Abstract
The MexXY components of the MexXY-OprM multidrug efflux system of Pseudomonas aeruginosa are encoded by a MexZ repressor-regulated operon that is inducible by antibiotics that target the ribosome. Mutant strains disrupted in a gene, PA5471, were shown to be compromised for drug-inducible mexXY expression and, therefore, MexXY-OprM-mediated antimicrobial resistance. The PA5471 gene was inducible by the same ribosome-targeting agents that induce mexXY expression. Moreover, vector-driven expression of cloned PA5471 was sufficient to promote mexXY expression and MexXY-mediated resistance in the absence of antibiotic exposure, consistent with PA5471 directly or indirectly activating mexXY expression following its own upregulation in response to antibiotics. The requirement for PA5471 for mexXY expression and antimicrobial resistance was, however, obviated in mutants lacking the MexZ repressor of mexXY expression, suggesting that PA5471 directly or indirectly modulates MexZ activity in effecting mexXY expression. While the recruitment of PA5471 and MexXY in response to ribosome disruption by antimicrobials is consistent with their genes playing a role in protecting cells from the adverse consequences of disrupting the translation process, reminiscent of trans-translation, these genes appear to operate independently in their contribution to resistance: mutants defective in trans-translation showed a much more modest (twofold) decrease in resistance to ribosome-targeting agents than those lacking PA5471 or MexXY, and this decrease was observed whether functional PA5471/MexXY was present or not.
Multidrug efflux systems of the resistance-nodulation-division (RND) family contribute significantly to intrinsic and acquired resistance to antimicrobials in a number of gram-negative bacteria (43, 45). Despite their significance as determinants of antibiotic resistance, however, RND-type multidrug exporters also, in many instances, accommodate biocides (42, 45), organic solvents (48), detergents (43), including bile salts (9, 18, 46, 60), toxic fatty acids/lipids (54), and in some instances, plant-derived antimicrobials (phytoalexins and isoflavonoids) (7, 39), metabolic inhibitors (52), organometallic compounds (tributyltin) (25), quorum-sensing effector molecules (13, 26, 40), and, possibly, virulence factors (21) in addition to antibiotics. Clearly, RND pumps can and do function as other than antibiotic exporters.
Pseudomonas aeruginosa expresses several three-component RND-type multidrug efflux systems, among which four, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM, are reported to be significant determinants of multidrug resistance in lab and clinical isolates (41, 44). A clear indication, however, that antimicrobial export may not be the intended function of many of these systems comes from the observation that while these pumps accommodate many of the same antimicrobials, each appears to be independently regulated by linked regulatory genes (44), but not (with the exception of MexXY [30]) in response to antibiotics.
The MexXY-OprM system is unique in P. aeruginosa in that the mexXY operon is induced by exposure to many of the antibiotics that this efflux system exports (30). While this is consistent with efflux of these agents being the intended function of the MexXY-OprM system, it is interesting that not all antibiotic substrates, but only those agents known to target the ribosome, induce mexXY expression (23). Moreover, in contrast to other drug-inducible multidrug efflux systems (e.g., QacA, an MF family exporter in Staphylococcus aureus), where drug binding to the cognate regulator (i.e., QacR) alleviates repression of the efflux gene (i.e., qacA) (16), providing some support for these systems as intended determinants of drug efflux, MexXY antimicrobial substrates that induce mexXY expression do not interact with or directly modulate the activity of the mexXY repressor, MexZ (32). Also, the observation that ribosome protection mechanisms compromise drug-inducible mexXY expression (23) supports this efflux system being recruited in response to ribosome disruption and not to antibiotics per se. One possibility is that the action of these agents on their ribosomal targets induces the expression of MexXY-OprM in order to counter/alleviate some stress or adverse effect resultant from ribosome disruption. Certainly, transcriptomic and proteomic studies confirm that agents that interfere with prokaryotic translation impact the expression of a myriad of genes (1, 5, 14, 27, 38, 47, 50, 55), in some instances including genes associated with stress responses (27, 38, 47, 50). In an effort to define MexXY's role in P. aeruginosa's response to translation inhibition, attempts were made to identify additional genes involved in MexXY-dependent antibiotic resistance by screening a transposon insertion mutant library for mutants compromised for resistance to MexXY substrate antibiotics. We report here the identification of a gene, PA5471, which, like mexXY, is drug inducible and is required for drug-inducible mexXY expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. Bacterial cells were cultured in Luria broth (L broth) and on Luria agar (8) with antibiotics, as necessary, at 37°C. Plasmid pEX18Tc and its derivatives were maintained in Escherichia coli with 10 μg/ml of tetracycline. Plasmids pUCP20T and pMMB190 and their derivatives were maintained in E. coli with 100 μg/ml ampicillin and in P. aeruginosa PAO1 strain K767 and its derivatives with 200 μg/ml carbenicillin.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 F− Δ(lacZYA-argF)U169 | 4 |
| S17-1 | thi pro hsdR recA Tra+ | 56 |
| KAM3 | ΔacrB Δ(lac-pro) supE thi hsdΔ5/F′ traD36 proA+B+lacIqlacZ ΔM15 | 36 |
| P. aeruginosa strains | ||
| K767 | PAO1, wild type | 29 |
| K1525 | K767 ΔmexXY | 58 |
| K2439 | K1525 ΔssrA | This study |
| K2440 | K1525 ΔsmpB | This study |
| K2413 | K767 ΔPA5471 | This study |
| K2414 | K767 ΔmexXY ΔPA5471 | This study |
| K2415 | K767 ΔmexZ | This study |
| K2416 | K767 ΔmexZ ΔPA5471 | This study |
| K2417 | K767 ΔPA5471-PA5470 | This study |
| K2437 | K767 ΔssrA | This study |
| K2438 | K767 ΔsmpB | This study |
| K2162 | Clinical isolate displaying MexXY-dependent pan-aminoglycoside resistance | 58 |
| K2418 | K2162 ΔPA5471 | This study |
| K2435 | K2162 PA5471::mini-Tn5-tet | This study |
| K2436 | K2162 PA5471::mini-Tn5-tet | This study |
| YM34 | PAO1 ΔmexAB ΔmexCD-oprJ ΔmexEF-oprN | 37 |
| YM44 | YM34 ΔmexXY | 37 |
| Plasmids | ||
| pEX18Tc | Broad-host-range gene replacement vector; sacB Tcr | 22 |
| pYM008 | pEX18Tc::ΔPA5471 | This study |
| pYM015 | pEX18Tc::ΔPA5471-PA5470 | This study |
| pYM021 | pEX18Tc::ΔmexZ | This study |
| pYM022 | pEX18Tc::ΔssrA | This study |
| pYM023 | pEX18Tc::ΔsmpB | This study |
| pUCP20T | Broad-host-range cloning vector; Mob+ Apr Cbr | 53 |
| pYM010 | pUCP20T::PA5471 | This study |
| pYM013 | pUCP20T::PA5471-PA5470 | This study |
| pYM017 | pUCP20T::mexZ | This study |
| pBC KS(+) | Phagemid cloning vector; Cmr | Stratagene |
| pTEM4 | pBR322::mexXY; Apr | 35 |
| pMMB190 | Broad-host-range, low-copy-number cloning vector; lacIq Apr Cbr | 34 |
| pYM004 | pMMB190::mexXY | This study |
Tcr, tetracycline resistance; Apr, ampicillin resistance; Cbr, carbenicillin resistance; Cmr, chloramphenicol resistance.
