Abstract
Pseudomonas sp. strain B13 is a bacterium known to degrade chloroaromatic compounds. The properties to use 3- and 4-chlorocatechol are determined by a self-transferable DNA element, the clc element, which normally resides at two locations in the cell's chromosome. Here we report the complete nucleotide sequence of the clc element, demonstrating the unique catabolic properties while showing its relatedness to genomic islands and integrative and conjugative elements rather than to other known catabolic plasmids. As far as catabolic functions, the clc element harbored, in addition to the genes for chlorocatechol degradation, a complete functional operon for 2-aminophenol degradation and genes for a putative aromatic compound transport protein and for a multicomponent aromatic ring dioxygenase similar to anthranilate hydroxylase. The genes for catabolic functions were inducible under various conditions, suggesting a network of catabolic pathway induction. For about half of the open reading frames (ORFs) on the clc element, no clear functional prediction could be given, although some indications were found for functions that were similar to plasmid conjugation. The region in which these ORFs were situated displayed a high overall conservation of nucleotide sequence and gene order to genomic regions in other recently completed bacterial genomes or to other genomic islands. Most notably, except for two discrete regions, the clc element was almost 100% identical over the whole length to a chromosomal region in Burkholderia xenovorans LB400. This indicates the dynamic evolution of this type of element and the continued transition between elements with a more pathogenic character and those with catabolic properties.
Genomic islands (GEIs) are a relatively newly recognized type of mobile element belonging to the class of integrative and conjugative elements (ICE) (5, 16). More and more members of this class have become known due to the large number of bacterial genome sequencing projects (3, 6, 25, 29, 33, 34, 38, 54, 57, 60, 65). GEIs have a size of between 10 and 502 kb and are characterized by three main features (27). First, they are located at one or a few specific sites in the bacterial chromosome, often nearby or inside a gene for a tRNA, and are flanked by direct repeats of between 16 and 79 bp, which are the result of the integration event. Second, GEIs harbor phage- and/or plasmid-like genes, one of which is coding for an integrase that is responsible for the integration and, in several cases, for the excision of the GEIs (67). Finally, GEIs are potentially self-transmissable and/or unstable. GEIs have been classified on the basis of the properties they invoke on the lifestyle of their bacterial hosts (i.e., genetic background and ecological habitat) (28). “Pathogenicity islands” constitute the most well-known subgroup and contribute, directly or indirectly, to the pathogenic properties of bacteria (43, 53). “Ecological islands” and “saprophytic islands” refer to those elements which confer specific advantages for the survival of their microbial hosts in the natural environment (42, 58, 63). Although GEIs are very diverse with respect to genetic structure and gene sequence, they share a similar modular genetic “outline” (5, 8, 38, 64, 68). Here we describe the complete sequence and structural analysis of a clc element, a GEI from Pseudomonas sp. strain B13, which is better known for its properties to enable the host bacterium to degrade chloroaromatic compounds.
The clc element was originally discovered in Pseudomonas sp. strain B13, the first described Pseudomonas able to metabolize 3-chlorobenzoate (3-CBA) (17). Essential for the metabolism of 3-CBA are the clcRABDE genes that encode the enzymes for 3- and 4-chlorocatechol degradation, which are two metabolic intermediates of 3-CBA (20). It had been known for a long time that the clc element is capable of self-transfer to other Beta- and Gammaproteobacteria (44, 51, 71). However, the self-transfer process could not be attributed to a conjugative plasmid (70). Instead, it was discovered that the clc element is normally integrated into the chromosome of its hosts but can excise at a low frequency and self-transfer to a new host in which it reintegrates (49). Southern hybridization analysis suggested that the clc element is present in two copies in the chromosome of Pseudomonas sp. strain B13 (49). Critical to the integration process is the integrase gene (intB13), which is situated near the right end of the clc element and near the integration site (50). Apart from the intB13 integrase gene and the clc genes, only one other region of the clc element had been characterized previously. This region at the outer left end was postulated to contain regulatory factors that are possibly involved in integrase expression control (56). This region has later been recognized as being part of a larger well-conserved core in other syntenic GEI sequences (38).
Even though ICE and GEIs share common features, such as the presence of integrase or conserved gene regions, many unanswered questions remain, particularly on their evolution and on their mode of mobilization. A GEI “core” structure was proposed on the basis of comparisons among a set of GEIs (38). In different GEIs, this core was interrupted by more variable regions, although the mechanisms governing such variability are not clear. As far as mobilization modes, clear hints exist on the nature of the transfer system of some GEIs and ICE. For example, the symbiosis island of Mesorhizobium loti strain R7A carries a trb operon, potential tra genes, and a type IV secretion system (60). Potential type IV secretion systems are carried on pKLC102 of Pseudomonas aeruginosa C (33), on ICEEc1 of E. coli strain ECOR31 (54), and on the biphenyl catabolic transposon Tn4371 (65), although the latter element is not self-transferable. Also, the SXT element carries tra-related genes and is an active mobile element (3, 7). This led to the hypothesis that ICE and GEI arose from merges between phages and plasmids during their transition to a chromosomal integrative element.
Because the clc element is one of the few genomic islands which is completely self-transferable and one of the only three ICEs currently known to contain catabolic gene functions, we decided to determine its complete nucleotide sequence in order to find out other specific catabolic properties of the clc element, to discover the possible nature of the self-transfer system, and to derive evolutionary relationships between the clc element and other GEIs or plasmids. Two other new catabolic gene clusters were detected on the element, reinforcing the general idea of the clc element being a specialized catabolic genomic island. By way of growth studies and gene expression analysis, we analyzed the newly discovered catabolic properties carried by the clc element. On the other hand, the origin of the presumed conjugative transfer system remained unclear.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α (Gibco Life Technologies, Gaithersburg, Md.) was routinely used for plasmid propagation and cloning experiments. Pseudomonas sp. strain B13 (17) is the original host of the clc element. The clc element was cloned from the chromosome of Pseudomonas putida strain RR221, a transconjugant that was previously obtained from mating between P. putida F1 and Pseudomonas sp. strain B13 (49). This transconjugant contains two copies of the clc element, which are integrated at two separate locations in the chromosome of P. putida F1. Pseudomonas aeruginosa strain 1999 is a transconjugant with a single integrated copy of the clc element and was obtained by conjugation between Pseudomonas strain B13 and P. aeruginosa PAO1-rif, a spontaneous rifampin-resistant mutant of the type strain PAO1 (obtained via Dieter Haas). Bacterial strains were stored at −80°C in spent Luria-Bertani (LB) medium containing 15% (vol/vol) glycerol. E. coli, P. aeruginosa, and Pseudomonas sp. strain B13 were routinely grown in LB medium (52). The type 21C mineral medium (MM) (21) was used to grow Pseudomonas strain B13, P. aeruginosa PAO1-rif, and P. aeruginosa 1999 under defined conditions. This medium was supplemented with 10 mM 3-CBA, 1 mM 2-aminophenol (2-AP), or 10 mM glucose as a carbon source. When necessary, the following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; rifampin, 50 μg/ml; and kanamycin, 50 μg/ml. Pseudomonas sp. strain B13 was grown at 30°C; P. aeruginosa and E. coli were grown at 37°C.
Molecular techniques.
The PCR, DNA cloning, plasmid or cosmid DNA isolations, DNA fragment recovery, DNA ligations, and restriction enzyme digestions were all carried out according to standard procedures (52) or according to specific recommendations by the suppliers.
Cosmid, plasmid, and transposon insertion libraries.
From the total DNA of P. putida RR221, a SuperCos 1 cosmid library with inserts ranging in size from 33 to 42 kb had been constructed previously (49). Cosmids 2B1, 1G3, 4H12, and 3G3 (49) were overlapping and covered the major part of the clc element. The complete sequence was then derived by the partial shotgun sequencing of plasmid sublibraries of cosmids 3G3 and 4H12, according to established procedures (4), and by sequencing a random transposon insertion library of cosmid 1G3, which was constructed with the help of the EZ::TN <oriV/KAN-2> insertion kit (Epicentre, Madison, Wis.). The outermost right end of the clc element was recovered as a 10-kb NotI fragment from cosmid 2B1 (resulting in plasmid pTCB172) and subcloned in different overlapping fragments in pUC28.
DNA sequencing on cosmids with transposon insertions was performed bidirectionally on double-stranded DNA templates with a Thermo Sequenase cycle sequencing kit with 7-deaza-dGTP (Amersham Biosciences AB) by using the suggested primers facing outwards from the kanamycin gene (Epicentre). Those sequencing reactions were analyzed on an automated DNA sequencer model 4200 IR2 (LI-COR, Lincoln, NE) as described previously (50). Plasmid DNA from sublibraries of the cosmids 3G3 and 4H12 was prepared by either alkaline lysis by applying the Montage Plasmid Miniprep96 kit (Millipore, Schwalbach, Germany) or in vitro amplification by applying the TempliPhi DNA sequencing template amplification kit (Amersham, Freiburg, Germany). Subsequently, the inserts of these plasmids were end sequenced by using M13 universal and reverse standard primers and the DYEnamic ET terminator cycle sequencing kit (Amersham, Freiburg, Germany) and were separated on MegaBACE 1000 (Amersham) or ABI PRISM 3700 capillary sequencers (PE Applied Biosystems). All plasmid DNA preparation and sequencing reactions were performed as recommended by the manufacturers. Multiple single sequences read in both directions covered the complete clc element region. After a first assembly and alignment, two gaps of 200 bp and 2.5 kb remained within cosmids 3G3 and 4H12, respectively. These two regions were amplified by PCR, cloned into pGEM-T Easy, and directly sequenced by primer walking using custom-made oligonucleotides as primers. Low-quality regions were resequenced with custom-made infrared-labeled oligonucleotides as primers and cosmid DNA as the template on the LI-COR system.
Assembly, correction, and annotation.
