Abstract
β-Defensins are a family of small cationic peptides involved in the innate response to microbial infection. Although their role in microbial killing is well established, the mechanisms through which this occurs remain largely undefined. Here, using protein array technology, we describe a role for human β-defensins in the induction of an inflammatory cytokine response by human peripheral blood mononuclear cells (PBMCs). Human β-defensins 1, 2, and 3 were examined for induction of an array of cytokines and chemokines. Some cytokines, such as interleukin 8 (IL-8) and monocyte chemoattractant protein 1, were up-regulated by all three defensins, while others, such as IL-6 and IL-10, were induced more selectively. It was notable that each defensin induced a unique pattern of cytokines. This report documents, for the first time, an analysis of the composite cytokine response of human PBMCs to β-defensins. The induction or up-regulation of a number of cytokines involved in the adaptive immune response suggests a possible role for these defensins in linking innate and acquired immunity.
Host defense peptides play an important role in the innate immune response of mammals (20), and among these, defensins seem to have a particularly prominent role in human antimicrobial defense (25).
Defensins are small (3.5 to 5.5 kDa), highly cationic peptides characterized by the presence of three intramolecular disulfide bonds among six distinctive and highly conserved cysteines (11). To date, three different classes of defensins have been described in primates: α-defensins, β-defensins, and θ-defensins. The θ-defensins, while evolutionarily derived from the α-defensins, are present only as pseudogenes in humans (19).
Four human β-defensins (hBDs) have been examined functionally thus far: hBD-1, -2, -3, and -4. However, computational methods, based on hidden Markov chain models linked to BLAST searches of the whole human genome, indicate the existence of various additional and as yet uncharacterized β-defensin-like molecules (15, 18, 27).
In common with all defensin classes, β-defensins have the ability to interact with microbial cell wall components, most often membrane lipids, leading to damage of biological membranes. Through this mechanism, defensins are able to control or kill a wide variety of potentially pathogenic organisms, including gram-positive and gram-negative bacteria as well as encapsulated viruses and fungi (11). However, it has also been observed that the bactericidal capacity of these peptides is strongly inhibited in physiological fluids and biological culture media, especially at higher salt concentrations (2, 12). Only hBD-3 retains its killing ability in a wide range of salt concentrations (6, 13). This apparent weak point in the direct antimicrobial activity within some of the biological fluids in which they are found suggests that defensins may exhibit other functions in such microenvironments. It has now been demonstrated that hBD-2 is a chemoattractant for immature dendritic cells, memory T cells, and neutrophils primed with tumor necrosis factor alpha (13, 24, 34). Furthermore, some β-defensins can help support the adaptive immune response, acting as potent adjuvants when coupled to nonimmunogenic tumor antigens (5). The mechanisms by which the defensins act in these situations are not, however, well understood. Indeed, very little is known about the effects that defensins have on immune cells.
Since the expression of β-defensins can be detected in blood after stimulation with lipopolysaccharide (LPS) or heat-inactivated Pseudomonas aeruginosa (10), we hypothesized that these molecules may play a role in this environment by acting on immune cells. We envisage that innate recognition of bacterial motifs resulting in β-defensin release may instruct immune cells to release cytokines and chemokines that play a role in inflammatory and/or adaptive immune responses. In order to address this hypothesis, we have investigated, for the first time, the stimulatory activities of three β-defensins (hBD-1, -2, and -3) on human peripheral blood mononuclear cells (PBMCs).
MATERIALS AND METHODS
Culture conditions.
For all experiments, the culture medium used was RPMI 1640 (Sigma, St. Louis, MO). Cultured cells were maintained at 37°C in a humidified atmosphere of 5% CO2. For cell culture, this medium was supplemented with 2 mM l-glutamine and 10% (vol/vol) heat-inactivated fetal calf serum (Sigma).
Isolation of mononuclear cells.
PBMCs were isolated from anonymous buffy coats or volunteer donor peripheral blood (collected with 10 IU preservative-free heparin for each 1 ml of blood; Sigma) by density gradient centrifugation over Histopaque-1077 (Sigma) according to the manufacturer's instructions.
Production of human β-defensins.