DNA methods.
Standard protocols were generally used for restriction endonuclease digestion, ligation, transformation, plasmid isolation, and agarose gel electrophoresis, as described by Sambrook and Russell (51). Plasmid DNAs were also prepared from E. coli or P. aeruginosa using a QIAprep Spin miniprep kit or QIAfilter Plasmid Midi kit (QIAGEN Inc., Mississauga, Ontario, Canada) according to the protocols provided by the manufacturer. Genomic DNA of P. aeruginosa was extracted following the protocol of Barcak et al. (6). DNA fragments used for cloning were extracted from agarose gels using a QIAquick gel extraction kit (QIAGEN). PCR products were purified using a QIAquick PCR purification kit (QIAGEN) and, when cloned, sequenced to verify that no mutations were introduced during PCR. Competent P. aeruginosa (10) and E. coli (51) cells were prepared as described previously. Chromosomal DNA flanking the mini-Tn5-tet element in aminoglycoside-susceptible K2162 insertion mutants was sequenced using the primer mini-Tn5-Right (8). Oligonucleotide synthesis was carried out by Cortec DNA Services (Kingston, Ontario, Canada), and nucleotide sequencing was carried out by ACGT Corp. (Toronto, Ontario, Canada). Once the flanking DNA sequences were obtained, disrupted genes were identified by BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) searches of the available P. aeruginosa genome sequence (59; http://www.pseudomonas.com).
Transposon insertion mutagenesis.
P. aeruginosa strain K2162, a MexXY-expressing, pan-aminoglycoside-resistant clinical isolate of P. aeruginosa (Table 1), was mutagenized with mini-Tn5-tet (12) as described previously (8), with mini-Tn5-tet-carrying K2162 mutants selected on L agar containing tetracycline (64 μg/ml) and imipenem (0.5 μg/ml; to counterselect donor E. coli used to mobilize plasmid-borne mini-Tn5-tet into K2162). Mutants showing increased aminoglycoside susceptibilities were screened initially for lack of growth on L agar containing either paromomycin (1,024 μg/ml) or spectinomycin (256 μg/ml) and later for increased susceptibilities to multiple aminoglycosides using a broth assay (see below).
RT-PCR.
Total bacterial RNAs were isolated from log-phase P. aeruginosa L broth cultures (with and without subinhibitory concentrations of antibiotics, as follows: kanamycin, cefotaxime, and norfloxacin at one-fourth the MIC and erythromycin, tetracycline, and chloramphenicol at one-eighth the MIC), using a QIAGEN RNeasy mini kit, RNase-free DNase (QIAGEN), and a protocol provided by the manufacturer. Reverse transcriptase PCR (RT-PCR) was performed with ca. 500 ng RNA and primer pairs internal to rpoD, mexX, PA5471, PA5470, and mexZ (Table 2), using a QIAGEN One Step RT-PCR kit according to a protocol provided by the manufacturer. To assess whether PA5470 and PA5471 were expressed from a polycistronic message, the primer pair PA5471-F and PA5470-R (Table 2) was used. RT-free (i.e., PCR) controls were carried out to ensure that there was no DNA contamination of RNA preparations.
TABLE 2.
Oligonucleotides used for this study
| Primer | Sequence (5′→3′)a | Purpose |
|---|---|---|
| RT-rpoD-F | GATCCGGAACAGGTGGAAGAC | RT-PCR of rpoD |
| RT-rpoD-R | TCAGCAGTTCCACGGTACCC | RT-PCR of rpoD |
| RT-mexX-F | CATCAGCGAACGCGAGTACAC | RT-PCR of mexX |
| RT-mexX-R | CAATTCGCGATGCGGATTG | RT-PCR of mexX |
| RT-PA5471-F | ACAGCACCTGGATCGAAGGC | RT-PCR of PA5471 |
| RT-PA5471-R | TTCGATGCAGTCGCTCCAG | RT-PCR of PA5471 |
| RT-PA5470-F | GATCCTGCTGCAACTCTCCG | RT-PCR of PA5470 |
| RT-PA5470-R | ACCAGTTCTTGCGCGCAT | RT-PCR of PA5470 |
| RT-mexZ-F | AAACCCGCGACGGCATACT | RT-PCR of mexZ |
| RT-mexZ-R | ACTGGCGGAGAAAGCCCAT | RT-PCR of mexZ |
| E-PA5471-F | GCTAGAATTCGATCTACCGTTTCAATCACATGGAT; EcoRI | PCR cloning of PA5471 |
| X-PA5471-R | GATCTCTAGAGGCCACCTCCTCGATTACCT; XbaI | PCR cloning of PA5471 |
| EH-mexX-F | GAATTCAAGCTTCAAGCTCGCGAGTTCACGA; EcoRI, HindIII | PCR cloning of mexXY |
| N-mexX-R | ACGTTGGACGAGGCGATCTC | PCR cloning of mexXY |
| mexZ-F | TCGTGAACTCGCGAGCTTG | PCR cloning of mexZ |
| mexZ-R | CACATCAGCGAGGAAGACGC | PCR cloning of mexZ |
| PA5471DU-F | GATCAAGCTTCCTGGGAAGGCTATACCAACGb | Deletion of PA5471; upstream fragment |
| PA5471DU-R | GCTAGGTACCGCCCATAATCCAATCCATGTGb; KpnI | Deletion of PA5471; upstream fragment |
| PA5471DD-F | GCTAGGTACCCGGAAGCCGGTGCTGACCTACA; KpnI | Deletion of PA5471; downstream fragment |
| PA5471DD-R | GCTAGAATTCGCTTCATCGGCACCATCAT; EcoRI | Deletion of PA5471; downstream fragment |
| XPA5470-F | GATCTCTAGATGCTCGACATCAACCACAACC | Deletion of PA5471-PA5470 |
| PA5470-DDR | GCTAGAATTCCCAACGACGCCTTCTACTAC; EcoRI | Deletion of PA5471-PA5470 |
| mexZDU-F | GATCAAGCTTAATTCGCGATGCGGATTG; HindIII | Deletion of mexZ; upstream fragment |
| mexZDU-R | GATCTCTAGACACTGAACGTCCTCACAAGGG; XbaI | Deletion of mexZ; upstream fragment |
| mexZDD-F | GATCTCTAGACGCAGTTCTCCCTCCTGTTG; XbaI | Deletion of mexZ; downstream fragment |
| mexZDD-R | GCTAGAATTCGAAGGAAATCTTGGTGGCGA; EcoRI | Deletion of mexZ; downstream fragment |
| PA0826.2DU-F | GATCAAGCTTTCGAATACCGCCTGCAAGC; HindIII | Deletion of ssrA; upstream fragment |
| PA0826.2DU-R | CTAGTCTAGACCGGCGTCGAATCCTAATC; XbaI | Deletion of ssrA; upstream fragment |
| PA0826.2DD-F | CTAGTCTAGAGCATGTAGAACCGATAGCGGA; XbaI | Deletion of ssrA; downstream fragment |
| PA0826.2DD-R | GCTAGGTACCCAGTCCTTCCTGGCGGCTAT; KpnI | Deletion of ssrA; downstream fragment |
| PA4768DU-F | GCTAGAATTCGCGGTAGATCTCCACCCGTT; EcoRI | Deletion of smpB; upstream fragment |
| PA4768DU-R | GATCTCTAGAGACCATAGGCGGCGCATTATAG; XbaI | Deletion of smpB; upstream fragment |
| PA4768DD-F | GATCTCTAGAGACTTCGACAAGCGCCACAC; XbaI | Deletion of smpB; downstream fragment |
| PA4768DD-R | GATCAAGCTTAGAGGCTTTGCGACGAAACTT; HindIII | Deletion of smpB; downstream fragment |
In some instances, restriction sites were introduced into oligonucleotides to be used for PCR, and these are underlined in the sequences, with the corresponding restriction endonucleases indicated.
