Abstract
Antibiotics with novel mechanisms of action are becoming increasingly important in the battle against bacterial resistance to all currently used classes of antibiotics. Bacterial DNA gyrase and topoisomerase IV (topoIV) are the familiar targets of fluoroquinolone and coumarin antibiotics. Here we present the characterization of two members of a new class of synthetic bacterial topoII ATPase inhibitors: VRT-125853 and VRT-752586. These aminobenzimidazole compounds were potent inhibitors of both DNA gyrase and topoIV and had excellent antibacterial activities against a wide spectrum of problematic pathogens responsible for both nosocomial and community-acquired infections, including staphylococci, streptococci, enterococci, and mycobacteria. Consistent with the novelty of their structures and mechanisms of action, antibacterial potency was unaffected by commonly encountered resistance phenotypes, including fluoroquinolone resistance. In time-kill assays, VRT-125853 and VRT-752586 were bactericidal against Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecalis, and Haemophilus influenzae, causing 3-log reductions in viable cells within 24 h. Finally, similar to the fluoroquinolones, relatively low frequencies of spontaneous resistance to VRT-125853 and VRT-752586 were found, a property consistent with their in vitro dual-targeting activities.
Antibiotic resistance in both hospital and community settings is steadily increasing, severely limiting the effectiveness of all currently used classes of antibiotics (4, 12, 17). Approximately 50% of Staphylococcus aureus hospital isolates are methicillin resistant, while 30% of enterococci are reported to be vancomycin resistant (4). The frequency of intermediate and high-level penicillin resistance in the community pathogen Streptococcus pneumoniae is approaching 50% (1, 12, 26). In addition, bacteria that are simultaneously resistant to more than one drug class are becoming increasingly prevalent (2). A significant problem with the majority of currently marketed antibiotics is that they are derived from structural classes that have been in widespread use for many years. For instance, resistance mechanisms acting on older ß-lactams, fluoroquinolones, and macrolides often impact newer generations of these commonly used classes of antibiotics. As a result, an actively pursued strategy of the antibiotic industry to thwart current drug resistance mechanisms is to create structurally novel class of antibiotics with new mechanisms of action. During the last 3 decades, only two truly novel classes of antibacterials with unique mechanisms of action, linezolid and daptomycin, have been developed to combat the problem of antibiotic resistance. Interestingly, there are already reports of linezolid resistance arising in the clinic (22), which underscores the need for new antibiotics with novel mechanisms of action.
Because of their essentiality and evolutionary conservation, DNA topoisomerases have become important antibiotic targets (11, 19). Both DNA gyrase and topoisomerase IV (topoIV) are highly homologous functional A2B2 heterotetramers (5). DNA gyrase is uniquely responsible for introducing negative supercoils into DNA, while the primary function of topoIV appears to be decatenation of replicated chromosomes and relaxation of DNA (6, 15, 31). The catalytic functions of both enzymes involve the breakage and rejoining of double-stranded DNA, with the intermediate passage of a second double strand of DNA through the break. Consistent with their high degree of structural and functional relatedness, both DNA gyrase and topoIV have been identified as the primary and secondary targets of the fluoroquinolone class of antibiotics which stabilize the enzyme-DNA complex in the double-strand break stage formed by gyrase and topoIV in bacteria (9). DNA gyrase and topoIV are also inhibited by members of the coumarin class of antibiotics, such as novobiocin and coumermycin, which target the ATP-binding sites of the corresponding B subunits (19), thereby inhibiting the energy source necessary for strand passage. At present, most coumarin resistance mutations in several organisms have mapped within the region of gyrB, encoding the ATP-binding site (10, 21, 29). However, recent in vitro studies have shown that novobiocin can also inhibit the ATPase activity of topoIV at higher concentrations than are necessary for inhibition of the gyrase enzyme (3). Therefore, the potential for “dual targeting” of both gyrase and topoIV exists at multiple steps during the catalytic pathway.