β-Defensins were chemically synthesized using optimized 9-fluorenylmethoxy carbonyl (Fmoc) chemistry protocols on a PE Biosystems Pioneer instrument with the column thermostated to 50°C. Chlorotrityl chloride resin was appropriately functionalized with insertion of the C-terminal amino acid carried out manually (substitution, 0.20 to 0.25 meq/g resin). A fourfold excess of 1:1:1.7 Fmoc-amino acid/TBTU/DIPEA [TBTU, 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate; DIPEA, diisopropylethylamine] and a version of the antiaggregation solvent “magic mixture” (DMF/NMP [3:1], 1% Triton X-100, 1 M ethylene carbonate) (DMF, dimethylformamide; NMP, N-methyl pyrrolidone) were used for each coupling step. Double coupling with HATU [O-(7-azabenzotriazole-1-yl)-1,1,3,3,-tetramethyluronium hexafluorophosphate] or PyBOP (benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate) instead of TBTU was carried out at sections predicted to be difficult by the PeptideCompanion software (CoshiSoft, AZ). The synthesis was interrupted regularly to check quality using electrospray ionization mass spectrometry (ESI-MS) (Sciex API I; Perkin-Elmer). Peptides were cleaved from the resin and deprotected with a mixture of trifluoroacetic acid, water, thioanisole, phenol, ethanedithiol, and triisopropylsilane (82.5:5:5:2.5:2.5:2.5) for 2 to 4 h at room temperature. The crude peptides were then treated with the reducing agent tris(2-carboxyethyl)-phosphine hydrochloride (TCEP · HCl) at pH 3 and directly purified on a preparative C18 reverse-phase high-performance liquid chromatography (RP-HPLC) column (Waters Delta-Pak C18; 15 μm, 300 Å, 25 by 100 mm), using N2-saturated solvents at 20 to 60% in a 40-min acetonitrile gradient. The fraction containing the correct hBD peptide, as determined by mass spectrum analysis, was used directly for oxidative folding. Before the oxidative folding step, a small quantity of the linear hBD-2 was collected and treated with iodoacetamide to alkylate cysteine residues, for use as a control for hBD-2 activity. The oxidative folding reaction was carried out in freshly prepared N2-saturated aqueous buffer (0.1 M ammonium acetate buffer, pH 7.8 to 8.5, in Milli-Q water, containing 2 mM EDTA and 1 M guanidine · HCl), in which cysteine (1 to 2 mM) and cystine (0.1 to 0.2 mM) were dissolved immediately prior to use. The RP-HPLC fraction containing the reduced and desalted peptide (30 to 40% acetonitrile) was added to this solution, so as to result in a 10 or 20 μM final peptide concentration, and the pH was adjusted with aqueous ammonia solution. The mixture was left at room temperature in a sealed container for 24 to 48 h, and oxidation was monitored using analytical RP-HPLC and ESI-MS. At the end of the reaction, the mixture was adjusted to pH 3, filtered, and purified directly using preparative RP-HPLC. The purified hBD was then lyophilized for later use.
Peptide concentrations in stock solutions were determined using Trp, Tyr, and Cys absorption at 280 nm, with molar extinction coefficients (ɛ, 280) of the peptides being calculated, using the ProtParam tools on the Expasy server (http://www.expasy.ch/tools/protparam.html) and verified using the bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Peptides were resuspended in Milli-Q water to a concentration of 640 μM, conditions in which the growth of microorganisms is completely prevented. Overall, synthesis, work-up, and storage conditions were such as to make LPS contamination highly unlikely. Experiments were also performed with hBD-2 purchased from Alpha Diagnostics.
Peptide characterization.
The purity, correct primary structure, and complete disulfide bridge formation of the peptides was confirmed by analytical HPLC followed by mass spectrometry (ESI-MS). The peptide with the correct folding and connectivities generally eluted as the principal band and was well separated from the fractions containing peptides with other connectivities. Partial limited proteolysis and/or circular dichroism helped individuate the correct folding of the peptides. The presence of endotoxin in stock solutions was verified using a commercially available Limulus amebocyte lysate assay kit (Cambrex Corporate, East Rutherford, NJ) according to the manufacturer's instructions and using control standard preparations of endotoxin as positive controls, and it was found to be negligible.
hBD activity on human PBMCs.
For dose-response experiments, PBMCs were cultured in 96-well, flat-bottomed plates (2 × 105 in 200 μl) for 18 h over a range of hBD concentrations. During protein array experiments, PBMCs were cultured at a density of 2 × 106 cells in 2 ml in the presence or absence of hBD-1, -2, or -3 at 20 μg/ml. Supernatants were harvested at both “early” (18 h) and “late” (6 days) time points previously shown to be optimal for analysis of both the afferent phase cytokine production mainly produced by monocytes and the later efferent phase cytokine production which includes autologous T-cell responses (17). Supernatants were frozen at −80°C for later cytokine analysis by enzyme-linked immunosorbent assay (ELISA) or protein array.
For quantitative PCR experiments, PBMCs were cultured at a density of 2 × 106 cells in 2 ml in the presence or absence of hBD-2 at 20 μg/ml. Cell pellets were harvested and washed in phosphate-buffered saline, and total RNA was extracted using a Stratagene Absolutely RNA kit (Stratagene, La Jolla, CA) for later quantitative real-time reverse transcription (RT)-PCR analysis.
Protein arrays.
Human protein cytokine arrays were purchased from RayBiotech (Norcross, GA), and supernatants from cell cultures were analyzed according to the manufacturer's instructions. Briefly, cytokine array membranes were first treated with a blocking buffer and then washed and incubated with 1 ml of culture supernatant from either hBD-treated or untreated PBMC cultures. The membranes were then incubated at room temperature for 2 h and washed, and 1 ml of biotin-conjugated detection antibodies was added. Following a further incubation of 2 h at room temperature, the membranes were again washed and finally incubated with 2 ml of streptavidin-horseradish peroxidase at room temperature for 30 min. Finally, the membranes were developed using the kit's enhanced chemiluminescence system, and the results were visualized on X-ray film. Two types of arrays were used. “Cytokine array V” allowed a semiquantitative analysis of the levels of 79 different cytokines or chemokines. “Cytokine array III” allowed the estimation of relative quantities with standard deviations for 42 cytokines.
Cytokine ELISAs.
Cytokine levels within supernatants were measured using ELISA. Paired antibodies consisting of purified capture and biotinylated secondary detection antibodies (BD Pharmingen) were used according to the manufacturer's recommendations. Standards were purchased from Peprotech. The ELISA protocols were performed as previously described (16).