The PCR product amplified with these primers and cloned into pCR-BluntII-TOPO was excised following PstI-KpnI digestion (a PstI site is present within the pCR-BluntII-TOPO multicloning site) prior to cloning into pEX18Tc.
Cloning of PA5471 and PA5470.
The PA5471 gene was amplified using primers EPA5471-F and XPA5471-R (Table 2) in a 50-μl PCR mixture containing 10 ng of chromosomal DNA, a 0.6 μM concentration of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1 mM MgSO4, 1 U of KOD Hot Start DNA polymerase (EMD Biosciences, Inc., Madison, WI), 1× KOD Hot Start polymerase buffer, and 4.0% (vol/vol) dimethyl sulfoxide. The mixture was heated for 2 min at 94°C, followed by 35 cycles of 0.25 min at 94°C, 0.5 min at 60°C, and 2 min at 68°C and a final step of 10 min at 68°C. The PA5471-containing PCR product was cloned into pSportI (Invitrogen, Carlsbad, CA), released by digestion with EcoRI and BamHI, and cloned into pUCP20T to yield pYM010. To clone the PA5471-PA5470 operon into pUCP20T, PA5470 was excised from pCR-PA5470+DD (see below) following digestion with XbaI and BamHI, cloned into pSportI, and subsequently released from this vector by BsiWI-BamHI digestion. This fragment and a PA5471-containing fragment released from pSportI (see above) following digestion with EcoRI and BsiWI were then jointly cloned into pUCP20T to yield pYM013.
Cloning of mexXY.
The mexX gene was amplified with primers EHmexX-F and NmexX-R (Table 2) from plasmid pTEM4 (1 ng). The reaction mixture was formulated as described above for the amplification of PA5471, using the same parameters. The blunt-ended PCR fragment was first cloned into plasmid pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA) before being released as an EcoRI-NotI fragment and cloned into pSportI. The mexY gene was subsequently excised from pTEM4 via digestion with NotI and BamHI and cloned into mexX-carrying pSportI, after which the mexXY gene pair was released via digestion with EcoRI and cloned into pMMB190 to yield pYM004.
Cloning of mexZ.
The mexZ gene was amplified with primers mexZ-F and mexZ-R (Table 2) in a reaction mixture formulated as described above for PA5471, with the exception that dimethyl sulfoxide was included at 4.0% (vol/vol) and Vent DNA polymerase (2 U; New England Biolabs, Ltd., Pickering, Ontario, Canada) in 1× ThermoPol buffer (New England Biolabs, Ltd., Pickering, Ontario, Canada) replaced the KOD enzyme. Reaction mixtures were heated for 3 min at 94°C, followed by 35 cycles of 0.5 min at 94°C, 0.75 min at 65°C, and 1 min at 72°C and a final 10-min elongation at 72°C. The mexZ-containing PCR product was first cloned into pCR-BluntII-TOPO as described above, excised from this vector using HindIII and XbaI, and then cloned into pUCP20T, yielding pYM017.
Construction of gene deletions in P. aeruginosa.
To introduce in-frame gene deletions into strains of P. aeruginosa, deletion constructs were first prepared in plasmid pEX18Tc by cloning PCR-amplified 1-kb DNA fragments corresponding to the regions upstream and downstream of the gene sequences to be deleted. Typically, these were amplified from the chromosome of P. aeruginosa PAO1 strain K767 and first cloned individually into pCR-BluntII-TOPO, from which they were sequenced to verify the absence of PCR-introduced mutations before being excised following restriction digestion (PCR primers were tagged with restriction sites [Table 2]) and sequentially cloned into pEX18Tc. While the upstream fragment used for construction of the PA5471 deletion served for construction of a ΔPA5471-PA5470 double deletion, attempts to amplify sequences 3′ of PA5470 failed to yield a correct product. Thus, the PA5470 gene together with sequences ca. 1 kb downstream of it were amplified using primers XPA5470-F and PA5470DD-R (Table 2) and cloned into pCR-BluntII-TOPO, and the PA5470 downstream fragment was then excised from this vector (pCR-PA5470+DD) via digestion with KpnI and EcoRI for cloning into pEX18Tc. PCR mixtures were formulated as described above for the amplification of mexZ and heated for 3 min at 95°C, followed by 35 (ΔPA5471, ΔPA547-PA5470, and ΔmexZ) or 40 (ΔssrA and ΔsmpB) cycles of 0.5 min at 95°C, 0.75 min at 60°C, and 2 min at 72°C and a final step of 10 min at 72°C. The resulting deletions lacked all but the first 2 (including the ATG start) and last 14 codons (ΔPA5471), all but the GTG start codon and the last 11 codons (ΔmexZ), all but the first 25 and last 61 bp (tmRNA gene ssrA), and all but the start codon and the last 27 codons (ΔsmpB). The ΔPA547-PA5470 construct lacked all but the first two codons of PA5471 and all of the PA5470 codons.
