 1992. Identification of the ferrioxamine B receptor, FoxB, in Escherichia coli K12. Biometals 5:37-46. [DOI] [PubMed] [Google Scholar]
- 41.Nelson, M., and P. J. Szaniszlo. 1992. TonB-independent ferrioxamine B-mediated iron transport in Escherichia coli K12. FEMS Microbiol. Lett. 79:191-196. [DOI] [PubMed] [Google Scholar]
- 42.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 43.Ochs, M., S. Veitinger, I. Kim, D. Welz, A. Angerer, and V. Braun. 1995. Regulation of citrate-dependent iron transport of Escherichia coli: fecR is required for transcription activation by FecI. Mol. Microbiol. 15:119-132. [DOI] [PubMed] [Google Scholar]
- 44.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 45.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 47.Palma, M., S. Worgall, and L. E. Quadri. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180:374-379. [DOI] [PubMed] [Google Scholar]
- 48.Postle, K. 1999. Active transport by customized beta-barrels. Nat. Struct. Biol. 6:3-6. [DOI] [PubMed] [Google Scholar]
- 49.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauer, M., K. Hantke, and V. Braun. 1990. Sequence of the fhuE outer-membrane receptor gene of Escherichia coli K12 and properties of mutants. Mol. Microbiol. 4:427-437. [DOI] [PubMed] [Google Scholar]
- 53.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 54.Tanabe, T., N. Takata, A. Naka, Y. H. Moon, H. Nakao, Y. Inoue, S. Narimatsu, and S. Yamamoto. 2005. Identification of an AraC-like regulator gene required for induction of the 78-kDa ferrioxamine B receptor in Vibrio vulnificus. FEMS Microbiol. Lett. 249:309-314. [DOI] [PubMed] [Google Scholar]
- 55.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 56.Visca, P., L. Leoni, M. J. Wilson, and I. L. Lamont. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177-1190. [DOI] [PubMed] [Google Scholar]
- 57.Yorgey, P., L. G. Rahme, M. W. Tan, and F. M. Ausubel. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol. Microbiol. 41:1063-1076. [DOI] [PubMed] [Google Scholar]