Gene expression analysis.
cDNA generation from total RNA and subsequent real-time PCR was performed using the TaqMan Gold RT-PCR kit with SYBR green I chemistry (ABI, Foster City, CA). Primers for cDNA amplification were as follows: interleukin-6(IL-6), CCCTGAGAAAGGAGACATGTAACA (sense) and ACCAGGCAAGTCTCCTCATTGA (antisense); IL-8, GCCAACACAGAAATTATTGTAAAGCTT (sense) and TTCTCAGCCCTCTTCAAAAACTTC (antisense); IL-10, CTACGGCGCTGTCATCGATTT (sense) and GTAGATGCCTTTCTCTTGGAGCTTATT (antisense); and hypoxanthine phospho-ribosyltransferase, CAGCCCTGGCGTCGTGATTAGT (sense) and GCAAGACGTTCAGTCCTGTCCATAA (antisense).
Gene amplification was performed using the MX4000 instrument (Stratagene, La Jolla, CA). Forty-five cycles of 95°C for 10 min, 95°C for 30 s, and 60°C for 1 min were performed. A melting temperature assay was performed on amplicons in order to verify the specificity of the amplification. Quantitative assessment of IL-6, IL-8, and IL-10 mRNA expression was obtained by measuring target mRNA cycle threshold (CT) values (IL-6 or IL-8 or IL-10) relative to CT values of the housekeeping gene (HPRT). Results were then expressed as n-fold increase over controls within each experiment (i.e., n-fold increase of cytokine mRNA expression in hBD-2-treated cultures in relation to untreated controls).
RESULTS
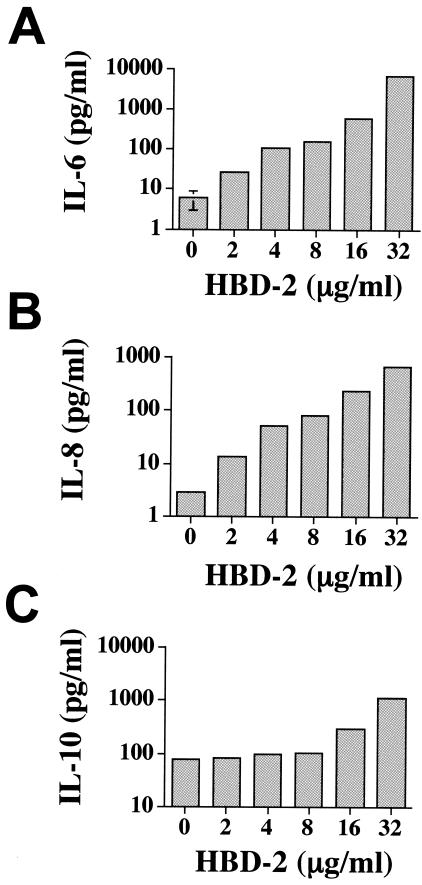
Initially, we examined whether hBD-2 was able to stimulate production of the cytokines IL-6, IL-8, and IL-10 by human PBMCs. Our data demonstrated that hBD-2 could induce secretion of all three of these cytokines in a dose-dependent manner (Fig. 1).
FIG. 1.
hBD-2 induces dose-dependent production of IL-6, IL-8, and IL-10 from human PBMCs. Whole PBMCs were exposed to a dose-response of hBD-2 over a period of 18 h. Supernatants were harvested and analyzed for the cytokines IL-6 (A), IL-8 (B), and IL-10 (C) by ELISA. The results shown represent the means of triplicate cultures and are representative of three experiments performed.
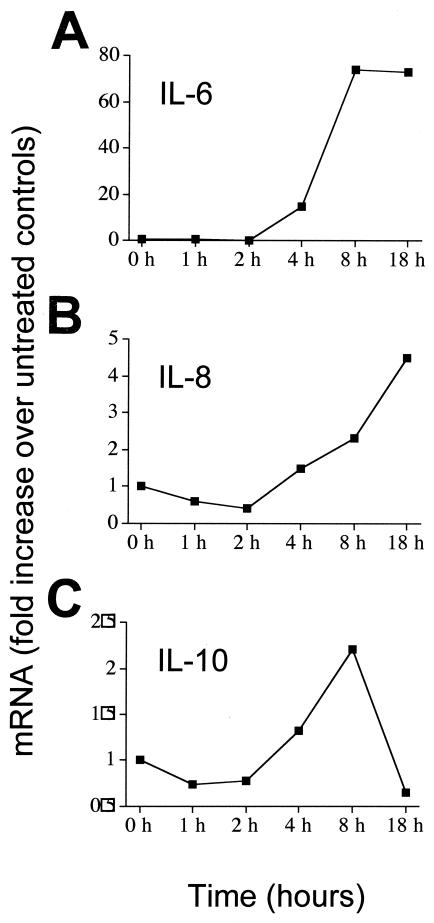
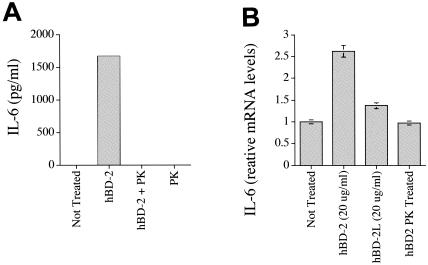
We went on to examine levels of mRNA for IL-6, IL-8, and IL-10 over an 18-h time course (Fig. 2). The results demonstrated that in each case, increases of cytokine protein in supernatants were associated with increased mRNA levels. Levels of IL-6 and IL-8 mRNA began to increase at 4 h poststimulation and increased thereafter. IL-10 mRNA elevation was more transient in nature, peaking at around 8 h and falling back down to baseline levels by 18 h poststimulation. To exclude the possibility that increases in cytokines could be caused by contaminating endotoxin, we examined the hBD preparations using a commercially available Limulus amebocyte lysate test kit. Results showed that endotoxin was present in insignificant quantities (less than 0.00025 endotoxin units/ml in all experiments). Further, we also examined whether denaturing the hBD-2 protein using proteinase K would result in a loss of cytokine-inducing activity at the level of both protein (Fig. 3A) and mRNA (Fig. 3B). Proteinase K treatment, which does not affect the biological function of endotoxin but disrupts protein activity, completely abrogated the ability of the hBD-2 to induce cytokines. However, proteinase K-treated LPS was still able to induce a strong cytokine response (data not shown). As further evidence of the purity of the preparation and acting as a negative control for the peptides, we also examined a linearized version of the hBD-2. Such denaturation resulted in loss of activity, demonstrating once again that preparations were not contaminated with endotoxin.