The deletion-carrying pEX18Tc derivatives were mobilized into P. aeruginosa from E. coli S17-1 (8). Briefly, 100 μl of log-phase E. coli S17-1 cultured in tetracycline-containing (10 μg/ml) L broth was transferred to L agar plates and immediately overlaid with an equal volume of a log-phase L broth culture of P. aeruginosa. Following incubation at 37°C for 18 h, the bacterial cells were resuspended in 1 ml of 0.85% NaCl and diluted 10-fold before being plated onto L agar plates containing tetracycline (75 μg/ml) and chloramphenicol (5 μg/ml; to counterselect E. coli S17-1). P. aeruginosa transconjugants harboring chromosomal inserts of the plasmid were recovered from these plates and streaked onto L agar containing sucrose (10% [wt/vol]). Sucrose-resistant colonies were then screened for the appropriate deletion using colony PCR (49).
Antimicrobial susceptibility testing.
The antimicrobial susceptibilities of the various P. aeruginosa strains were assessed in microtiter plates by a twofold serial dilution technique (24). In some experiments, MgCl2 was included in the growth medium (5 mM) since this appears to enhance MexXY-mediated antimicrobial resistance (28).
RESULTS
Involvement of PA5471 in MexXY-mediated antimicrobial resistance.
Inducible (by antibiotics) (23, 30) and mutational (23, 58, 64) up-regulation of MexXY is associated with resistance to multiple antimicrobials in P. aeruginosa, although the details of MexXY expression in each instance remain obscure. A recent paper, however, highlights the significance of drug-ribosome interactions in ultimately stimulating mexXY expression (23), a finding consistent with earlier observations that while MexXY accommodates and thus provides resistance to a variety of antimicrobials (30, 31), only those targeting the ribosome (e.g., aminoglycosides) actually induce mexXY expression (23). To gain some insights into the details of drug-inducible mexXY expression, including the identity of any additional gene(s) needed for this, a P. aeruginosa clinical strain in which MexXY is expressed and implicated in antimicrobial resistance (i.e., K2162) (58) was subjected to random transposon insertion mutagenesis (with mini-Tn5-tet) and screened for a loss of resistance to representative MexXY antimicrobial substrates. A library of mini-Tn5-tet mutants of K2162 was thus constructed, and mutants initially showing enhanced susceptibility to the aminoglycosides paromomycin and streptomycin (good MexXY substrates [58]) were selected. Subsequent screening for increased susceptibilities to the aminoglycosides spectinomycin and gentamicin, but not to antimicrobials known not to be MexXY substrates (e.g., carbenicillin and imipenem), identified seven mutants with generalized increased susceptibilities to aminoglycosides. Of these, only two (K2435 and K2436; see Table 3, for susceptibility data) lacked mini-Tn5-tet insertions in mexXY. Cloning and sequencing of the disrupted genes in each instance revealed that the mini-Tn5-tet element had inserted in the putative promoter region of (K2435) or within (K2436) an opening reading frame dubbed PA5471 by the Pseudomonas Genome Project (http://www.pseudomonas.com). PA5471 encodes a predicted product of 43,508 Da identified as a conserved hypothetical protein and a member of the UPF0027 uncharacterized protein family (Protein Families Database of Alignments and HMMS accession number pfam01139 [http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF01139]), which has numerous members broadly distributed among bacteria (gram-positive and gram-negative) and archaea.
TABLE 3.
Involvement of PA5471 in antimicrobial resistance in P. aeruginosa
| Strain | Relevant phenotype | MIC (μg/ml)a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ami | Gen | Kan | Neo | Par | Spc | Str | Tob | Cef | Cam | Car | Ery | Nor | Tet | ||
| K2162 | WT | 128 | 256 | 1,024 | 512 | >2,048 | 1,024 | 256 | 128 | 64 | 256 | 512 | 512 | 1 | 16 |
| K2435 | PA5471− | — | 32 | — | — | 256 | 128 | 32 | — | — | — | 512 | — | — | — |
| K2436 | PA5471− | — | 32 | — | — | 256 | 128 | 32 | — | — | — | 512 | — | — | — |
| K2418 | PA5471− | 32 | 64 | 512 | 256 | 512 | 128 | 64 | 64 | 64 | 256 | 512 | 512 | 1 | 16 |
| K767 | WT | 4 | 8 | 128 | 64 | 256 | 512 | 32 | 4 | 16 | 32 | 32 | 512 | 0.5 | 16 |
| K2413 | PA5471− | 2 | 4 | 64 | 64 | 64 | 128 | 8 | 4 | 16 | 32 | 32 | 128 | 0.5 | 8 |
| K1525 | MexXY− | 2 | 4 | 64 | 32 | 32 | 64 | 4 | 4 | 16 | 32 | 32 | 64 | 0.5 | 8 |
| K2414 | PA5471− MexXY− | 2 | 4 | 64 | 32 | 32 | 64 | 4 | 4 | 16 | 32 | 32 | 64 | 0.5 | 8 |
| K2413(pUCP20T) | PA5471− | 2 | 4 | 64 | 64 | 64 | 128 | 8 | 4 | 8 | 32 | — | 128 | 0.5 | 8 |
| K2413(pYM010) | PA5471+ | 8 | 8 | 128 | 128 | 512 | 512 | 64 | 4 | 8 | 32 | — | 512 | 1 | 8 |
| K2413(pMMB190) | PA5471− | 2 | 4 | 64 | 64 | 64 | 128 | 8 | 4 | 8 | 32 | — | 128 | 0.5 | 8 |
| K2413(pYM004) | PA5471− MexXY++ | 8 | 8 | 128 | 128 | 512 | 512 | 64 | 4 | 8 | 32 | — | 512 | 1 | 8 |
| K2417(pUCP20T) | PA5471− PA5470− | 2 | 2 | 64 | 64 | 64 | 128 | 8 | 2 | 16 | 32 | — | 128 | 0.5 | 8 |
| K2417(pYM010) | PA5471+ PA5470− | 8 | 8 | 256 | 128 | 512 | 1,024 | 64 | 4 | 16 | 32 | — | 512 | 1 | 16 |
Ami, amikacin; Gen, gentamicin; Kan, kanamycin; Neo, neomycin; Par, paromomycin; Spc, spectinomycin; Str, streptomycin; Tob, tobramycin; Cef, cefotaxime; Cam, chloramphenicol; Car, carbenicillin; Ery, erythromycin; Nor, norfloxacin; Tet, tetracycline. —, not determined.