For each cosmid, the individual sequences were assembled into contigs by using the Staden software package (59), including the base-calling program phred (18, 19) and the assembly program phrap. For the finishing phase, the graphical tool Consed (22) was used to correct assembly errors and resolve low-quality regions. The Autofinish program (23) suggested primers to close gaps, improve sequence quality in regions of high error rates, and eliminate any single subclone regions. Two sequences that were submitted previously, AJ004950 and AJ536665, corresponding to the right and left extremity of the clc element, respectively, were included in the final assembly. Open reading frames (ORF) were identified with the application MapDraw of the DNAStar software (DNAStar, Madison, Wis.) and compared with sequences in GenBank using the BLASTP search tools (1). Assignment of ORFs was based on additional contextual information, such as the proximity of ribosome binding sequence motifs. The BLASTN algorithm was used to determine pairwise GEI homologies. Graphical comparison between GEIs was generated with the Artemis comparison tool (ACT) (9), which was downloaded from the website http://www.sanger.ac.uk/Software/ACT. Cumulative TA skew (i.e., the number of T residues minus the number of A residues per 100 bp) and G+C content were analyzed by previously described methods (25, 26).
Growth and induction experiments and RNA isolation.
Pseudomonas sp. strain B13 was grown at 30°C in 30 ml MM supplemented with 10 mM glucose to an optical density at 600 nm of 0.5. Cells were then washed and resuspended into 30 ml fresh preheated (30°C) MM. To measure catabolic gene expression, various compounds were added (a single compound per assay): 10 mM glucose, 10 or 1 mM 3-CBA, 10, 1 or 0.1 mM 2-AP, 10 or 1 mM anthranilate, 10 or 1 mM salicylate, 10 or 1 mM benzoate, 10 or 1 mM nitrobenzene, 10 or 1 mM 4-hydroxybenzoate, or 10 or 1 mM 4-CBA. Cultures were incubated for an additional 1 h at 30°C with rotary shaking before isolating total RNA. For RNA isolation, 30 ml of culture was immediately harvested by centrifugation (13,000 × g, 1 min) in 1.5-ml tubes and the supernatant was decanted. Cell pellets were resuspended in 50 μl of RNAprotect bacteria reagent (QIAGEN GmbH) in order to stabilize the RNA. Suspensions from 6 or 7 pellets were pooled together and centrifuged again, the supernatant was discarded, and the pellet was stored at −80°C for a maximum of 1 month. Prior to RNA isolation, pellets were thawed and resuspended in 0.5 ml of TES buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl), after which the total RNA was extracted and purified according to a protocol described previously (2).
Growth on 2-AP was tested on Pseudomonas sp. strain B13, P. aeruginosa strain 1999 (containing one integrated copy of clc), and P. aeruginosa PAO1. All strains were grown during 16 h in 5 ml LB and subsequently diluted 1:500 (vol/vol) into fresh minimal medium with 1 mM 2-AP. Higher 2-AP concentrations resulted in the toxicity and retardation of cell growth (not shown). Flasks were completely covered to avoid exposure to light. At regular time intervals, samples were taken from the culture, serially diluted in sterile physiological salt solution (i.e., 0.9% NaCl, 1 mM MgCl2), and plated on LB medium to count the number of CFU. Regular measurements of culture turbidity were unreliable due to the formation of dark photooxidation products from 2-AP.
Probe preparation.
Probes for mRNA analysis were generated by digoxigenin-labeling of DNA templates in the PCR by using a mixture of deoxyribonucleotides containing DIG-11-dUTP (Roche Diagnostics AG). The following probes were prepared: (i) a 425-bp fragment of the clcA gene, amplified with primers 5′-AGGTGGCCCAGCAGACGCC-3′ at position 13304 and 5′-CACCGAGGTCGCACAGACGC-3′ at position 13729; (ii) an 808-bp amnB fragment, amplified with primers 5′-CGCATCTGGTCTATGGGGA-3′ at position 23997 and 5′-GTTGCCAGTGCCGATGAC-3′ at position 24805 with plasmid pCBA253 as a template; and (iii) a 1-kb fragment coding for the large subunit of the putative aromatic dioxygenase (ORF5994), amplified with primers 5′-TCGCAGGAAGTGTATGACC-3′ at position 7142 and 5′-GCGATGTACGAATGGCTGT-3′ at position 6109 of the total sequence. The labeling quality as well as the specificity of probes was judged satisfactory after Southern hybridization against Pseudomonas sp. strain B13 genomic DNA digested with different restriction enzymes (data not shown).
Dot blot hybridization and relative mRNA quantification.
For gene expression analysis, the purified total RNAs were centrifuged, dried, and dissolved in diethyl pyrocarbonate-treated water to achieve a concentration of ∼1 μg per μl. Volumes of 15 μl of each RNA sample and dilutions thereof containing 1, 0.3, and 0.1 μg RNA were dot blotted onto positively charged nylon transfer membranes (Hybond-N+, Amersham Biosciences AG) in a 96-well dot blot manifold (Gibco Life Technologies). Dilutions of denatured plasmid DNAs containing the targeted catabolic gene inserts were included on the same blot. Membranes were hybridized, and the digoxigenin-label was detected by an antidigoxigenin alkaline phosphatase-conjugated antibody and chemiluminescence according to the supplier's instructions (Roche Diagnostics AG). Signal intensities of hybridization spots on film were measured densitometrically and calculated as intensity volumes (i.e., mean pixel intensity times the total number of pixels per spot area) and then standardized for the area of an ideal spot (i.e., with a diameter of 6 mm) similar to that described by Leveau et al. (35). Signal intensities of RNA samples were then interpolated on the standard curve, expressed as “equivalent number of DNA copies,” and divided by the amount of total RNA that was blotted on that spot. RNA copy numbers were then divided by the value that was derived under incubation conditions with glucose to obtain an induction factor. All experiments were performed with independent biological triplicates.
Nucleotide sequence accession number.
The DNA sequence of the clc element of strain P. putida RR221 is deposited in the NCBI/EMBL database under accession number AJ617740.
RESULTS AND DISCUSSION
Sequence analysis and annotation of the clc element.
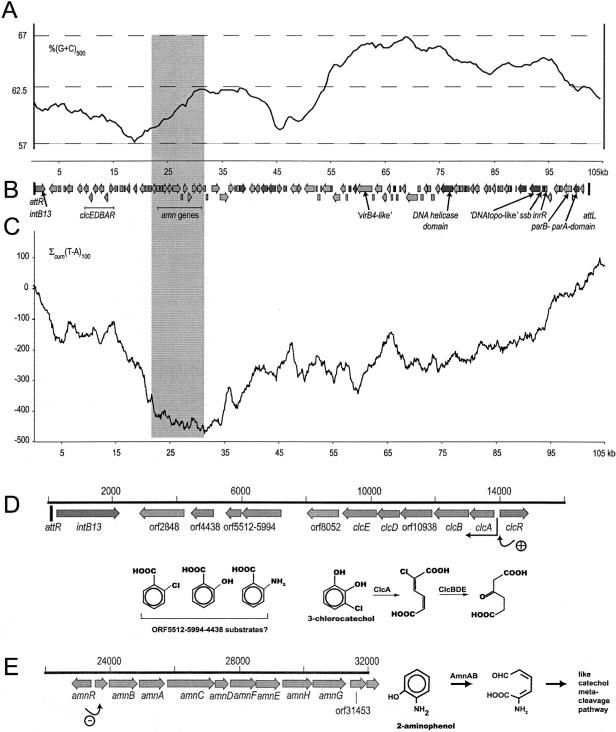
The total sequence derived from the overlapping cosmids 1G3, 4H12, and 3G3 as well as from the plasmid pTCB172 and the already submitted sequences AJ004950 and AJ536665 had a length of 105,027 bp. The clc element itself has a length of 102,784 bp, which was defined from the previously determined boundaries of the clc element (49). The right end (attR), formed by the 18 most 3′ bp of the tRNAGly (GTCTCGTTTCCCGCTCCA), was found at position 60 of the sequenced region. The left end (attL) is formed by a repetition of these 18 bp and was found at position 102826 of the sequence. The 2,184 bp outside the left end and the 77 bp upstream of the right end originate from the genome of P. putida RR21. Sequences of attR and attL had been derived previously from inverse PCR (49) and were found to be exactly the same as those determined here. A total of 107 ORFs was annotated on the clc element, of which 25 were found on the plus strand (with respect to the orientation of the intB13 gene) and 82 were found on the minus strand. ORF prediction was based on the following criteria: (i) the largest predicted ORF without overlap to its neighbors or to those on the other strand, (ii) the presence of a recognizable ribosome binding site at between 6 and 16 bp upstream of the start codon (ATG or GTG), and (iii) homologies to other entries in GenBank. Since about 55% of the designated ORFs putatively encoded polypeptides with high homologies to conserved hypothetical proteins only, we emphasized the first two criteria. Table 1 shows the name, size, direction of transcription, location of the coding region, and possible function for each ORF. The average G+C content of the clc element was 62.5%, but values for individual ORFs varied between 52.1 and 72.0% (Table 1). The G+C distribution was not homogenous along the clc element, with the first 50 kb below and the second half above the G+C average (Fig. 1A). The overall ORF gene organization on the clc element is presented in Fig. 1B. Annotation to ORFs was based on BLASTP searches and illustrates the ORFs' putative functions and their assignment to clusters of orthologous groups, protein families, or their closest relatives in the database (Table 1).
TABLE 1.