FIG. 2.
Effect of hBD-2 stimulation on cytokine mRNA levels in PBMC. Whole PBMCs were stimulated in the presence or absence of hBD-2, and cells were harvested at various time points between 1 and 18 h poststimulation. RT-PCR analysis was then used to determine the levels of IL-6 (A), IL-8 (B), and IL-10 (C) mRNA over time. Results are expressed as n-fold increase over expression levels in unstimulated controls.
FIG. 3.
Effects of hBD-2 are abrogated through linearization or treatment with proteinase K. Whole PBMCs were incubated in either the presence or absence of hBD-2, proteinase K-denatured hBD-2 (hBD+PK), proteinase K only (PK), or no defensin, and IL-6 protein levels were measured using ELISA (A). mRNA levels of IL-6 produced by PBMCs following incubation with hBD-2, no defensin control, proteinase K-treated hBD-2, and linearized hBD-2 (hBD-2L) (B).
Having shown that hBD-2 could induce cytokine secretion by PBMC, we then went on to study β-defensin-induced cytokine regulation in greater detail, examining hBD-1, -2, and -3. In order to obtain a more detailed picture of the composite cytokine response induced by these defensins, we utilized a newly developed human cytokine-protein array methodology allowing semiquantitative analysis of many cytokines in a single experiment.
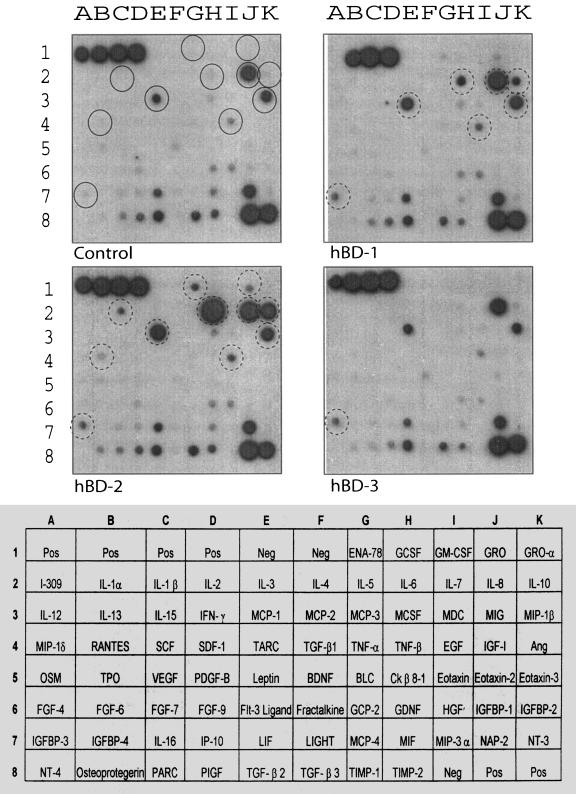
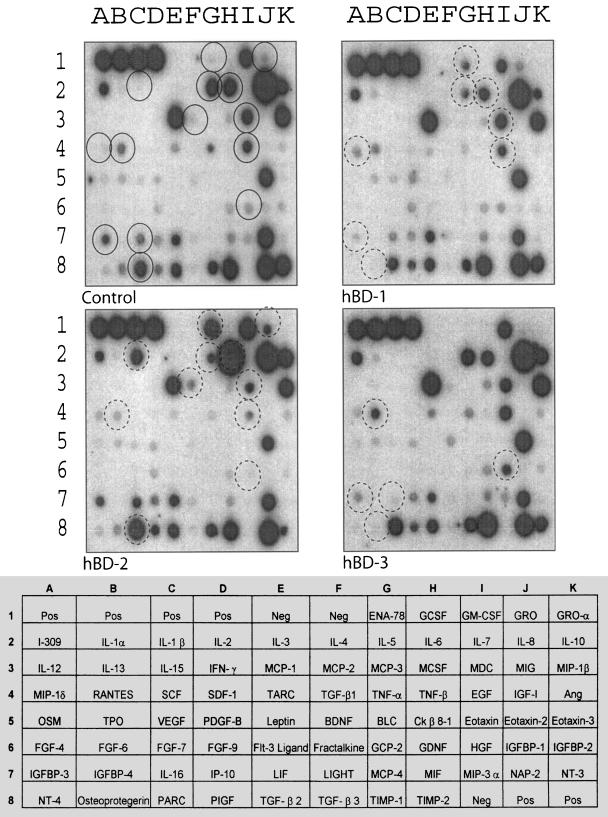
Cytokine production after 18 h in response to treatment with 20 μg/ml of hBD-1, -2, and -3 is shown in Fig. 4, and the results are summarized in Table 1. hBD-2 produced the most-potent cytokine response and confirmed the previously observed effects on IL-6, IL-8, and IL-10. In addition, it was clear that hBD-2 strongly up-regulated monocyte chemoattractant protein 1 (MCP-1) and moderately up-regulated MIP-1-β. hBD-2 also induced the production of RANTES, IL-1-β, ENA-78 and GRO. hBD-1 up-regulated or induced IL-8, MIP-1-β, and MCP-1 as well as low levels of IGFBP-3 and EGF. The induction of IL-6 and IL-10 by hBD-1 was also evident, but to a lesser extent than that observed in response to hBD-2. In contrast, hBD-3 had little effect on cytokine expression. After 18 h, some induction of IGFBP-3, and some up-regulation of IL-8 and MCP-1, was observed. IP-10 appeared to be reduced by hBD-3, as did MIF.