The creation of an in-frame deletion of PA5471 in K2162 also compromised resistance to a variety of aminoglycosides, but not to MexXY antimicrobial substrates known not to induce this efflux system (e.g., cefotaxime and carbenicillin; see strain K2418 in Table 3), confirming the contribution of this gene to pan-aminoglycoside resistance. The elimination of PA5471 in wild-type PAO1 strain K767 also increased the susceptibilities to aminoglycosides as well as other ribosome-targeting agents, such as erythromycin and tetracycline (see strain K2413 in Table 3), reminiscent of the impact of a mexXY deletion on resistance in this strain (see strain K1525 in Table 3). These data suggested that PA5471 plays a role in MexXY-mediated antimicrobial resistance, and consistent with this, deletion of PA5471 in strains already lacking MexXY had no impact on antimicrobial resistance (Table 3, compare K2414 with K1525). As expected, the cloned PA5471 gene (on plasmid pYM010) restored antimicrobial resistance in the PA5471 deletion strain K2413 (Table 3).
Requirement for PA5471 for drug-inducible mexXY expression.
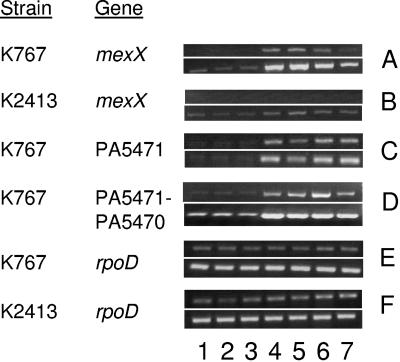
One way in which PA5471 could contribute to MexXY-mediated antimicrobial resistance is via involvement in the process of drug induction of mexXY expression. Indeed, the observation that expression of the cloned mexXY genes from a vector-borne promoter on plasmid pYM004 was sufficient to reverse the drug susceptibility of the PA5471 deletion in strain K2413 (Table 3) is consistent with PA5471 being required only for the expression of mexXY. To assess a contribution of PA5471 to drug-inducible expression of mexXY, the impact of PA5471 loss on mexX (as a measure of mexXY) gene expression was examined. As expected, agents that target the ribosome, including chloramphenicol, tetracycline, erythromycin, and kanamycin, induced the expression of mexXY in log-phase cells of P. aeruginosa strain K767 (Fig. 1A, cf. lanes 4 to 7 and lane 1), while those that do not (e.g., norfloxacin and cefotaxime, a fluoroquinolone and a β-lactam, respectively) did not (Fig. 1A, lanes 2 and 3). Elimination of PA5471 in K767, however, severely compromised drug-inducible mexXY expression (Fig. 1B, lanes 4 to 7; compare with Fig. 1A, lanes 4 to 7). The mexXY message was still detectable in these mutants (Fig. 1B, lower panel), at levels comparable to or minimally above that seen in cells not exposed to antibiotics (Fig. 1A and B, lanes 1), consistent with PA5471 having a specific involvement in drug induction of mexXY expression. The fact that the PA5471 mutant K2413 still expressed some mexXY was also consistent with observations that it was more resistant to some MexXY antimicrobials than was the ΔmexXY knockout K1525 (Table 3). As expected, given its involvement in drug induction of mexXY expression, the expression of PA5471 was stimulated in log-phase cells of K767 (Fig. 1C, cf. lanes 4 to 7 and lane 1) by the same ribosome-targeting antimicrobials that induce mexXY, but not by agents that do not induce mexXY expression (Fig. 1C, lanes 2 and 3).
FIG. 1.
Requirement for PA5471 for antibiotic-inducible mexXY expression. The expression of mexX, PA5471, and rpoD was assessed in P. aeruginosa strains K767 and K2413 (K767 ΔPA5471) grown without antibiotics (lane 1) or with norfloxacin (lane 2), cefotaxime (lane 3), chloramphenicol (lane 4), erythromycin (lane 5), tetracycline (lane 6), and kanamycin (lane 7) by semiquantitative RT-PCR. The rpoD reaction served as an internal control that ensured that equal amounts of RNA were employed in all of the RT-PCRs shown. The PCR portion of the reactions was carried out for 28 (top panel) and 30 (bottom panel) cycles for mexX, 19 (top panel) and 21 (bottom panel) cycles for PA5471 and rpoD, and 31 (top panel) and 33 (bottom panel) cycles for PA5471-PA5470. Data are representative of two or three replicates.
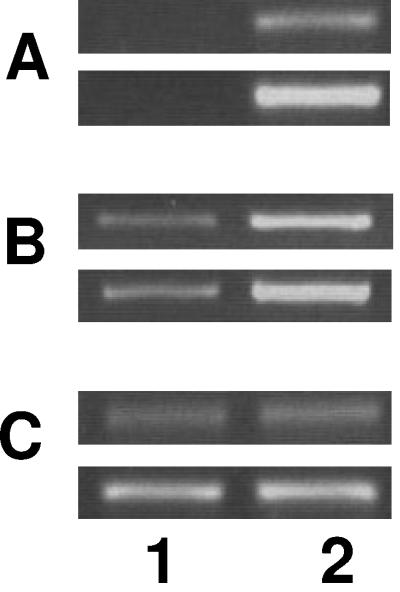
PA5471, but not PA5470, is required for mexXY expression.
Examination of the P. aeruginosa genome reveals that PA5471 occurs upstream of and in a possible operon with an open reading frame dubbed PA5470. PA5470 is predicted to encode a peptide chain release factor of 22,282 Da (http://www.pseudomonas.com). RT-PCR confirmed both the drug inducibility of PA5470 and its expression from a polycistronic message that also contains PA5471 (Fig. 1D, lanes 4 to 7). Still, the observation that resistance to MexXY antimicrobial substrates was restored with the cloned PA5471 gene alone (on plasmid pYM010) in a PA5471-PA5470 double deletion mutant (K2417) (Table 3) suggested that PA5471 alone was needed for drug-inducible mexXY expression. Moreover, expression of the cloned PA5471 gene from a vector-borne promoter on plasmid pYM010 (Fig. 2A, lane 2) stimulated mexXY expression in strain K767 in the absence of antibiotic (Fig. 2B, cf. lane 2 and lane 1), indicating that drug induction of mexXY is a consequence, directly or indirectly, of PA5471 upregulation in response to antibiotic exposure. The cloned PA5471 gene also promoted resistance to norfloxacin, a noninducing MexXY substrate (Fig. 1A, lane 2), in MexXY+ (i.e., YM34) but not MexXY− (i.e., YM44) P. aeruginosa (Table 4), consistent with PA5471 positively affecting mexXY expression.
FIG. 2.
PA5471 stimulates mexXY expression in the absence of antibiotics. The expression of PA5471 (A), mexX (B), and rpoD (C) was assessed in P. aeruginosa strain K767 carrying pUCP20T (lane 1) or pYM010 (pUCP20T::PA5471), using semiquantitative RT-PCR. The rpoD reaction served as an internal control that ensured that equal amounts of RNA were employed in all of the RT-PCRs shown. The PCR portion of the reactions was carried out for 15 (top panel) and 17 (bottom panel) cycles for PA5471, 19 (top panel) and 21 (bottom panel) cycles for rpoD, and 30 (top panel) and 32 (bottom panel) cycles for mexX. Data are representative of two or three replicates.