Localization and annotation of open reading frames and other features of the clc element
| ORF no. or feature | Gene name | Coding region | Orientation | Size (aa) | Ribosome binding site | Putative product | Homology (source)a | Accession no. | Amino acid identity
|
E valueb | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | Range (aa) | ||||||||||
| tRNA-Gly | glyV | 1-77 | + | 26 | tRNA-Gly | tRNA-Gly | |||||
| Repeated region | attR | 60-77 | 18 | Right end attachment site | |||||||
| ORF262 | intB13 | 262-2235 | + | 658 | GGGAA | Integrase | Phage-related integrase (Xylella fastidiosa 9a5c) | AAF84527 | 91 | 613 | 0e-00 |
| ORF2848 | 2848-4233 | − | 462 | GGAGAA | Permease | COG0477: permeases of the major facilitator superfamily (Ralstonia eutropha JMP134) | ZP_00166365 | 61 | 436 | 1e-151 | |
| ORF4438 | 4438-5157 | − | 240 | GAGGGGGA | Oxidoreductase | COG1018: flavodoxin reductases (ferredoxin- NADPH reductases) family 1 (Burkholderia cepacia R18194) | ZP_00214263 | 56 | 239 | 1e-75 | |
| ORF5512 | 5512-5991 | − | 160 | GATGGGG | Putative ring dioxygenase small subunit | Anthranilate dioxygenase small subunit (Burkholderia cepacia) | AAO83640 | 57 | 157 | 1e-48 | |
| ORF5994 | 5994-7256 | − | 421 | GGAGA | Large subunit aromatic dioxygenase | Ortho-halobenzoate 1,2-dioxygenase alpha-ISPc protein OhbB (Burkholderia mallei ATCC 23344) | YP_105030 | 67 | 424 | 1e-167 | |
| ORF8052 | 8052-9035 | − | 328 | GAGG | Hypothetical protein | Unknown (Pseudomonas aeruginosa) | AAC69479 | 97 | 327 | 0e-00 | |
| ORF9151 | clcE | 9151-10209 | − | 353 | AAGAAG | Maleylacetate reductase | Maleylacetate reductase | AAB71540 | 99 | 352 | 0e-00 |
| ORF10206 | clcD | 10206-10916 | − | 237 | GGAGAG | Dienelactone hydrolase | Dienelactone hydrolase (Pseudomonas aeruginosa) | AAC69477 | 99 | 236 | 1e-134 |
| ORF10938 | 10938-11921 | − | 328 | GGGAA | Hypothetical protein | Hypothetical UPF0065 protein in clcB-clcD intergenic region precursor | P0A177 | 99 | 326 | 1e-180 | |
| ORF11948 | clcB | 11948-13060 | − | 371 | GGAGA | Chloromuconate cycloisomerase | Chloromuconate cycloisomerase (Pseudomonas aeruginosa) | AAC69475 | 99 | 370 | 0e-00 |
| ORF13057 | clcA | 13057-13839 | − | 261 | GGAGA | Chlorocatechol 1,2-dioxygenase | Chlorocatechol 1,2-dioxygenase (Ralstonia sp. JS705) | CAA06968 | 99 | 258 | 1e-148 |
| ORF14009 | clcR | 14009-14893 | + | 295 | AGAGG | lysR family transcriptional regulator | Lys-R type regulatory protein (Pseudomonas aeruginosa) | AAC69473 | 99 | 292 | 1e-160 |
| ORF15037 | 15037-15387 | − | 117 | No | Threonine efflux protein | COG1280: putative threonine efflux protein (Burkholderia cepacia R18194) | ZP_00214880 | 79 | 115 | 2e-44 | |
| ORF15405 | 15405-15647 | − | 81 | GGAT | Hypothetical protein | COG1280: putative threonine efflux protein (Burkholderia cepacia R18194) | ZP_00214880 | 84 | 73 | 1e-28 | |
| ORF15962 | 15962-16603 | − | 214 | No | Hypothetical protein | Hypothetical protein XF1719 (Xylella fastidiosa 9a5c) | NP_299008 | 68 | 141 | 2e-65 | |
| ORF16775 | 16775-17023 | + | 83 | GGCA | Hypothetical protein | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62096 | 86 | 50 | 2e-17 | |
| ORF17162 | 17162-17959 | − | 266 | GGGAA | Transcriptional regulator | Putative transcriptional regulator (Pseudomonas aeruginosa) | AAN62138 | 79 | 261 | 1e-113 | |
| ORF18502 | 18502-19188 | − | 229 | GGGAA | Transcriptional regulator | Probable transcription regulator protein (Ralstonia solanacearum GMI1000) | NP_522004 | 57 | 202 | 2e-59 | |
| ORF19619 | 19619-20563 | − | 315 | GGGAA | Hypothetical protein | Hypothetical protein SMc01405 (Sinorhizobium meliloti 1021) | NP_386189 | 30 | 285 | 3e-27 | |
| ORF20709 | 20709-21128 | − | 140 | GGAG | Hypothetical protein | COG2259: predicted membrane protein (Pseudomonas aeruginosa UCBPP-PA14) | ZP_00137687 | 58 | 136 | 2e-39 | |
| ORF21241 | 21241-21900 | − | 220 | GGAGCA | tetR-type transcriptional regulator | Transcriptional regulator (Xanthomonas oryzae pv. oryzae KACC10331) | YP_199568 | 30 | 198 | 2e-21 | |
| ORF21922 | 21922-22674 | − | 251 | GGAGCA | Hypothetical protein | COG2259: predicted membrane protein (Pseudomonas aeruginosa UCBPP-PA14) | ZP_00137687 | 38 | 131 | 2e-13 | |
| ORF22813 | amnR | 22813-23430 | − | 206 | GGTGA | Aminophenol repressor | NbzR, aminophenol operon repressor (Pseudomonas putida) | AAK26517 | 57 | 146 | 8e-39 |
| ORF23526 | 23526-23939 | + | 138 | AAAGGA | Ferredoxin-like protein | Putative ferredoxin (Pseudomonas putida) | AAK26518 | 49 | 136 | 2e-28 | |
| ORF23951 | amnB | 23951-24865 | + | 305 | GAGGAGAA | 2-Aminophenol 1,6-dioxygenase beta subunit | 2-Aminophenol 1,6-dioxygenase beta subunit (Comamonas testosteroni) | AAT35226 | 79 | 298 | 5e-163 |
| ORF24910 | amnA | 24910-25722 | + | 271 | AGGAGA | 2-Aminophenol 1,6-dioxygenase alpha subunit | 2-Aminophenol 1,6-dioxygenase alpha subunit (Comamonas testosteroni) | AAT35227 | 59 | 269 | 7e-91 |
| ORF25781 | amnC | 25781-27259 | + | 493 | AAGAAGG | 2-Aminomuconic semialdehyde dehydrogenase | 2-Aminomuconic semialdehyde dehydrogenase (Comamonas testosteroni) | AAT35228 | 72 | 485 | 0e-00 |
| ORF27249 | amnD | 27249-27701 | + | 151 | GCATCC | 2-Aminomuconate deaminase | 2-Aminomuconate deaminase (Pseudomonas fluorescens) | BAC65310 | 72 | 136 | 1e-50 |
| ORF27716 | amnF | 27716-28525 | + | 270 | AAGAGG | 2-Keto-4-pentenoate hydratase | Putative hydratase protein (Ralstonia solanacearum GMI1000) | NP_522452 | 60 | 259 | 3e-82 |
| ORF28522 | amnE | 28522-29286 | + | 255 | GGAG | 4-Oxalocrotonate decarboxylase | Probable 4-oxalocrotonate decarboxylase protein (Ralstonia solanacearum GMI1000) | NP_522453 | 66 | 250 | 4e-87 |
| ORF29347 | amnH | 29347-30288 | + | 314 | GGAG | Acetylating aldehyde dehydrogenase | Acetaldehyde dehydrogenase oxidoreductase (Ralstonia oxalatica) | CAD61138 | 85 | 312 | 5e-163 |
| ORF30304 | amnG | 30304-31341 | + | 346 | GAGGAG | 4-Hydroxy-2-ketovalerate aldolase | 4-Hydroxy-2-ketovalerate aldolase (Comamonas testosteroni) | BAA82884 | 87 | 332 | 1e-164 |
| ORF31453 | 31453-31953 | + | 167 | AGCGGTT | Hypothetical protein | COG0657: esterase/lipase (Nostoc punctiforme PCC 73102) | ZP_00105982 | 35 | 139 | 3e-15 | |
| ORF31950 | 31950-32345 | + | 132 | CAAG | Hypothetical protein | COG0657: esterase/lipase (Nostoc punctiforme PCC 73102) | ZP_00105982 | 47 | 126 | 2e-25 | |
| ORF32963 | 32963-34498 | + | 512 | GGGTGA | Outer membrane protein or channel-forming component | Probable channel-forming component of a multidrug resistance efflux pump protein (Ralstonia solanacearum GMI1000) | NP_522003 | 54 | 460 | 1e-134 | |
| ORF34495 | 34495-36069 | + | 525 | GAAGG | Permease of the major facilitator superfamily | Probable inner membrane multidrug resistance transmembrane protein (Ralstonia solanacearum GMI1000) | NP_522002 | 57 | 478 | 1e-159 | |
| ORF36077 | 36077-37111 | + | 345 | AAGGA | Multidrug efflux pump | Putative multidrug resistance homolog transmembrane protein (Ralstonia solanacearum GMI1000) | NP_522001 | 57 | 342 | 1e-103 | |
| ORF37143 | 37143-37445 | + | 101 | AGGAGA | Hypothetical protein | Hypothetical protein YPTB3109 (Yersinia pseudotuberculosis IP 32953) | YP_071613 | 43 | 101 | 8e-16 | |
| ORF37489 | 37489-38133 | + | 215 | AAAGGA | Hypothetical protein | Hypothetical protein Raeut03005309 (Ralstonia eutropha JMP134) | ZP_00166494 | 47 | 117 | 8e-18 | |
| ORF38184 | 38184-39365 | + | 394 | AGGC | Esterase of the alpha-beta hydrolase superfamily | COG1752: predicted esterase of the alpha-beta hydrolase superfamily (Rubrivivax gelatinosus PM1) | ZP_00245180 | 58 | 391 | 1e-127 | |
| ORF40894 | 39860-40894 | − | 345 | GGAGGA | Hypothetical protein | COG0823: periplasmic component of the Tol biopolymer transport system (Cytophaga hutchinsonii) | ZP_00310748 | 25 | 300 | 4e-08 | |
| ORF41917 | 40922-41917 | − | 332 | AGGA | Amidohydrolase/nitrilase | NitA (Pseudomonas fluorescens) | AAW79573 | 75 | 313 | 1e-137 | |
| ORF41973 | 41973-43385 | − | 471 | No | Acyl-CoA synthetase | Putative long-chain-fatty-acid-CoA ligase (Rhodopseudomonas palustris CGA009) | NP_947491 | 28 | 446 | 2e-31 | |
| ORF43387 | 43387-44967 | − | 527 | AGAGGAAG | Acyl-CoA synthetase | Putative long-chain-fatty-acid-CoA ligase (Rhodopseudomonas palustris CGA009) | NP_947491 | 28 | 518 | 1e-39 | |