FIG. 4.
Protein array analysis demonstrating the effects of hBD-1, -2, and -3 on PBMC cytokine profile after 18 h of stimulation. Whole PBMCs were either left untreated or treated with 20 μg/ml of hBD-1, hBD-2, or hBD-3 for 18 h. Supernatants were then analyzed using the RayBiotech protein array. Spots with the most prominently differentially regulated cytokines are identified by circles. Circles correspond to IGFBP-3 (A7), RANTES (B4), IL-1-β (C2), MCP-1 (E3), ENA-78 (G1), IL-6 (H2), EGF (E4), GRO (J1), IL-8 (J2), IL-10 (K2), and MIP-1-β (K3).
TABLE 1.
Regulation of cytokines on exposure to different human β-defensins
| Defensin | Day | Cytokine (array position)a
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-1β (C2) | IL-5 (G2) | IL-6 (H2) | IL-8 (J2) | IL-10 (K2) | IL-16 (C7) | EGF (I4) | ENA-78 (G1) | GRO (J1) | HGF (I6) | IGFBP-3 (A7) | MCP-1 (E3) | MCP-2 (F3) | MDC (I3) | MIP-1-β (K3) | MIP-1-δ (A4) | OPG (B8) | RANTES (B4) | ||
| hBD-1 | 1 | − | − | U+ | U+ | U+ | − | U+ | U− | U− | − | U+ | U+ | − | − | U+ | − | − | − |
| 6 | − | D+ | − | − | − | − | − | U+ | − | − | D+ | − | − | − | − | U+ | D+ | − | |
| hBD-2 | 1 | U+ | − | U++ | U− | U++ | − | U+ | U+ | U− | − | U+ | U++ | − | − | U− | − | − | U− |
| 6 | U++ | D+ | U++ | − | − | − | D+ | U++ | U+ | D+ | − | − | U− | D+ | − | − | − | D− | |
| hBD-3 | 1 | − | − | − | U− | − | − | − | − | − | − | U+ | U− | − | − | − | − | − | − |
| 6 | − | − | − | − | D+ | − | − | − | U++ | D− | − | − | − | − | − | D+ | U+ | ||
U, up-regulated; D, down-regulated; +, moderate effect; ++, strong effect. A minus (−) with U or D indicates a weak effect; a minus alone indicates no effect.
On day 6 of exposure (Fig. 5 and Table 1), several cytokines were differentially regulated by the three β-defensins. hBD-2-induced secretion of ENA-78, GRO, and IL-1-β and IL-6 remained up-regulated as it had been at 18 h. MCP-2 was now being induced by hBD-2 at this later time point. Unlike the earlier time point, by day 6 several cytokines were now being down-regulated by hBD-2. This was most apparent for IL-5, although MDC, EGF, and HGF were also being reduced. hBD-1-stimulated cultures showed continued elevation of ENA-78 (although not to the same degree as with hBD-2), while an induction of MIP-1 was also now apparent. Like hBD-2, this peptide also down-regulated IL-5 but was also able to reduce levels of IGFBP-3 and OPG. hBD-3 up-regulated HGF and down-regulated OPG and IL-16. Thus, by day 6 of culture, IL-6 remains up-regulated only by hBD-2 but not by hBD-1 or -3.
FIG. 5.
Protein array analysis demonstrating the effects of hBD-1, -2, and -3 on PBMC cytokine profile after 6 days of stimulation. Whole PBMCs were either left untreated or treated with 20 μg/ml of hBD-1, hBD-2, and hBD-3 for 6 days. Spots with differentially regulated cytokines are identified by circles. Circles correspond to MIP-1-δ (A4), IGFBP-3 (A7), RANTES (B4), OPG (B8), IL-1-β (C2), IL-16 (C7), MCP-2 (F3), ENA-78 (G1), IL-5 (G2), IL-6 (H2), MDC (I3), EGF (I4), HGF (I6), and GRO (J1). Minor changes were suggested in MCP-1, PARC, and TIMP-2.