TABLE 4.
Influence of plasmid-expressed PA5471 on norfloxacin resistance in P. aeruginosa
| Strain | Relevant property | MIC (μg/ml) of norfloxacina
|
|
|---|---|---|---|
| −Mg2+ | +Mg2+ | ||
| YM34(pUCP20T)b | MexXY+ | 0.25 | 2 |
| YM34(pYM010) | MexXY+ PA5471++c | 0.5 | 8 |
| YM44(pUCP20T) | MexXY− | 0.12 | 0.25 |
| YM44(pYM010) | MexXY− PA5471++c | 0.12 | 0.25 |
Norfloxacin MICs were determined in the presence and absence of 5 mM MgCl2. Mg2+ enhances MexXY-mediated antimicrobial resistance, possibly by enhancing the activity of this efflux system (28).
Lacks MexAB-OprM, MexCD-OprJ, and MexEF-OprN.
PA5471 was overexpressed from plasmid pYM010.
PA5471 acts via the MexZ repressor in mediating drug-inducible mexXY expression.
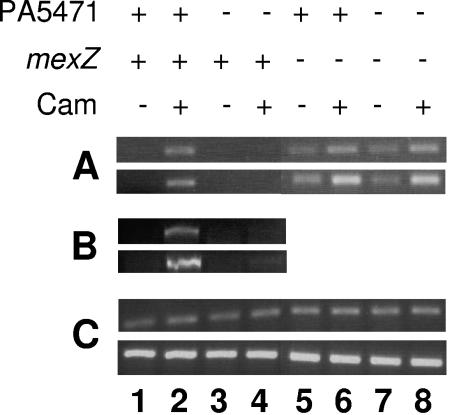
The gene mexZ occurs upstream of mexXY and encodes a repressor of mexXY expression (3, 32). While MexZ does not directly mediate the drug inducibility of mexXY (i.e., antibiotics do not bind to MexZ and modulate its repressor activity [32]), it may respond to PA5471 or the activity of this protein in ultimately effecting drug-inducible mexXY expression. To address this possibility, the impact of PA5471 loss on antimicrobial resistance and mexXY expression in a strain carrying a mexZ deletion was assessed. If PA5471 should act, directly or indirectly, to modulate MexZ repressor activity, such that PA5471 expression in response to antimicrobials leads to derepression of mexXY, then the loss of PA5471 in a mutant already lacking mexZ should have no adverse impact on mexXY expression or resistance. Conversely, and in light of previous observations that mexZ knockouts do not demonstrate maximal mexXY expression (23), if drug-inducible/PA5471-dependent mexXY expression is independent of MexZ, then the loss of PA5471 would compromise drug-inducible mexXY expression and thus resistance, even in a mexZ mutant expected to already demonstrate an increase in mexXY expression and resistance relative to its MexZ+ counterpart (i.e., increased mexXY expression in a mexZ knockout would not mask an additional contribution of PA5471 and thus a negative impact of PA5471 loss on mexXY expression and resistance). As expected, the loss of mexZ (in K767 derivative K2415) increased the resistance to multiple antimicrobials, though only modestly (Table 5), consistent with the increase in mexXY expression seen in this mutant even without antibiotic (e.g., chloramphenicol) induction (Fig. 3A, cf. lane 5 and lane 1). Indeed, without drug exposure, the mexZ deletion mutant K2415 expressed this efflux system at levels comparable to that seen for the drug-exposed MexZ+ parental strain K767 (Fig. 3A, compare lanes 2 and 5). In contrast to the adverse impact of a PA5471 deletion on the resistance of otherwise wild-type cells (Table 3), however, the loss of PA5471 in the mexZ deletion strain K2415 had no effect on resistance (Table 5, compare strains K2416 and K2415). Consistent with this, mexXY expression in the mexZ mutant strain was not adversely impacted by the loss of PA5471 (Fig. 3A, cf. lane 7 and lane 5).
TABLE 5.
Influence of mexZ on PA5471-dependent antimicrobial resistance
| Strain | Relevant genotype | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ami | Gen | Kan | Neo | Par | Spc | Str | Tob | Ery | Nor | Tet | ||
| K767 | 4 | 8 | 128 | 64 | 256 | 512 | 32 | — | 512 | 0.5 | 16 | |
| K2415 | ΔmexZ | 4 | 8 | 128 | 128 | 512 | 1,024 | 64 | — | 512 | 1 | 16 |
| K2416 | ΔmexZ ΔPA5471b | 4 | 8 | 128 | 128 | 512 | 1,024 | 64 | — | 512 | 1 | 16 |
Antibiotic abbreviations are defined in Table 3. —, not determined. The loss of PA5471 did not impact the MICs of these agents in K767 (Table 3), and thus the impact of PA5471 loss on the MICs of these agents was not studied here. Agents for which resistance was increased upon loss of mexZ are indicated in bold.
In contrast to the lack of any impact of PA5471 loss on resistance of the MexZ− strain K2415, the loss of PA5471 in the MexZ+ strain K767 compromised resistance to all agents listed here, except for neomycin and norfloxacin (Table 3, compare K2418 and K767).
FIG. 3.
Requirement for PA5471 for drug-inducible mexXY expression in MexZ+ but not MexZ− P. aeruginosa. The expression of mexX (A), mexZ (B), and rpoD (C) was assessed in strain K767 (PA5471+ MexZ+) and its derivatives K2413 (PA5471− MexZ+), K2415 (PA5471+ MexZ−), and K2416 (PA5471− MexZ−) grown without (−) and with (+) chloramphenicol (Cam; 8 μg/ml), using semiquantitative RT-PCR. The rpoD reaction served as an internal control that ensured that equal amounts of RNA were employed in all of the RT-PCRs shown. The PCR portion of the reactions was carried out for 28 (mexX), 40 (mexZ), or 20 (rpoD) cycles (top) and 30 (mexX), 42 (mexZ), or 22 (rpoD) cycles (bottom). Data are representative of two or three replicates.
Intriguingly, drug (chloramphenicol)-exposed K2415 (K767 ΔmexZ) still showed some increase in mexXY expression (Fig. 3A, cf. lane 6 and lane 5), and this drug inducibility of mexXY in the absence of mexZ was also not compromised by a subsequent loss of PA5471 (Fig. 3A, cf. lane 8 and lane 7). Clearly, then, PA5471 is required for drug-inducible mexXY expression only in strains expressing MexZ, consistent with it functioning to directly or indirectly modulate the activity of this repressor. In agreement with this, too, antibiotics or PA5471 did not adversely impact mexZ expression (data not shown), i.e., did not increase mexXY expression via a negative influence on mexZ expression, and thus must act at the level of MexZ activity. mexZ expression was, in fact, antibiotic (e.g., chloramphenicol) inducible (Fig. 3B, cf. lane 2 and lane 1), and this was dependent on PA5471 (Fig. 3B, lane 4), exactly mirroring the antibiotic and PA5471 dependence of mexXY expression. This is consistent with mexZ being subject to autoregulation, as for repressors of other multidrug efflux systems (41, 44), and given the low levels of mexZ mRNA detected (it took a minimum of 40 cycles to detect mexZ using RT-PCR), it is not inconsistent with antibiotics and/or PA5471 positively impacting mexXY expression via modulation of MexZ repressor activity.