Antibiotics that inhibit multiple targets, such as some fluoroquinolones, tend to exhibit lower spontaneous-resistance frequencies, and their antibacterial activity is less affected by individual, target-based mutations (7). This potentially delays the emergence of high-level resistance during use, a major cause of treatment failure. In order to exploit the dual-targeting potential of the GyrB and ParE ATP-binding sites, a series of compounds belonging to the aminobenzimidazole class were optimized by using structure-guided drug design. In this report, we describe the characterization of two representative compounds of this class, VRT-125853 and VRT-752586 (Fig. 1), with respect to their abilities to inhibit the essential ATPase activities of Escherichia coli and S. aureus gyrase and topoIV, their antibacterial potencies and spectrum of activity against clinically prevalent resistant isolates, their bactericidal effectiveness in time-kill assays, and the frequency of the emergence of resistance in vitro.
FIG. 1.
Chemical structures of dual-targeting aminobenzimidazoles VRT-125853 and VRT-752586.
MATERIALS AND METHODS
Compounds.
VRT-125853 and VRT-752586 (Fig. 1) were synthesized at Vertex Pharmaceuticals Incorporated, and dry powder was dissolved in 100% dimethyl sulfoxide (DMSO) at a concentration of 25.6 mg/ml. Aliquots of DMSO stock solutions were stored at −20°C prior to use. Novobiocin (Sigma, St. Louis, MO), linezolid, (Zyvox, resuspended oral suspension; Pfizer, New York, NY), and ciprofloxacin (US Biological, Swampscott, MA) stocks were prepared and stored as described above. All of the other antibiotics used in this study were from standard commercial sources.
Enzyme assays and Ki determinations. (i) DNA gyrase and topoIV ATPase assays.
Enzymes were recombinantly expressed and purified as previously described for the E. coli enzymes (3, 10). Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min with a Molecular Devices plate reader. The rate of enzymatic hydrolysis was plotted against a serial dilution of inhibitor to determine potency, and the data were fitted to the Morrison equation for tight-binding inhibition (20). Standard coupled enzyme reactions were carried out at 30°C in a final volume of 100 μl. E. coli gyrase A2B2 assay reaction mixtures contained 100 mM Tris-HCl (pH 7.5), 1.5 mM MgCl2, 150 mM KCl, 2.5 mM phosphoenolpyruvate (PEP), 0.2 mM NADH, 1 mM dithiothreitol (DTT), 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% DMSO, 0.9 mM ATP (equal to the Km), and 40 nM (active enzyme) E. coli gyrase A2B2. E. coli topoIV C2E2 assay reaction mixtures contained 100 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 20 mM KCl, 2.5 mM PEP, 0.2 mM NADH, 10 mM DTT, 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% DMSO, 0.05 mg of bovine serum albumin per ml, 5 μg of HindIII-linearized pBR322 plasmid DNA per ml, 0.7 mM ATP (equal to the Km), and ∼20 nM E. coli topoIV C2E2 (active enzyme). S. aureus gyrase A2B2 assay reaction mixtures contained 100 mM Tris-HCl (pH 7.5), 1.5 mM MgCl2, 150 mM KCl, 2.5 mM PEP, 0.2 mM NADH, 1 mM DTT, 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% DMSO, 0.55 mM ATP (equal to the Km), and ∼60 nM S. aureus gyrase A2B2 (active enzyme). For S. aureus topoIV C2E2 (encoded by the grlA [C subunit] and grlB [E subunit] genes) assay, reaction mixtures contained 100 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 200 mM potassium glutamate, 2.5 mM PEP, 0.2 mM NADH, 1 mM DTT, 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% DMSO, 0.05 mg of bovine serum albumin per ml, 5 μg of HindIII-linearized pBR322 plasmid DNA per ml, 0.24 mM ATP (equal to the Km), and ∼21 nM S. aureus topoIV C2E2 (active enzyme).