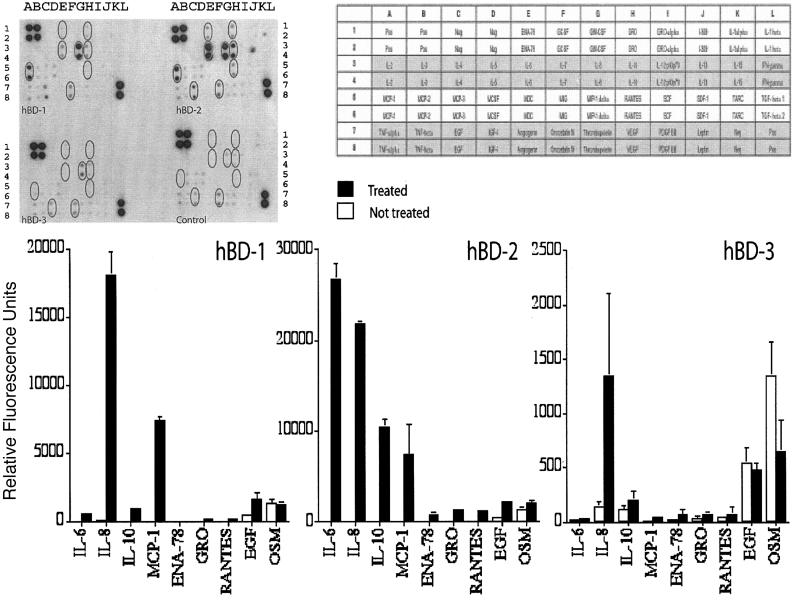
In order to confirm these preliminary data, we performed a second array analysis using a cytokine array with duplicate points for each cytokine, thus helping confirm that the results were not anomalies and allowing for better semiquantification. Experiments were performed on PBMCs from two different healthy donors, and supernatants were analyzed after 18 h. These data are illustrated in Fig. 6, along with semiquantitative analysis following densitometric analysis of spot size.
FIG. 6.
Semiquantitative analysis of the effects of hBD-1, -2, and -3 on PBMC cytokine profile after 18 h of stimulation. Relative levels of cytokines were determined using densitometry on the cytokine array III (RayBiotech). Cytokines that were most strongly regulated by the human β-defensins are illustrated in the accompanying bar chart. Mean relative quantities and standard deviations are based on two different experiments performed on two healthy subjects.
The data gained confirmed the previous array analysis. hBD-2 was again the most active peptide, significantly up-regulating IL-6, IL-8, IL-10, and MCP-1 and inducing some expression of ENA-78, GRO, RANTES, and EGF. hBD-1 stimulated the production of IL-8 and MCP-1 along with some IL-6 and IL-10. Again, hBD-3 was quite inactive after 18 h, with only the IL-8 expression being significantly up-regulated.
Overall, the data demonstrated that human β-defensins are active on PBMCs, inducing cytokines that would be expected to contribute to the early phases of response to bacterial and other pathogens. Furthermore, our data demonstrate that the three major human β-defensins act upon these cells to different extents and with distinct patterns.
DISCUSSION
Since the isolation of human β-defensin 1 from human plasma in 1995 (3), the principal activity of β-defensins has been thought to be the direct killing of pathogens by damaging their membranes, leading to microbial inactivation (23, 32). However, the fact that this particular function is inhibited in some conditions where defensins are often secreted has raised the possibility that defensins have novel, as yet undiscovered functions in host defense. Recently, it has been shown that β-defensins exert a chemokine-like activity (34), and a number of different cell surface receptors have been described for these molecules (4, 34). Reports have also demonstrated that hBD-2 is able to stimulate cells by interacting with surface receptors and that α-defensins up-regulate the expression of several cytokines in a mouse model (29). We hypothesized that hBD-2 could be able to induce cytokine responses known to be involved in microbial defense.
Our initial observations demonstrated that hBD-2 is active at inducing the classic inflammatory cytokine IL-6 and the IL-8 chemokine, as well as IL-10, a strongly anti-inflammatory cytokine. Our experiments with synthetic hBDs were performed at a concentration of 20 μg/ml, a level which is often reached in physiological conditions by hBDs at sites of infection (14, 22). It is also worthy of note that the concentration of synthetic peptides in the medium may be drastically reduced both by their interactions with plastic walls of the plates and by their binding to serum fetuin (31).
Using a cytokine-protein detection approach capable of examining many cytokines within a single experiment, we confirmed and extended our initial findings, demonstrating that hBD-2 induces or up-regulates various other cytokines/chemokines such as MCP-1, MIP-1-β, RANTES, IL-1-β, ENA-78, GRO, IGFBP-3, and EGF. The lack of tumor necrosis factor alpha production was surprising, although it is possible that it may be transient and observable at time points not investigated. Furthermore, we have shown that all three of the human β-defensins studied here behave in quite different ways: hBD-1 affects a more limited spectrum, which includes IL-8, IL-10, IL-6, and MIP-1-β, while hBD-3 had only modest effects on the PBMC cytokine profiles, but nevertheless stimulated some IL-8, MCP-1, and IGFBP-3 production.
These data support previous findings demonstrating the ability of hBD-2 to bridge innate and adaptive immunity and suggest a possible similar role for at least one of the other two studied. One might hypothesize that this particular ability of hBD-2 is due to its structure, in particular to the α-helical segment near the N terminus, which is more stable than in the other defensins (26). It is equally likely that they reflect real differential functions between these molecules.