PA5471-dependent MexXY operates independently of trans-translation.
Aminoglycosides and other ribosome-targeting agents promote mistranslation and stop codon readthrough, the latter of which results in ribosome stalling at the 3′ ends of mRNAs and thus in the depletion of free tRNAs and ribosomes needed for translation (2, 19, 61, 62). Stalled ribosomes are rescued in bacteria by a process known as trans-translation that requires a specialized RNA species termed tmRNA (which functions as both a tRNA and an mRNA) and a small accessory protein, SmpB (17, 65). To assess, then, whether MexXY functions as part of a trans-translation process in P. aeruginosa that serves to counter the adverse effects of ribosome-targeting antimicrobials, homologues of the tmRNA (i.e., ssrA, or PA0826.2) and smpB (PA4768) genes were disrupted in MexXY+ (K767) and MexXY− (K1525) strains, and the impact on antimicrobial resistance was assessed. The loss of ssrA or smpB had a modest (twofold decrease) but reproducible impact on resistance to aminoglycosides (amikacin, gentamicin, kanamycin, neomycin, paromomycin, and spectinomycin) and chloramphenicol in the MexXY+ K767 derivatives K2437 and K2438, respectively, and this was seen even in the absence of MexXY (in strains K2439 and K2440) (data not shown), consistent with tmRNA/SmpB and PA5471/MexXY operating independently of one another in promoting resistance to these agents. The observation, too, that the loss of ssrA or smpB in K767 did not adversely impact resistance to erythromycin (data not shown), while the loss of mexXY clearly did (Table 3, K1525), further supports these systems operating independently in P. aeruginosa, with PA5471/MexXY apparently playing no role in the process of trans-translation.
DISCUSSION
PA5471 is a member of a family of proteins (UPF0027) that are broadly conserved in bacteria and archaea, consistent with it playing a basic, housekeeping function in P. aeruginosa. Interestingly, however, a linkage of PA5471-like genes to a putative release factor gene is seen in a more limited number of organisms that include a variety of enterobacteria (Erwinia carotovora subsp. atroseptica SCRI1043, Salmonella enterica serovar Typhi, Salmonella enterica serovar Paratyphi, Salmonella enterica serovar Choleraesuis, Escherichia coli CFT073, Escherichia coli K-12, and Shigella flexneri 2a) and only three pseudomonads (Pseudomonas fluorescens Pf-5, Ralstonia solanacearum, and Burkholderia cepacia R18154). Whether this reflects a specific and unique function of PA5471 in these organisms or simply a lack of linkage of PA5470/PA5471 homologues in most bacteria harboring PA5471-like genes is unclear. Interestingly, a homologue of PA5471 from E. coli, ykfJ (b0235; GenBank accession numbers NP_414770 and CAH19161), was also shown to be inducible by an agent, 4-azaleucine, known to interfere with translation (50), and it too is linked to a putative peptide release factor gene (GenBank accession numbers NP_414771 and CAH19162).
Ribosome-targeting antibiotics, including those shown here to induce PA5471-PA5470 and mexXY expression, typically cause mistranslation and/or stop codon readthrough, leading to an accumulation of aberrant polypeptides or stalling of ribosomes at the 3′ ends of mRNAs (e.g., aminoglycosides and chloramphenicol [2, 19]), dissociation of incomplete peptidyl-tRNAs from the ribosome (e.g., macrolides [20]), or an accumulation of truncated peptidyl-tRNAs and ribosome stalling (e.g., tetracycline and chloramphenicol [61]). Stalled ribosomes pose a serious problem in that they deplete pools of free tRNAs and ribosomes, and the accumulation of peptidyl-tRNAs is toxic to cells (33). While genes for a trans-translation system implicated in the alleviation of drug-induced ribosome stalling were identified in P. aeruginosa and were shown here to contribute, albeit modestly, to aminoglycoside and chloramphenicol resistance, as for other organisms (e.g., E. coli [2] and Synechocystis sp. [11]), MexXY clearly does not participate in this process.
The PA5470 gene is present on a polycistronic message that also encodes PA5471, which is annotated as a peptide chain release factor and carries signature sequences of peptidyl-tRNA hydrolases (http://www.tigr.org/tigr-scripts/CMR2/GenePage.spl?db=ntpa03&locus=PA5470). Intriguingly, a peptidyl-tRNA hydrolase (Pth) in E. coli is responsible for recycling of peptidyl-tRNAs formed, for example, as a result of antibiotic action (57). One possibility, then, is that PA5470 participates in the release of aberrant peptides from peptidyl-tRNAs that accumulate in response to drug treatment. Still, this gene is dispensable with regards to MexXY recruitment and MexXY-mediated antibiotic resistance, arguing that while PA5470 and PA5471 (and thus MexXY) may function in a common process that is initiated by ribosome disruption, antibiotic resistance promoted by PA5471/MexXY is independent of this common function. Should PA5470 function in the release of aberrant peptides, PA5471 and MexXY (and possibly others) may play a role in downstream processing of these peptides or the recruitment of components responsible for this. In such a scenario, MexXY may play an intended role in export of these anomalous peptides or processed products thereof and, given the anticipated variation in amino acid sequence and composition of these components (drugs will be targeting ribosomes translating a myriad of different mRNAs and disrupting them at various stages of translation, producing a very heterogeneous mixture of aberrant peptidyl-tRNAs), may need to be flexible with regards to substrate recognition. Such flexibility might then explain the ability of MexXY-OprM to accommodate a diverse array of unintended antimicrobial substrates. Certainly, the observations that substantial mexXY expression has only a modest positive impact on antimicrobial resistance (Tables 4 and 5) and that the loss of this efflux mechanism only modestly increases susceptibilities to many antimicrobials (Table 3) suggest that antimicrobials are not the intended or preferred substrates. Consistent with this, too, a recent DNA array study demonstrated that both PA5471 and PA5470 are inducible (two- to threefold) under anaerobic conditions, in parallel with several ribosome-related genes, possibly due to some adverse impact of anaerobiosis on ribosome function (15; unpublished data).