| ORF45180 | 45180-46136 | + | 319 | GGAG | Transcriptional regulator (AraC-type DNA binding domain-containing protein) | COG2207: AraC-type DNA binding domain- containing proteins (Pseudomonas syringae pv. syringae B728a) | ZP_00205730 | 30 | 323 | 8e-36 | |
| ORF46315 | 46315-46698 | − | 128 | GGCAGG | Hypothetical protein | COG1249: pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes (Polaromonas sp. JS666) | ZP_00360452 | 77 | 118 | 3e-47 | |
| ORF46777 | 46777-46932 | − | 52 | GAAGACGG | Transposase (fragment) | COG2801: transposase and inactivated derivatives (Polaromonas sp. JS666) | ZP_00360815 | 74 | 51 | 5e-16 | |
| ORF47630 | 47630-48811 | − | 394 | AGGAA | Transport protein | COG2807: cyanate permease (Pseudomonas aeruginosa UCBPP-PA14) | ZP_00140706 | 35 | 391 | 5e-47 | |
| ORF48922 | 48922-49725 | + | 268 | GGAGA | Transcriptional regulator (AraC-type DNA binding domain-containing protein) | Transcriptional regulator, AraC family (Pseudomonas putida KT2440) | NP_742746 | 41 | 244 | 2e-42 | |
| ORF50240 | 50240-52087 | − | 616 | AGGA | Hypothetical protein | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62129 | 80 | 616 | 0e-00 | |
| ORF52324 | 52324-52710 | + | 129 | GGAGAG | Hypothetical protein | Hypothetical protein NE0293 (Nitrosomonas europaea ATCC 19718) | NP_840380 | 75 | 105 | 1e-39 | |
| ORF52710 | 52710-53168 | + | 153 | GAAGTGGAA | Hypothetical protein | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62268 | 82 | 150 | 1e-71 | |
| ORF53196 | 53196-53573 | + | 126 | AGGAGA | Hypothetical protein | Hypothetical protein Reut02005849 (Ralstonia metallidurans CH34) | ZP_00271404 | 60 | 122 | 9e-33 | |
| ORF53587 | 53587-55104 | − | 506 | GGGGAGA | Hypothetical protein | Hypothetical protein Reut02005848 (Ralstonia metallidurans CH34) | ZP_00271403 | 85 | 502 | 0e-00 | |
| ORF55120 | 55120-55479 | − | 120 | GGGAGG | Hypothetical protein | Hypothetical protein Reut02005847 (Ralstonia metallidurans CH34) | ZP_00271402 | 77 | 117 | 2e-48 | |
| ORF55476 | 55476-56873 | − | 466 | GAGGTGG | Hypothetical protein | Hypothetical protein Reut02005846 (Ralstonia metallidurans CH34) | ZP_00271401 | 85 | 462 | 0e-00 | |
| ORF56883 | 56883-57830 | − | 316 | GGAGGG | Hypothetical protein | COG1154: Deoxyxylulose-5-phosphate synthase (Ralstonia metallidurans CH34) | ZP_00271400 | 90 | 313 | 1e-170 | |
| ORF57827 | 57827-58273 | − | 149 | AGGGG | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62137 | 75 | 148 | 6e-61 | |
| ORF58432 | 58432-58926 | − | 165 | GGAGG | DNA repair protein | COG2003: DNA repair proteins (Ralstonia metallidurans CH34) | ZP_00271398 | 82 | 164 | 1e-72 | |
| ORF59110 | 59110-59874 | − | 255 | GGAGAA | Protein-disulfide isomerase | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62139 | 82 | 254 | 1e-122 | |
| ORF59888 | 59888-62755 | − | 956 | GGAGGA | Conserved hypothetical protein VirB4 domain | Hypothetical protein (Pseudomonas aeruginosa), COG3451 | AAN62141 | 95 | 885 | 0e-00 | |
| ORF62755 | 62755-63195 | − | 147 | AGGAGAAA | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62142 | 90 | 146 | 3e-71 | |
| ORF63176 | 63176-64594 | − | 473 | AAGGAG | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62143 | 92 | 472 | 0e-00 | |
| ORF64584 | 64584-65516 | − | 311 | AAGGAGGAAG | Hypothetical protein | Hypothetical protein Reut02005836 (Ralstonia metallidurans CH34) | ZP_00271391 | 84 | 310 | 1e-141 | |
| ORF65513 | 65513-66205 | − | 231 | AAGGGGG | Hypothetical protein | Hypothetical protein Reut02005835 (Ralstonia metallidurans CH34) | ZP_00271390 | 94 | 230 | 1e-129 | |
| ORF66202 | 66202-66612 | − | 137 | AGGCGAGG | Hypothetical protein | Hypothetical protein Reut02005834 (Ralstonia metallidurans CH34) | ZP_00271389 | 96 | 130 | 4e-70 | |
| ORF66625 | 66625-66984 | − | 120 | GAAAGG | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62147 | 96 | 119 | 2e-59 | |
| ORF67001 | 67001-67234 | − | 78 | GGAGAACAAG | Hypothetical protein | Hypothetical protein Reut02005832 (Ralstonia metallidurans CH34) | ZP_00271387 | 100 | 68 | 5e-32 | |
| ORF67231 | 67231-67614 | − | 128 | AGGAATGG | Hypothetical protein | COG0643: chemotaxis protein histidine kinase and related kinases (Ralstonia metallidurans CH34) | ZP_00271386 | 85 | 126 | 2e-52 | |
| ORF67800 | 67800-68204 | + | 135 | GGGGAGA | Hypothetical protein | Hypothetical protein Npun02008345 (Nostoc punctiforme PCC 73102) | ZP_00106053 | 38 | 91 | 1e-07 | |
| ORF68241 | 68241-68990 | − | 250 | AACGAGG | Hypothetical protein | Hypothetical protein Reut02005852 (Ralstonia metallidurans CH34) | ZP_00271362 | 95 | 249 | 1e-133 | |
| ORF68987 | 68987-71173 | − | 729 | GAAG | Hypothetical protein, VirD4 domain | Conserved hypothetical protein (Pseudomonas aeruginosa), COG3505 | AAN62159 | 96 | 730 | 0e-00 | |
| ORF71178 | 71178-71726 | − | 183 | AGGAGA | Hypothetical protein | Hypothetical protein Reut02005854 (Ralstonia metallidurans CH34) | ZP_00271364 | 87 | 182 | 2e-84 | |
| ORF71723 | 71723-72313 | − | 197 | GAGGTGAA | Hypothetical protein | COG0741: soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) (Ralstonia metallidurans CH34) | ZP_00271365 | 87 | 196 | 5e-95 | |
| ORF72295 | 72295-73014 | − | 240 | GGAGCA | Hypothetical protein | COG0695: glutaredoxin and related proteins (Ralstonia metallidurans CH34) | ZP_00271366 | 88 | 245 | 1e-121 | |
| ORF73029 | 73029-73679 | − | 217 | GGGAGG | Hypothetical protein | COG0845: membrane-fusion protein (Ralstonia metallidurans CH34) | ZP_00271367 | 80 | 217 | 2e-88 | |
| ORF73676 | 73676-74296 | − | 207 | GAGG | Hypothetical protein | Hypothetical protein Reut02005858 (Ralstonia metallidurans CH34) | ZP_00271368 | 87 | 199 | 1e-96 | |
| ORF74436 | 74436-75305 | − | 290 | AAGGAGAG | Hypothetical protein | Hypothetical protein Rgel02003074 (Rubrivivax gelatinosus PM1) | ZP_00242828 | 30 | 108 | 8e-03 | |
| ORF75419 | 75419-77698 | − | 760 | GGAG | DNA/RNA helicase | Conserved hypothetical plasmid protein (Pseudomonas aeruginosa) | AAN62165 | 96 | 759 | 0e-00 | |
| ORF77798 | 77798-78907 | − | 370 | AGGAGA | Hypothetical protein | Conserved hypothetical plasmid protein (Pseudomonas aeruginosa) | AAN62168 | 97 | 369 | 0e-00 | |
| ORF78972 | 78972-79622 | − | 217 | GAGGA | Hypothetical protein | Hypothetical protein Reut02005863 (Ralstonia metallidurans CH34) | ZP_00271373 | 95 | 216 | 1e-116 | |
| ORF79699 | 79699-79959 | − | 87 | AGGAGGAA | Hypothetical protein | Hypothetical protein XF1757 (Xylella fastidiosa 9a5c) | NP_299046 | 90 | 86 | 2e-39 | |
| ORF79976 | 79976-80383 | − | 136 | AGGAGG | Hypothetical protein | Hypothetical protein XF1758 (Xylella fastidiosa 9a5c) | NP_299047 | 91 | 134 | 3e-68 | |
| ORF80480 | 80480-80812 | − | 111 | GGAGG | Hypothetical protein | Conserved plasmid protein (Xylella fastidiosa 9a5c) | NP_299048 | 65 | 106 | 2e-33 | |
| ORF80908 | 80908-81597 | − | 230 | AAGGAGAA | Hypothetical protein | Hypothetical protein Reut02005867 (Ralstonia metallidurans CH34) | ZP_00271377 | 87 | 229 | 1e-111 | |
| ORF81655 | 81655-82572 | − | 306 | GGAGA | Hypothetical protein | Hypothetical protein XF1761 (Xylella fastidiosa 9a5c) | NP_299050 | 94 | 305 | 1e-160 | |
| ORF83350 | 83350-84192 | − | 281 | GGAGACGA | Hypothetical protein | Hypothetical protein XF1763 (Xylella fastidiosa 9a5c) | NP_299052 | 94 | 280 | 1e-158 | |
| ORF84338 | 84338-84691 | − | 118 | AGGAGA | Hypothetical protein | Hypothetical protein XF1764 (Xylella fastidiosa 9a5c) | NP_299053 | 83 | 117 | 7e-53 | |
| ORF84835 | 84835-85647 | − | 271 | AAGGAGA | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62182 | 87 | 274 | 1e-134 | |
| ORF85934 | 85934-86212 | − | 93 | AGGAG | Hypothetical protein | COG0528: uridylate kinase (Ralstonia metallidurans CH34) | ZP_00271279 | 89 | 92 | 1e-43 | |
| ORF86310 | 86310-87047 | − | 246 | GGAGAAA | Hypothetical protein | COG0834: ABC-type amino acid transport/signal transduction systems, periplasmic component/ domain (Ralstonia metallidurans CH34) | ZP_00271280 | 86 | 243 | 1e-120 | |
| ORF87127 | 87127-87939 | − | 271 | GAGAGGGA | Hypothetical protein | Hypothetical protein (Pseudomonas aeruginosa) | AAN62185 | 80 | 243 | 1e-108 | |