It may be significant that evolutionary studies on orthologues of these three defensins in several other primate species have indicated positive selection for variation for hBD-2, neutral selection for hBD-1, and purifying selection for hBD-3 (1, 6, 7, 9). In fact, the stable helical segment at the N terminus of hBD-2 seems to be a positively selected characteristic of only great apes and humans (1). Interestingly, the strongly conserved β-defensin 3, which is the only one to show a broad, potent, and salt-insensitive direct antimicrobial activity also at physiological salt concentrations (6), is the least active in stimulating cytokine or chemokine production.
Only IL-8 was significantly up-regulated by all three defensins studied here. This result coincides with the findings of van Wetering et al. (28), who showed IL-8 up-regulation by three human neutrophil defensins (HNP-1, -2, and -3) in human airway epithelial cells. Similarly, using a mouse model, Lillard et al. (21) demonstrated that α-defensins can enhance the immunoglobulin G systemic response against ovalbumin in a manner assisted by CD4+ Th1- and Th2-type cytokines, such as gamma interferon, IL-5, IL-6, and IL-10. These findings were confirmed and extended by Brogden et al. (8), who demonstrated that administration of ovalbumin with hBD-1, but not hBD-2, significantly up-regulates IL-10.
At present we do not know which receptors are involved in stimulation of PBMC cytokine production by β-defensins, although candidates include TLR-4 and CCR-6, which are known receptors for hBD-2. Cellular receptors have not yet been identified for hBD-1 and hBD-3. Likewise, the interaction between epithelial cells and α-defensins is thought to occur via an as yet unidentified membrane receptor (29). The fact that each defensin induced a unique pattern of cytokines and chemokines may reflect the existence of different receptors for the three peptides, or perhaps different modes or extents of interaction with shared receptors.
The data presented here develop the growing research illustrating the links between antimicrobial peptides and chemoattractants. The induction of IL-1-β, IL-6, IL-8, and IL-10 may be particularly important in triggering the physiological changes associated with sepsis (30). In addition, Yang et al. (33) have shown that certain chemokines are microbicidal on both gram-positive and gram-negative bacteria, including GRO, MCP-2, and MDC, which we have shown here to be regulated by human β-defensins. Not only do β-defensins act as chemoattractants themselves, but here we show that different human β-defensins induce unique patterns of cytokine induction, cytokines which would be crucial in the development and expansion of the early response to bacterial pathogens. Thus, it would appear that a host defense function for β-defensins is to influence cells within the immediate microenvironment to their site of induction, to recruit and activate other cells, perpetuating the response to pathogenic microorganisms. IL-8 attraction of neutrophils would lead to further defensin production, thereby maintaining the antimicrobial environment, which is broadened by the action of certain chemokines. In this context, it is worth noting that hBD-2 up-regulated IL-1-β, which in turn is able to promote hBD-2 expression in epithelial cells, thus triggering an amplification circuit.
In summary, we describe the composite inflammatory response of human PBMCs to hBD-1, hBD-2, and hBD-3. The data presented should be of considerable use in the further study of the action of human β-defensins both in vitro and in vivo.
Acknowledgments
This study was supported by a Department of Defense appropriation to the UMDNJ Center for BioDefense, New Jersey Dental School, a COFIN 2003 grant from the Italian Ministry of Research (MIUR), and the RC 03/04 grant from IRCCS “Burlo Garofolo.” W. J. Jordan was supported by the National Institutes of Health (grant no. R1DE14997A) and J. Eskdale by the Department of Oral Biology, New Jersey Dental School.
REFERENCES
- 1.Antcheva, N., M. Boniotto, I. Zelezetsky, S. Pacor, M. V. Verga Falzacappa, S. Crovella, and A. Tossi. 2004. Effects of positively selected sequence variations in human and Macaca fascicularis β-defensins 2 on antimicrobial activity. Antimicrob. Agents Chemother. 48:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensch, K. W., M. Raida, H. J. Magert, P. Schulz-Knappe, and W. G. Forssmann. 1995. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 368:331-335. [DOI] [PubMed] [Google Scholar]
- 4.Biragyn, A., P. A. Ruffini, C. A. Leifer, E. Klyushnenkova, A. Shakhov, O. Chertov, A. K. Shirakawa, J. M. Farber, D. M. Segal, J. J. Oppenheim, and L. W. Kwak. 2002. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science 298:1025-1029. [DOI] [PubMed] [Google Scholar]
- 5.Biragyn, A., M. Surenhu, D. Yang, P. A. Ruffini, B. A. Haines, E. Klyushnenkova, J. J. Oppenheim, and L. W. Kwak. 2001. Mediators of innate immunity that target immature, but not mature, dendritic cells induce antitumor immunity when genetically fused with nonimmunogenic tumor antigens. J. Immunol. 167:6644-6653. [DOI] [PubMed] [Google Scholar]