While there are as yet no definitive clues to the function of PA5471 in P. aeruginosa or how it effects mexXY upregulation, directly or indirectly, it does not impact mexZ expression and thus clearly works to modulate the activity of the MexZ repressor of mexXY expression—the loss of PA5471 only compromises drug-inducible mexXY expression in MexZ+ and not MexZ− strains. The observation that antibiotics still enhance mexXY expression in a mexZ knockout mirrors previous results (23) and is consistent with the presence of additional pathways in P. aeruginosa by which mexXY can be unregulated in response to antibiotics. Mutations in a gene(s) other than mexZ (as yet unidentified) are, in fact, also associated with mexXY upregulation (63), although whether these play a role in drug-inducible mexXY expression independent of MexZ is unknown. In any case, these additional pathways must be masked by MexZ repressor activity, inasmuch as drug-inducible mexXY expression is not observed in MexZ+ PA5471 deletion mutants.
Acknowledgments
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation. M.L.S. was supported by a studentship from the Natural Sciences and Engineering Research Council (Canada).
REFERENCES
- 1.Aakra, A., H. Vebo, L. Snipen, H. Hirt, A. Aastveit, V. Kapur, G. Dunny, B. E. Murray, and I. F. Nes. 2005. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 49:2246-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abo, T., K. Ueda, T. Sunohara, K. Ogawa, and H. Aiba. 2002. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 7:629-638. [DOI] [PubMed] [Google Scholar]
- 3.Aires, J. R., T. Köhler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 7.Burse, A., H. Weingart, and M. S. Ullrich. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant-Microbe Interact. 17:43-54. [DOI] [PubMed] [Google Scholar]
- 8.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, A., S. Chaudhuri, G. Saha, S. Gupta, and R. Chowdhury. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 186:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. Benchtop and microcentrifuge preparation of Pseudomonas aeruginosa competent cells. BioTechniques 33:760-763. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz, J., and A. Vioque. 2001. Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA 7:1708-1716. [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum-sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evers, S., K. Di Padova, M. Meyer, H. Langen, M. Fountoulakis, W. Keck, and C. P. Gray. 2001. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 1:522-544. [DOI] [PubMed] [Google Scholar]
- 15.Filiatrault, M. J., V. E. Wagner, D. Bushnell, C. G. Haidaris, B. H. Iglewski, and L. Passador. 2005. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 73:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haebel, P. W., S. Gutmann, and N. Ban. 2004. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr. Opin. Struct. Biol. 14:58-65. [DOI] [PubMed] [Google Scholar]
- 18.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 19.Harms, J. M., H. Bartels, F. Schlunzen, and A. Yonath. 2003. Antibiotics acting on the translational machinery. J. Cell Sci. 116:1391-1393. [DOI] [PubMed] [Google Scholar]
- 20.Hermann, T. 2005. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 15:355-366. [DOI] [PubMed] [Google Scholar]
- 21.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 23.Jeannot, K., M. L. Sobel, F. El Garch, K. Poole, and P. Plesiat. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jude, F., C. Arpin, C. Brachet-Castang, M. Capdepuy, P. Caumette, and C. Quentin. 2004. TbtABM, a multidrug efflux pump associated with tributytlin resistance in Pseudomonas stutzeri. FEMS Microbiol. Lett. 232:7-14. [DOI] [PubMed] [Google Scholar]
- 26.Köhler, T., C. Van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J. T., M. B. Connelly, C. Amolo, S. Otani, and D. S. Yaver. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 49:1915-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao, W., M. S. Warren, A. Lee, A. Mistry, and O. Lomovskaya. 2001. MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuo, Y., S. Eda, N. Gotoh, E. Yoshihara, and T. Nakae. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238:23-28. [DOI] [PubMed] [Google Scholar]
- 33.Menninger, J. R. 1979. Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol. 137:694-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 35.Morita, Y., N. Kimura, T. Mima, T. Mizushima, and T. Tsuchiya. 2001. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO. J. Gen. Appl. Microbiol. 47:27-32. [DOI] [PubMed] [Google Scholar]
- 36.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein of Vibrio parahaemolyticus, and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, Y., Y. Komori, T. Mima, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202:139-143. [DOI] [PubMed] [Google Scholar]
- 38.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palumbo, J. D., C. I. Kado, and D. A. Phillips. 1998. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180:3107-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole, K. 2003. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms, p. 273-298. In I. T. Paulsen and K. Lewis (ed.), Microbial multidrug efflux. Horizon Scientific Press, Norwich, United Kingdom.
- 42.Poole, K. 2004. Acquired resistance, p. 170-183. In A. P. Fraise, P. A. Lambert, and J.-Y. Maillard (ed.), Russell, Hugo & Ayliffe's principles and practice of disinfection, preservation and sterilization. Blackwell Publishing, Oxford, United Kingdom.
- 43.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 44.Poole, K. 2004. Efflux pumps, p. 635-674. In J.-L. Ramos (ed.), Pseudomonas, vol. 1. Genomics, life style and molecular architecture. Kluwer Academic/Plenum Publishers, New York, N.Y. [Google Scholar]
- 45.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56:20-51. [DOI] [PubMed] [Google Scholar]
- 46.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775-783. [DOI] [PubMed] [Google Scholar]
- 47.Qiu, J., D. Zhou, Y. Han, L. Zhang, Z. Tong, Y. Song, E. Dai, B. Li, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, and R. Yang. 2005. Global gene expression profile of Yersinia pestis induced by streptomycin. FEMS Microbiol. Lett. 243:489-496. [DOI] [PubMed] [Google Scholar]
- 48.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 49.Redly, G. A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabina, J., N. Dover, L. J. Templeton, D. R. Smulski, D. Soll, and R. A. LaRossa. 2003. Interfering with different steps of protein synthesis explored by transcriptional profiling of Escherichia coli K-12. J. Bacteriol. 185:6158-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Schweizer, H. P. 1998. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob. Agents Chemother. 42:394-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweizer, H. P., and C. Po. 1996. Regulation of glycerol metabolism in Pseudomonas aeruginosa: characterization of the glpR repressor gene. J. Bacteriol. 178:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shafer, W. M., J. T. Balthazar, K. E. Hagman, and S. A. Morse. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to fecal lipids. Microbiology 141:907-911. [DOI] [PubMed] [Google Scholar]
- 55.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 56.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 57.Singh, N. S., and U. Varshney. 2004. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 32:6028-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 60.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts in Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J., M. O'Connor, J. A. Mills, and A. E. Dahlberg. 2002. The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. J. Mol. Biol. 322:273-279. [DOI] [PubMed] [Google Scholar]
- 62.Vioque, A., and J. de la Cruz. 2003. Trans-translation and protein synthesis inhibitors. FEMS Microbiol. Lett. 218:9-14. [DOI] [PubMed] [Google Scholar]
- 63.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plesiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Withey, J. H., and D. I. Friedman. 2003. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 57:101-123. [DOI] [PubMed] [Google Scholar]