| ORF87986 | 87986-88378 | − | 131 | GGAGGAA | Hypothetical protein | Hypothetical protein XF1771 (Xylella fastidiosa 9a5c) | NP_299060 | 98 | 130 | 9e-71 | |
| ORF88400 | 88400-88612 | − | 71 | AAGGAG | Hypothetical protein | Hypothetical protein XF1772 (Xylella fastidiosa 9a5c) | NP_299061 | 100 | 70 | 1e-33 | |
| ORF89247 | 89247-89501 | − | 85 | AGGA | Hypothetical protein | Hypothetical protein XF1773 (Xylella fastidiosa 9a5c) | NP_299062 | 96 | 84 | 5e-40 | |
| ORF89746 | 89746-91347 | − | 534 | AGGAGA | DNA methyltransferase | DNA methyltransferase (Xylella fastidiosa 9a5c) | NP_299063 | 89 | 539 | 0e-00 | |
| ORF91884 | 91884-93896 | − | 671 | GGATGGA | DNA topoisomerase IA | Putative DNA topoisomerase III (Pseudomonas aeruginosa) | AAN62194 | 90 | 676 | 0e-00 | |
| ORF94175 | 94175-94615 | − | 147 | GGAGA | Single-stranded-DNA binding protein | COG0629: single-stranded DNA-binding protein (Ralstonia metallidurans CH34) | ZP_00271286 | 89 | 146 | 1e-75 | |
| ORF94689 | inrR | 94689-95216 | − | 176 | GAGGACGAA | Transcriptional regulator | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62196 | 86 | 175 | 5e-82 |
| ORF95213 | 95213-95992 | − | 260 | GGGCGG | Hypothetical protein | Conserved hypothetical protein (Pseudomonas aeruginosa) | AAN62197 | 91 | 263 | 1e-131 | |
| ORF96323 | 96323-97567 | − | 415 | AAGGA | Hypothetical protein | Hypothetical protein Reut02005948 (Ralstonia metallidurans CH34) | ZP_00271289 | 81 | 417 | 0e-00 | |
| ORF97571 | 97571-98131 | − | 187 | GAGGG | Hypothetical protein | COG0635: coproporphyrinogen III oxidase and related Fe-S oxidoreductases (Ralstonia metallidurans CH34) | ZP_00271290 | 90 | 186 | 3e-93 | |
| ORF98147 | 98147-99799 | − | 551 | AAAGGAA | Hypothetical protein | Conserved hypothetical protein (Pseudomonas aeruginosa), COG1475 ParB domain | AAN62200 | 82 | 559 | 0e-00 | |
| ORF99792 | 99792-100049 | − | 86 | GGGAGG | Hypothetical protein | Hypothetical protein Reut02004806 (Ralstonia metallidurans CH34) | ZP_00272227 | 78 | 89 | 3e-33 | |
| ORF100033 | 100033-100908 | − | 292 | GGAGAG | Chromosome partioning-related protein | COG1192: ATPases involved in chromosome partitioning (Ralstonia metallidurans CH34) | ZP_00272228 | 93 | 291 | 1e-48 | |
| ORF100952 | 100952-101164 | − | 71 | AGGAGTGA | Transcriptional regulator | Phage-related protein (Pseudomonas aeruginosa) | AAN62202 | 90 | 70 | 4e-30 | |
| ORF101284 | 101284-102039 | − | 252 | AAGGAGA | Hypothetical protein | Hypothetical protein XF1787 (Xylella fastidiosa 9a5c) | NP_299075 | 83 | 249 | 1e-117 | |
| Repeat region | attL | 102826-102843 | Left end attachment site | ||||||||
Due to an almost 100% sequence conservation between the clc element and a chromosomal region in B. xenovorans, homologies between the two are not listed.
E values are based on BLASTP results of the nonredundant NCBI database.
ISP, intracellular serine protease.
FIG. 1.
Sequence characteristics of the clc element. (A) G+C content variation calculated from a 500-bp window. Horizontal dotted lines indicate the minimum, maximum, and mean values of the G+C content. (B) Gene map of the clc element. For ORF details, please see Table 1. (C) Cumulative TA skew analysis with a window of 100 bp. (D) Enlargement of the gene organization near the clc genes and schematic illustration of the chlorocatechol pathway and possible substrates for the putative aromatic ring dioxygenase. (E) Gene organization of the amn region and conceptual illustration of the 2-AP pathway. Rounded arrows in (D) and (E) indicate transcriptional activation (+, by ClcR) and repression (−, assumed for AmnR). For location and scale, see Table 1.
The annotation revealed two very distinct regions on the clc element. The first region of approximately 50 kb, extending from the tRNAGly (attR) to about half-way into the clc element, consisted mainly of relatively clearly identifiable genes encoding catabolic properties. The second half of the clc element was composed of a large number of co-oriented genes, encoding predominantly hypothetical proteins, although the region itself carried a high percentage of nucleotide sequence identity to other GEIs and to several genomic regions in other bacteria (see below). Interestingly, not a single transposase or insertion element, except for a truncated one (ORF46777), was detected on the clc element. This is in contrast to many other GEIs like PAGI-2 (34), PAGI-4 (33), the GEI present in Ralstonia sp. strain JS705 (40), the SXT pathogenicity island (3), and catabolic plasmids.
The first gene next to the attR sequence was coding for the IntB13 integrase. This enzyme belongs to the bacteriophage-type integrases of the phage P4 subfamily and has been shown to be implicated in the site-specific chromosomal integration of the clc element (50). Downstream of the intB13 gene in the direction of attL, a gene coding for a putative permease of the major facilitator superfamily (ORF2848) is situated. This was followed by a cluster of genes (ORFs 4438, 5512, and 5994) putatively encoding an aromatic ring dioxygenase enzyme complex (Table 1, Fig. 1D). Products of these genes had strong homologies to the anthranilate 1,2-dioxygenase from Burkholderia cepacia DB01 (57% amino acid [aa] identity for the small subunit and 67% for the large subunit, AY223539) (10) as well as to the PhnA1A2 salicylate dioxygenase present in Sphingomonas (50% amino acid identity to PhnA1a and 34% to PhnA2a, AJ633532) (15). At positions 9151 to 14893, the previously identified clcRABDE genes were found (20, 30, 32). The next easily recognizable region (positions 22813 to 31341, Fig. 1E) consisted of a complete catabolic operon for 2-AP degradation via meta cleavage (31) that was similar to the amn gene cluster of Pseudomonas sp. strain AP-3 (61, 62) and to parts of the nitrobenzene pathway genes of Pseudomonas fluorescens strain KU-7 (41) and P. putida HS12 (45). The amn operon on the clc element is formed by a putative transcriptional repressor (amnR, ORF22813), a ferredoxin-like protein (ORF23526), and eight amn genes, amnBACDFEHG (Fig. 1E). The amn genes of the clc element were in slightly different order than in the amn operon of Pseudomonas sp. strain AP-3 (AB020521), in which the amnD gene is located downstream of amnE. The amino acid sequence identity levels between orthologous amn partners of the clc element and those of strain AP-3 were as follows: AmnB, 76%; AmnA, 54%; AmnC, 66%; AmnD, 66%; AmnF, 56%; AmnE, 49%; AmnH, 68%; and AmnG, 77%. Next, we found three components of a putative multidrug efflux pump (ORF32963, ORF34495, and ORF36077) that were closely related to those present in the plant pathogen Ralstonia solanacearum (Table 1). Among other putative genes with a distinguishable function, we found a nitrilase (ORF41917).
Functionality of the 2-AP pathway.
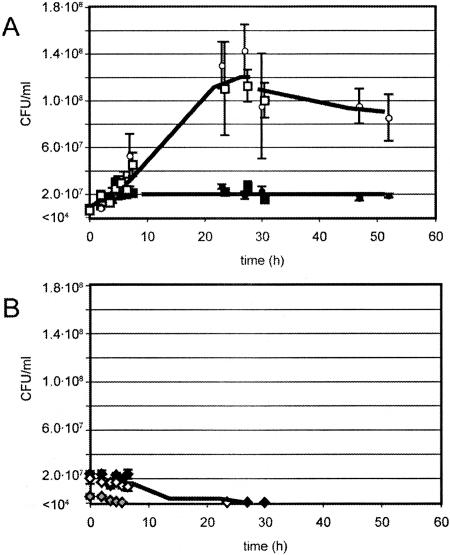
Growth experiments and gene expression analysis were used to reveal whether the newly discovered catabolic genes on the clc element were functional. Since the clc element is present in two copies in Pseudomonas sp. strain B13, a functional analysis was more easily performed in a transconjugant carrying only one copy. The clc element was transferred by conjugation from Pseudomonas sp. strain B13 into P. aeruginosa PAO1-rif. As expected, PAO1 transconjugants could grow on 3-CBA, whereas PAO1 itself could not. This confirmed functionality of the clc genes for chlorocatechol degradation in PAO1 as well. Some PAO1 transconjugants (e.g., strain 1999) carried only a single clc insertion as determined by Southern hybridization and PCR analysis (not shown). In contrast to PAO1-rif, strain 1999 formed a 10-fold higher biomass on 2-AP (Fig. 2A). Due to the toxicity and rapid photooxidation of 2-AP, growth experiments could be conducted with only 1 mM. Strangely enough, Pseudomonas sp. strain B13 itself could not grow on 1 mM 2-AP, which might be the result of a metabolic misrouting or production of a toxic intermediate from 2-AP (Fig. 2B). Since PAO1 does not carry the clc element, whereas strain 1999 does, we can conclude that the clc element indeed confers the ability to grow on 2-AP (in some strains). Most likely, this is the result of the presence of the amn genes.