- 6.Boniotto, M., N. Antcheva, I. Zelezetsky, A. Tossi, V. Palumbo, M. V. Verga Falzacappa, S. Sgubin, L. Braida, A. Amoroso, and S. Crovella. 2003. A study of host defence peptide beta-defensin 3 in primates. Biochem. J. 374:707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boniotto, M., A. Tossi, M. DelPero, S. Sgubin, N. Antcheva, D. Santon, J. Masters, and S. Crovella. 2003. Evolution of the beta defensin 2 gene in primates. Genes Immun. 4:251-257. [DOI] [PubMed] [Google Scholar]
- 8.Brogden, K. A., M. Heidari, R. E. Sacco, D. Palmquist, J. M. Guthmiller, G. K. Johnson, H. P. Jia, B. F. Tack, and P. B. McCray. 2003. Defensin-induced adaptive immunity in mice and its potential in preventing periodontal disease. Oral Microbiol. Immunol. 18:95-99. [DOI] [PubMed] [Google Scholar]
- 9.Del Pero, M., M. Boniotto, D. Zuccon, P. Cervella, A. Spano, A. Amoroso, and S. Crovella. 2002. Beta-defensin 1 gene variability among non-human primates. Immunogenetics 53:907-913. [DOI] [PubMed] [Google Scholar]
- 10.Fang, X. M., Q. Shu, Q. X. Chen, M. Book, H. G. Sahl, A. Hoeft, and F. Stuber. 2003. Differential expression of alpha- and beta-defensins in human peripheral blood. Eur. J. Clin. Investig. 33:82-87. [DOI] [PubMed] [Google Scholar]
- 11.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 12.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 13.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 14.Hiratsuka, T., M. Nakazato, Y. Date, J. Ashitani, T. Minematsu, N. Chino, and S. Matsukura. 1998. Identification of human beta-defensin-2 in respiratory tract and plasma and its increase in bacterial pneumonia. Biochem. Biophys. Res. Commun. 249:943-947. [DOI] [PubMed] [Google Scholar]
- 15.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray, Jr. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 16.Jordan, W. J., P. A. Brookes, R. M. Szydlo, J. M. Goldman, R. I. Lechler, and M. A. Ritter. 2004. IL-13 production by donor T cells is prognostic of acute graft-versus-host disease following unrelated donor stem cell transplantation. Blood 103:717-724. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, W. J., and M. A. Ritter. 2002. Optimal analysis of composite cytokine responses during alloreactivity. J. Immunol. Methods 260:1-14. [DOI] [PubMed] [Google Scholar]
- 18.Kao, C. Y., Y. Chen, Y. H. Zhao, and R. Wu. 2003. ORFeome-based search of airway epithelial cell-specific novel human β-defensin genes. Am. J. Respir. Cell Mol. Biol. 29:71-80. [DOI] [PubMed] [Google Scholar]
- 19.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 20.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 21.Lillard, J. W., Jr., P. N. Boyaka, O. Chertov, J. J. Oppenheim, and J. R. McGhee. 1999. Mechanisms for induction of acquired host immunity by neutrophil peptide defensins. Proc. Natl. Acad. Sci. USA 96:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, A. Y., D. Destoumieux, A. V. Wong, C. H. Park, E. V. Valore, L. Liu, and T. Ganz. 2002. Human beta-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J. Investig. Dermatol. 118:275-281. [DOI] [PubMed] [Google Scholar]
- 23.Lohner, K., A. Latal, R. I. Lehrer, and T. Ganz. 1997. Differential scanning microcalorimetry indicates that human defensin, HNP-2, interacts specifically with biomembrane mimetic systems. Biochemistry 36:1525-1531. [DOI] [PubMed] [Google Scholar]
- 24.Niyonsaba, F., H. Ogawa, and I. Nagaoka. 2004. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology 111:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oppenheim, J. J., A. Biragyn, L. W. Kwak, and D. Yang. 2003. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 62(Suppl. 2):17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawai, M. V., H. P. Jia, L. Liu, V. Aseyev, J. M. Wiencek, P. B. McCray, Jr., T. Ganz, W. R. Kearney, and B. F. Tack. 2001. The NMR structure of human beta-defensin-2 reveals a novel alpha-helical segment. Biochemistry 40:3810-3816. [DOI] [PubMed] [Google Scholar]
- 27.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wetering, S., S. P. Mannesse-Lazeroms, M. A. van Sterkenburg, and P. S. Hiemstra. 2002. Neutrophil defensins stimulate the release of cytokines by airway epithelial cells: modulation by dexamethasone. Inflamm. Res. 51:8-15. [DOI] [PubMed] [Google Scholar]
- 29.van Wetering, S., G. S. Tjabringa, and P. S. Hiemstra. 2004. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J. Leukoc. Biol. 77:444-450. [DOI] [PubMed] [Google Scholar]
- 30.Vedrine, C., C. Caraion, C. Lambert, and C. Genin. 2004. Cytometric bead assay of cytokines in sepsis: a clinical evaluation. Cytometry Ser. B 60:14-22. [DOI] [PubMed] [Google Scholar]
- 31.Wang, W., S. M. Owen, D. L. Rudolph, A. M. Cole, T. Hong, A. J. Waring, R. B. Lal, and R. I. Lehrer. 2004. Activity of alpha- and theta-defensins against primary isolates of HIV-1. J. Immunol. 173:515-520. [DOI] [PubMed] [Google Scholar]
- 32.Wimley, W. C., M. E. Selsted, and S. H. White. 1994. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 3:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, D., Q. Chen, D. M. Hoover, P. Staley, K. D. Tucker, J. Lubkowski, and J. J. Oppenheim. 2003. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74:448-455. [DOI] [PubMed] [Google Scholar]
- 34.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]