FIG. 2.
Growth of P. aeruginosa strain 1999 (open symbols), carrying one copy of the clc element, and P. aeruginosa PAO1-rif (closed symbols) (A) or Pseudomonas sp. strain B13 (diamonds) (B) on 1 mM 2-aminophenol. Growth is indicated as CFU per ml of culture. Different symbols correspond to individual growth experiments.
Catabolic gene expression analysis.
We then tested whether any of the catabolic genes were actively transcribed in Pseudomonas sp. strain B13 after exposure to various aromatic substrates. Total RNA was isolated from batch cultures of Pseudomonas sp. strain B13 that were induced in exponential phase with different aromatic compounds and hybridized with probes specific for the clc, amn, and dioxygenase (ORF5994) gene regions. Cells of Pseudomonas sp. strain B13 that were induced with 1 mM 2-AP showed a 50-fold increase of amnB mRNA compared to that for cells cultivated in the presence of 10 mM glucose (Table 2). At 0.1 mM 2-AP, only a twofold induction of amn mRNA compared to that for glucose-grown cells was measured. On the contrary, no induction was observed in cells exposed to 10 mM 2-AP, suggesting cellular intoxification by 2-AP or its metabolites at this concentration (not shown).
TABLE 2.
Relative induction of three catabolic genes on the clc element after exposure of Pseudomonas sp. strain B13 to different substrates
| Substrate |
amnB
|
clcA
|
orf5994
|
|||
|---|---|---|---|---|---|---|
| I.F.a | %b | I.F. | % | I.F. | % | |
| Glucose (10 mM) | 1.00 | 39.87 | 1.00 | 4.18 | 1.00 | 26.30 |
| 3-Chlorobenzoate (10 mM) | 23.22c | 14.99 | 2.03 | 26.12 | 0.82 | 64.20 |
| 3-Chlorobenzoate (1 mM) | 3.50 | 44.91 | 5.53 | 47.42 | 17.76 | 35.87 |
| 2-Aminophenol (1 mM) | 49.15 | 8.78 | 1.39 | 49.70 | 18.83 | 45.43 |
| Anthranilate (10 mM) | 4.94 | 19.57 | 3.74 | 6.23 | 0.69 | 56.78 |
| Anthranilate (1 mM) | 29.20 | 24.55 | 6.52 | 35.95 | 25.79 | 20.33 |
| Salicylate (10 mM) | 1.05 | 36.58 | 0.72 | 43.90 | 0.65 | 59.40 |
| Salicylate (1 mM) | 2.91 | 48.12 | 3.43 | 50.90 | 6.59 | 23.31 |
| Benzoate (10 mM) | 1.37 | 59.15 | 0.54 | 47.18 | 0.75 | 75.97 |
| Benzoate (1 mM) | 2.58 | 33.74 | 10.04 | 29.80 | 2.92 | 33.71 |
| Nitrobenzene (10 mM) | 1.31 | 58.49 | 0.52 | 32.78 | 0.77 | 49.31 |
| Nitrobenzene (1 mM) | 2.48 | 59.11 | 1.04 | 35.50 | 1.04 | 33.38 |
| 4-OH-benzoate (10 mM) | 65.85 | 39.12 | 0.61 | 54.92 | 1.73 | 66.83 |
| 4-OH-benzoate (1 mM) | 2.58 | 47.24 | 0.54 | 19.78 | 1.05 | 59.38 |
| 4-Chlorobenzoate (10 mM) | 1.31 | 54.03 | 0.55 | 46.23 | 0.70 | 58.18 |
| 4-Chlorobenzoate (1 mM) | 1.32 | 33.06 | 2.40 | 38.71 | 8.04 | 48.72 |
I.F., induction factor (calculated mRNA amount relative to that of glucose-exposed cultures).
%, standard deviation calculated as percentage of the average mRNA amount in triplicate determinations.
Values in boldface type indicate induction values significantly different (P < 0.05) from that of the glucose-exposed culture.
Expression of the amnB gene (ORF23951) was analyzed in cells exposed to various other aromatic compounds at a 10 mM or 1 mM concentration (Table 2). Compared to that for 1 mM 2-AP, 1 mM anthranilate provoked a similar strong induction of the amnB gene. Exposure to 10 mM anthranilate resulted in a fivefold increase of amnB mRNA content compared to that for the culture grown in glucose only. Induction levels similar to those with 1 mM 2-AP and anthranilate were observed with 10 mM 3-CBA and 10 mM 4-hydroxybenzoate (23- and 65-fold, respectively), whereas at 1 mM, stimulation of amnB expression was about 10-fold lower.
Expression of the putative large subunit dioxygenase (ORF5994) increased 26-fold in the presence of 1 mM anthranilate and 18-fold with 1 mM 3-CBA compared to that in the presence of glucose alone. In contrast to amnB, a concentration of 10 mM anthranilate or 10 mM 3-CBA provoked no induction of ORF5994. Strong (19-fold) induction of ORF5994 was also observed with 1 mM 2-AP, and slightly weaker induction was observed with 1 mM but not with 10 mM of benzoate, 4-chlorobenzoate, and salicylate (Table 2).
Exposure of Pseudomonas sp. strain B13 to 1 mM 3-CBA provoked a fivefold induction of clcA compared to that for the culture grown in glucose only. Similar amounts of clcA mRNA were observed with 1 mM anthranilate, and twofold less was observed after exposure to 1 mM salicylate or 4-chlorobenzoate. As for ORF5994 expression, less clcA mRNA was produced after exposure to 10 mM 3-CBA and 10 mM anthranilate. The strongest clcA induction (10-fold) was detected in bacteria exposed to 1 mM benzoate. In contrast to amnB and ORF5994, 1 mM 2-AP and 4-hydroxybenzoate did not induce clcA expression relative to glucose. The clear expression observed for all three gene clusters confirms (clc or amn) or suggests (orf5994) that the genes are functional and implicated in aromatic compound metabolism. However, as mentioned above, Pseudomonas sp. strain B13 could not grow on 2-AP despite amn gene expression.
Expression patterns were much more complex than expected from the nature of the metabolic pathways themselves and suggested cross-activation by nonnative regulators. Only two specific regulatory proteins for regulating catabolic gene expression can be deduced from the clc element's sequence: ClcR, a Lys-type transcriptional regulator activating the clc genes in response to 2-chloro-cis,cis-muconate (arising from 3-chlorocatechol metabolism) (Fig. 1D) (12), and AmnR, a repressor for the 2-AP pathway (in analogy to NbzR, a transcriptional repressor for the nbz genes of which the effector compound is unknown) (Fig. 1E) (45). In contrast to what we expected, we observed that the clc genes were induced in the presence of not only 3-CBA and 4-chlorobenzoate but also anthranilate, benzoate, and salicylate. Anthranilate, benzoate, and salicylate can typically be converted by pseudomonads to catechol and cis,cis-muconate, an analog to 2-chloro-cis,cis-muconate. However, ClcR is not effectively induced by cis,cis-muconate (45). Therefore, activation of the clcA promoter by anthranilate, benzoate, and salicylate seems to take place via cross-activation by a non-clc-element-encoded activator such as CatR (46). ORF5994 (taken as representative for the four-gene cluster in this region) showed a similar expression profile as clcA (i.e., induction with 3-CBA, 4-chlorobenzoate, anthranilate, salicylate, and benzoate), but this was in addition to 2-AP as well. This suggests that there is a separate regulatable promoter in front of ORF5994 which is different from the clcA promoter. Several potential LysR-type binding regions (i.e., ATAC-N7-GTAT) are located upstream of ORF5994, but it is presently unclear whether these are involved in binding ClcR, CatR, or even another transcription regulator. To complicate matters further, the amn genes were not only induced with 2-AP (as expected for a functional 2-AP pathway) but also with 3-CBA (at 10 mM), anthranilate, and 4-hydroxybenzoate. Of these, only anthranilate might theoretically be converted into 2-AP, thus leading to the effector needed for derepression of AmnR. The experiments showed that, whereas we usually think of metabolic pathways as “linear” (i.e., induced in the presence of only the true pathway substrate or intermediate), many of them are actually part of a cross-induced network. This, in turn, may be the cause for the misrouting of certain metabolites and formation of toxic intermediates and may be one reason for the absence of growth on 2-AP by Pseudomonas sp. strain B13.
A highly conserved, yet unknown, left end.
The other half of the clc element, from ORF50240 to the 18-bp, left-end repeat, encoded mostly proteins of unknown function, some of which have been recognized previously to be conserved in various different bacterial strains (38) (Table 1). Due to the absence of recognizable transfer or mobilization functions in the integrase-containing first half of the clc element, we hypothesize that the second half must encode such functions, given the self-transferable nature of the clc element (49, 51). The size of the region (∼50 kb) would be sufficiently large to harbor a complete set of genes for plasmid relaxosome and mating-pair formation (∼20 kb) (11). However, no overall DNA or protein homologies to known plasmid conjugative systems or the type IV secretion systems were detected in this area. Hence, we can conclude that the clc element is not a hybrid between a phage and a known tra-like conjugative plasmid like the SXT-element (8) or Tn4371 (65). Only a few putative ORF products encoded in this region showed significant similarity to gene products involved in conjugative DNA transfer. For example, ORF59888 encoded a 956-aa peptide with a 43.7% alignment to the COG3451 VirB4 domain between amino acids 400 and 800 (including 15 gaps). The overall similarity of the predicted ORF59888 peptide to VirB4 of Agrobacterium tumefaciens, however, was less than 20%. At the same time, orthologs of ORF59888 with between 47% and 99% amino acid sequence identity over the full length of the peptide were detected in (currently) 28 complete bacterial genomes, including Xanthomonas campestris, P. aeruginosa, Rubrivivax gelatinosus, Azotobacter vinelandii, and Photorabdus luminescens. This strongly suggests an important functional conservation of this protein. Next, ORF68987 weakly resembled another component of DNA transfer/type IV secretion system in gram-negative bacteria. The region of aa 191 to 662 aligned 86.5% to the pfam02534 TraG/TraD family (E value 8 · 10−7) and 64.4% among 596 residues to the COG3505 VirD4 domain (E-value 6 · 10−5). The hypothetical protein encoded by ORF75419 (760 aa) contained two domains with putative DNA helicase function, between amino acid positions 112 and 186 (DExH box, 53.8% alignment without gaps) and 599 and 721 (HELICc motif, 90.8% alignment without gaps). Helicases are implicated in the unwinding of the nicked plasmid in relaxosome formation during conjugation (13, 37). Thirteen highly identical orthologs (49% to 99%) were currently detected in the NCBI database for the ORF75419 peptide by BLAST searches.
ORF91884 putatively encodes a type IA DNA topoisomerase (671 aa, 99.8% alignment to the COG0550 TOP1Ac domain between positions 150 and 600), and it matched more than 30 entries in the NCBI database with as much as 50% amino acid identity. Topoisomerases of this type are capable of reversible cleavage of double- and single-stranded DNA and function in DNA replication, conjugation, and cointegrate resolution. Finally, a putative single-stranded DNA binding protein (SSB) was predicted from ORF94175 (79.6% alignment with COG0629, SSB), which may be implicated in the primase complex during the nicking of a double-stranded circular intermediate and its transport into the new cell. No regions with significant similarities to the oriT sequence of IncP1-β plasmids were found on the clc element. Two genes which previously had been implicated in the regulation of integrase expression, inrR (ORF94689) and ORF98147 (previously named ORF3) (56), were situated at the left extremity of the clc element. ORF100033 (previously named ORF1) and ORF98147 carry 70-aa domains with significant similarity to ParA and ParBc, respectively (56).
If we thus assume an analogy in the clc element's transfer process to known plasmid conjugative systems, clc transfer might possibly look like the following. At a certain low frequency under stationary-phase conditions, the clc element excises and forms a circular double-stranded intermediate, which has been detected by PCR and Southern hybridization (55). In some cells, the transfer of this circular intermediate may be initiated analogously to conjugative plasmids by the nicking, unwinding, and presentation of a single-stranded DNA at a mating-pair complex in the membrane, which is exemplified by such proteins as the ORF59888-encoded VirB4-domain containing peptide, the ORF75419 DNA helicase, the DNA topoisomerase Ia (ORF91884 peptide), and the ORF94175 SSB-like peptide. Interestingly, similarly to the expression of IntB13, the expression of ORF75419 (putative helicase) but not ORF59888 (putative VirB4) was induced under stationary-phase conditions in Pseudomonas sp. strain B13 (M. Gaillard, unpublished data), suggesting that excision and unwinding are regulated independently from the formation of a mating-pair complex. Further confirmation for the conjugative transfer hypothesis, however, has to await functional analysis of the many conserved hypothetical genes in this region.
Comparison and possible evolution of the clc element.
The clc element was related to a number of GEIs found in P. aeruginosa (e.g., pathogenicity islands PAGI-2 and PAGI-3) (34), suspected GEIs (e.g., in Burkholderia xenovorans LB400, Ralstonia metallidurans, and Xylella fastidiosa), and nonclassified chromosomal regions (e.g., in Xanthomonas axonopodis pv. citri strain 306, R. gelatinosus PM1, and Azoarcus sp. strain EbN1). Various “core” functions shared between the clc element and an even larger set of syntenic genomic islands have been recognized before (38). Two more or less strongly conserved regions between GEIs related to the clc element are interspersed with a more variable region (Fig. 3). The first highly conserved region solely encompassed attR and the integrase gene (85% nucleotide identity with PAGI-2, 82% with PAGI-3, 88% to 92% with X. fastidiosa, 88% with Azoarcus sp., 85% with R. gelatinosus, and 99% with B. xenovorans). The second highly conserved region comprised the circa 50 kb at the left end of the clc element (Fig. 3). In between those regions, the GEIs differed, and in the case of the clc element, the variable part contained the catabolic genes. The most highly related partner to the clc element was a 125-kb chromosomal region detected in the unpublished genome sequence of B. xenovorans LB400, previously B. fungorum LB400, a microorganism known for its capabilities to degrade polychlorobiphenyls (39). The boundaries of the GEI present in B. xenovorans LB400 (AAAJ00000000) may be formed by two direct repeats of 79 bp encompassing two tRNAGly genes. The clc element and the chromosomal region on B. xenovorans shared an overall nucleotide identity of 99% to 100%, except for two distinct regions which were absent in the clc element (Fig. 3A). The first one is a 20-kb fragment occupying the area between the ORFs 5994 and 8052 on the clc element, containing genes for two subunits of an ortho-halobenzoate 1,2-dioxygenase, for two transmembrane proteins, and for a general secretion pathway protein A. The 20-kb region is flanked by two insertion element copies (3.3 kb, with 99% nucleotide sequence identity to ISPpu12, one of which was subject to a secondary 1.5-kb insertion), suggesting that it might have become inserted into an ancestor clc element present in or transferred to B. xenovorans. The second 2-kb region contains a gene coding for a putative reverse transcriptase maturase and is present between homologs of ORFs 83350 and 84338 (clc element annotation). The clc element is also very similar to a GEI present in Ralstonia sp. strain JS705 (AJ006307) (40) which has been only partly sequenced. The GEI of strain JS705 has acquired an additional 10-kb region (40) containing the mcb genes for chlorobenzene dioxygenase and dihydrodiol dehydrogenase at a site between ORF15405 and ORF15962 of the clc element (Fig. 3A). The cumulative TA skew distribution along the clc element (Fig. 1C) showed a region of 9.5 kb with a distinctly lower value compared to that for the rest of the clc element. This region corresponded to the location of the amn genes and suggests that an ancestor of the clc element had acquired the amn gene fragment in a distinctive single insertion. This suggestion is further supported by the ORF transcription direction in this area (Table 1). Although the majority of ORFs on the clc element are transcribed in a direction opposite to that of the integrase gene, the amn genes and a few other downstream-located genes are oriented like the integrase gene. These data demonstrate that the evolution of the clc element and its close relatives (as in B. xenovorans and Ralstonia) primarily takes place by acquisition of new gene fragments into the variable region.
FIG. 3.
Large scale comparisons of the clc element with seven other (suspected) GEIs and genomic regions. (A) Comparison between clc element (middle segment) and a 123-kb suspected GEI present in the chlorobiphenyl-degrading bacterium B. xenovorans LB400 (accession no. AAAJ00000000, between ORF5425 and ORF5534 in contig 482). Uppermost segment shows the 10-kb insertion observed in a clc-like element in Ralstonia sp. strain JS705 (40). (B) Comparison with PAGI-2 (AF440523) from the clinical isolate P. aeruginosa strain C and PAGI-3 (AF440524) from the environmental P. aeruginosa isolate strain SG17M (34). (C) Comparison with a 67-kb GEI found in the plant pathogen X. fastidiosa 9a5c (AE003849, sections 141 to 147) (57) and with a 73-kb chromosomal region of R. gelatinosus PM1, located between positions 634242 and 707481 in contig 562 (NZ_AAEM00000000). (D) Comparisons with a 134-kb chromosomal region in the phytopathogen X. axonopodis pv. citri strain 306 (AE008923, positions 2540723 to 2675160) (14) and a 141-kb region in the genome of the aromatic hydrocarbon-degrading strain Azoarcus sp. strain EbN1 (CR555306, positions 1383280 to 1523958) (47). Gray-shaded areas indicate significant sequence identity by BLASTN analysis (75 to 100%) as represented in the ACT software (9). The degree of grayness in the homologous regions differs from pale gray (75 to 80% nucleotide sequence identity) to black (95 to 100% nucleotide sequence identity), with intermediate steps of 5% (80 to 85%, 85 to 90, and 90 to 95% nucleotide sequence identity). Annotated ORFs are shown in white (direction of transcription from left to right) or shaded gray (right to left). Black and white triangles indicate the direct repeats present at each extremity of the GEI (where present). The presence of a tRNAGly insertion site is symbolized by a black star. The scales mark 19,500-bp distances.
Other GEIs were found without any relationship to the catabolic genes of the clc element but with relatively high conservation of the 50-kb left end region. Some of these relationships have been recognized before (34, 38) but have not been shown in great detail in comparison to the clc element. PAGI-2 does not carry any specific catabolic functions and was isolated from a pathogenic P. aeruginosa strain C (34). Yet, the 50-kb conserved region is highly similar (85 to 100% nucleotide sequence identity) between the clc element and PAGI-2 (Fig. 3B). Almost all ORFs present in this part of the clc element are also present in PAGI-2 with general conservation of the gene order. PAGI-3 is present in the environmental P. aeruginosa isolate SG17 (34) and shares less extensive (79 to 94% nucleotide sequence identity) and more “patchy” homologies with the clc element than PAGI-2 (Fig. 3B). Thirteen ORFs present in this part of the clc element are not present in PAGI-3, among which is the putative DNA methyltransferase (ORF89746) as well as a putative protein-disulfide isomerase (ORF59110). A further relative to a clc element was found in the X. fastidiosa clone 9a5c sequence (57). This suspected GEI has an overall smaller size (67,011 bp) and a smaller region homologous to the clc element (82 to 95% nucleotide sequence identity) but is still flanked by two 18-bp direct repeat sequences and has a distinct variable region (Fig. 3C).
Newer members of this family of GEI may include a chromosomal region of 73 kb in the R. gelatinosus PM1 genome (NZ_AAEM00000000), although no repeat sequences and no nearby tRNA gene were detected (Fig. 3C). This region almost completely lacked a variable part and consisted basically of only the genes present in the conserved region of the clc element. Ten ORFs from the clc element were absent in the R. gelatinosus region, including the putative DNA methyltransferase (ORF89746). On the other hand, the R. gelatinosus region had about 18 kb of DNA different from the clc element (between ORF81655 and ORF91884), coding among others for a cation efflux system, for a mercury resistance regulatory protein (MerR), and for an arsenate reductase. In contrast to those mentioned above, two chromosomal regions in X. axonopodis pv. citri strain 306 (14) and in Azoarcus sp. strain EbN1 (47) of 134 kb and 141 kb, respectively, showed strong similarity (79 to 89% nucleotide sequence identity) to the clc element but differed in gene organization (Fig. 3D). Both regions were not flanked by repeated sequences, and no gene for tRNA was found nearby. In addition, the X. axonopodis region did not contain an integrase gene. On the contrary, the putative GEI in Azoarcus sp. strain EbN1 carried a duplication of the integrase and of a 16-kb region corresponding to the segment extending from ORF50240 to ORF68987 on the clc element. The Azoarcus region contained several catabolic gene functions, such as an ethylbenzene dehydrogenase and an acetophenone carboxylase.
Thirteen ORFs were uniformly present in the conserved regions between the GEIs presented here, including the putative DNA topoisomerase III/Ia (ORF91884), the single-stranded-DNA binding protein (ORF94175), the probable transcriptional regulator inrR (ORF94689), and the chromosome partitioning-related protein (ORF100033). Parts of those functions have been recognized previously as core among an even larger set of GEIs (38, 56). This conservation may indicate that potentially more of the suspected GEIs than just the clc element of Pseudomonas sp. strain B13 and Ralstonia sp. strain JS705 are functionally self-transferable entities. However, although it is presumed that GEIs have arisen from a merge of plasmid and phage functions (8, 33), it is curious that no fully replicative plasmid counterparts have so far been detected for the clc and ICEHin1056 type of elements.
In summary, our results present the first complete sequence of a self-transmissible GEI from Pseudomonas implicated in aromatic compound degradation. Herewith, therefore, an important paradigm in aromatic compound degradation, namely, that of pertinent association of self-transmissibility, aromatic compound degradation, and plasmid conjugation, has to be changed. Although the recent past has seen various reports of complete catabolic plasmid sequences, including pWW0 (xylene degradation in P. putida) (24), pEST4011 (69), and pJP4 (2,4-dichlorophenoxyacetic acid degradation in Achromobacter xylosoxidans and Cupriviadus necator JMP134) (66), or pADP-1 from Pseudomonas sp. strain ADP for atrazine degradation (36), the clc element sequence demonstrates that genes for aromatic degradation may become part of a different class of mobile elements, namely that of ICE. In this respect, the clc element is similar to the biphenyl transposon Tn4371 of Ralstonia oxalatica A5 (65), which, however, is defective for self-transfer but carries recognizable tra and trb genes similar to those of IncP1-β plasmids. A second important realization from the work presented here, which has been stated before in a slightly different form (38), is that GEIs with a similar core structure can display various more or less pronounced functions, e.g., antibiotic resistance, aromatic compound metabolism, or toxin production. However, there is a certain danger when subsequently classifying GEI on the basis of these “pronounced” functions (16). For example, although the clc element may seem “specialized” for aromatic compound metabolism (and, thus, would represent an “ecological” GEI), it is not exempt of putative pathogenicity functions, such as a potential multidrug or solvent efflux system (ORFs 32963, 34495, and 36077) (48). Hence, rather than maintaining absolute categories, it has to be stressed that there is a continuum of different functional characteristics among GEIs (such as from more pathogenic to more pronounced catabolic character) and a further evolution by gene acquisition and rearrangement by which GEIs contribute to adaptation and selection of bacteria in changing environments.
Acknowledgments
We thank Lionel Guy and Claude-Alain Roten for their help with the TA skew analysis.
The work of M.G. was supported by the Swiss National Science Foundation, grant no. 3100-67229.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, B., M. Snozzi, A. J. B. Zehnder, and J. R. van der Meer. 1996. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 7.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 9.Carver, T. J., K. M. Rutherford, M. Berriman, M.-A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics. 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H. K., P. Mohseni, and G. J. Zylstra. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185:5871-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B. 1993. Bacterial conjugation. Plenum Press, New York, N.Y.
- 12.Coco, W. M., M. R. Parsek, and A. M. Chakrabarty. 1994. Purification of the LysR family regulator, ClcR, and its interaction with the Pseudomonas putida clcABD chlorocatechol operon promoter. J. Bacteriol. 176:5530-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csitkovits, V. C., D. Dermic, and E. L. Zechner. 2004. Concomitant reconstitution of TraI-catalyzed DNA transesterase and DNA helicase activity in vitro. J. Biol. Chem. 279:45477-45484. [DOI] [PubMed] [Google Scholar]
- 14.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 15.Demaneche, S., C. Meyer, J. Micoud, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 17.Dorn, E., M. Hellwig, W. Reineke, and H. J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 18.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-198. [PubMed] [Google Scholar]
- 20.Frantz, B., and A. M. Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerhardt, P., R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.). 1981. Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 22.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence fishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with Autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greated, A., L. Lambertsen, P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 25.Greub, G., F. Collyn, L. Guy, and C. A. Roten. 2004. A genomic island present along the bacterial chromosome of the Parachlamydiaceae UWE25, an obligate amoebal endosymbiont, encodes a potentially functional F-like conjugative DNA transfer system. BMC Microbiol. 4:48 [Online.] doi: 10.1186/1471-2180-1184-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guy, L., D. Karamata, P. Moreillon, and C.-A. H. Roten. 2005. Genometrics as an essential tool for the assembly of whole genome sequences: the example of the chromosome of Bifidobacterium longum NCC2705. BMC Microbiol. 5:60 [Online.] doi: 10.1186/1471-2180-1185-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 28.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickey, W. J., G. Sabat, A. S. Yuroff, A. R. Arment, and J. Perez-Lesher. 2001. Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, G. R., and J. C. Spain. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110-123. [DOI] [PubMed] [Google Scholar]
- 32.Kasberg, T., D. L. Daubaras, A. M. Chakrabarty, D. Kinzelt, and W. Reineke. 1995. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J. Bacteriol. 177:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klockgether, J., O. Reva, K. Larbig, and B. Tummler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leveau, J. H. J., F. König, H.-P. Füchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matson, S. W., and H. Ragonese. 2005. The F-plasmid TraI protein contains three functional domains required for conjugative DNA strand transfer. J. Bacteriol. 187:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeño-Tárraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller, T. A., C. Werlen, J. Spain, and J. R. van der Meer. 2003. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol. 5:163-173. [DOI] [PubMed] [Google Scholar]
- 41.Muraki, T., M. Taki, Y. Hasegawa, H. Iwaki, and P. C. Lau. 2003. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishi, A., K. Tominaga, and K. Furukawa. 2000. A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 182:1949-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oelschlaeger, T. A., and J. Hacker. 2004. Impact of pathogenicity islands in bacterial diagnostics. APMIS 112:930-936. [DOI] [PubMed] [Google Scholar]
- 44.Oltmanns, R. H., H. G. Rast, and W. Reineke. 1988. Degradation of 1,4-dichlorobenzene by constructed and enriched strains. Appl. Microbiol. Biotechnol. 28:609-616. [Google Scholar]
- 45.Park, H. S., and H. S. Kim. 2001. Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process. J. Bacteriol. 183:5074-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsek, M. R., S. M. McFall, D. L. Shinabarger, and A. M. Chakrabarty. 1994. Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc. Natl. Acad. Sci. USA 91:12393-12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabus, R., M. Kube, J. Hieder, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 48.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 49.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravatn, R., S. Studer, A. J. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravatn, R., A. J. Zehnder, and J. R. van der Meer. 1998. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl. Environ. Microbiol. 64:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schubert, S., S. Dufke, J. Sorsa, and J. Heesemann. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51:837-848. [DOI] [PubMed] [Google Scholar]
- 55.Sentchilo, V., R. Ravatn, C. Werlen, A. J. Zehnder, and J. R. van der Meer. 2003. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 185:4530-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 57.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. Nhani, Jr., F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. Pereira, Jr., J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. M. de Rosa, V. E. de Rosa, Jr., R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 58.Springael, D., and E. M. Top. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12:53-58. [DOI] [PubMed] [Google Scholar]
- 59.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. De Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takenaka, S., S. Murakami, Y. J. Kim, and K. Aoki. 2000. Complete nucleotide sequence and functional analysis of the genes for 2-aminophenol metabolism from Pseudomonas sp. AP-3. Arch. Microbiol. 174:265-272. [DOI] [PubMed] [Google Scholar]
- 62.Takenaka, S., S. Murakami, R. Shinke, K. Hatakeyama, H. Yukawa, and K. Aoki. 1997. Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J. Biol. Chem. 272:14727-14732. [DOI] [PubMed] [Google Scholar]
- 63.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 64.Toussaint, A., and C. Merlin. 2002. Mobile elements as a combination of functional modules. Plasmid 47:26-35. [DOI] [PubMed] [Google Scholar]
- 65.Toussaint, A., C. Merlin, S. Monchy, M. A. Benotmane, R. Leplae, M. Mergeay, and D. Springael. 2003. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl. Environ. Microbiol. 69:4837-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trefault, N., R. de la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Pérez-Pantoja, M. A. Sánchez, M. Stuardo, and B. Gonzàlez. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 67.van der Meer, J. R., R. Ravatn, and V. Sentchilo. 2001. The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch. Microbiol. 175:79-85. [DOI] [PubMed] [Google Scholar]
- 68.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:248-254. [DOI] [PubMed] [Google Scholar]
- 69.Vedler, E., M. Vahter, and A. Heinaru. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weisshaar, M.-P., F. C. H. Franklin, and W. Reineke. 1987. Molecular cloning and expression of the 3-chlorobenzoate-degrading genes from Pseudomonas sp. strain B13. J. Bacteriol. 169:394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, J. Z., and J. M. Tiedje. 1995. Gene transfer from a bacterium injected into an aquifer to an indigenous bacterium. Mol. Ecol. 4:613-618. [DOI] [PubMed] [Google Scholar]