(ii) Human topoII DNA decatenation assay.
Human topoII was assayed for its enzymatic activity, the ability to convert catenated kinetoplast DNA to decatenated DNA, by using a kit and the human topoII enzyme, which were both obtained from TopoGEN (Columbus, OH). After 20 min, the reaction was stopped with 5 μl of quench buffer (250 mM EDTA, 50% glycerol, 25 μg of bromophenol blue per ml). The DNA was separated and visualized on a 1% agarose gel stained with ethidium bromide. Standard reactions were carried out at 37°C (reaction mixture volume, 25 μl) with the following components: 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 120 mM KCl, 1 mM DTT, 20 μg of kinetoplast DNA per ml, 4% DMSO, 1.5 mM ATP, and 0.08 U of human topoII activity per μl. The EC50 is the compound concentration where approximately 50% inhibition of decatenation activity was observed, as determined by densitometric quantitation.
Bacterial strains.
All bacteria, as indicated, were either from our laboratory stocks, the proprietary clinical isolate collection of Focus Technologies (Herndon, VA), the American Type Culture Collection (ATCC; Manassas, VA), or the E. coli Genetic Stock Center (Yale University, New Haven, CT). Haemophilus influenzae strain Rd 0894 acrA::kan was a generous gift from H. Nikaido (27). S. aureus strains SA-1199, SA-1199B (NorA overproducer, GrlA A116E mutant), SA-8325-4, and SA-K2068 (MepA overproducer) were generous gifts from G. Kaatz (13, 14).
Susceptibility testing.
Susceptibility testing was performed at either Vertex Pharmaceuticals Incorporated or Focus Technologies (Herndon, VA). For aerobic organisms, MIC determinations were performed in liquid medium in 96-well microtiter plates as described by the Clinical and Laboratory Standards Institute (CLSI [formerly the National Committee for Clinical and Laboratory Standards]; 23). Liquid medium was supplemented as appropriate for optimal growth (23). Serial twofold dilution series of 200×-concentrated compound stocks were prepared in 100% DMSO, and 0.5 μl of each compound dilution was added to 50 μl of liquid medium in 96-well microtiter assay plates. An additional 50 μl of liquid medium containing the appropriate bacterial cells was then added to the assay plate. The final DMSO concentration in the assay was 0.5%. Compounds and cells were mixed, and plates were incubated at 35°C for at least 18 h prior to reading the susceptibility result. A twofold variation in the MIC was the standard error of the assay and hence considered insignificant. To measure the relative effects of serum on compound susceptibility, human serum (US Biological, Swampscott, MA) was added to liquid medium to a final concentration of 50% in assays performed with S. aureus ATCC 29213. For determining the MICs of anaerobic organisms, the agar proportion method was used according to CLSI recommendations (24). Susceptibility testing of Mycoplasma pneumoniae was performed by a reference broth microdilution method established by Waites et al. (30). MICs for Legionella pneumophila were determined by the standardized CLSI method (23). Mycobacterium tuberculosis susceptibility testing was performed with the BACTEC 460 TB System manufactured by Becton Dickinson, Cockeysville, MD. MICs for the Mycobacterium avium complex (MAC) were determined by the agar proportion method recommended for testing slow-growing mycobacterium species (25). All of the test methods used met acceptable standards based on recommended quality control ranges for all comparator antibiotics and the appropriate ATCC strain.
Determination of spontaneous-resistance frequencies.
The microtiter MIC of a given compound stock was verified prior to determination of spontaneous-resistance frequencies. Resistance frequencies were minimally determined in duplicate with two independently inoculated cultures. Fresh overnight cultures or glycerol seed stocks of mid-log-phase cultures were diluted 100-fold into appropriate fresh medium and grown at 37°C with a floor shaker rotating at 300 rpm, until late-log phase (approximately 108 to 109 CFU/ml). The inoculum was serially diluted and plated to confirm the starting number of CFU. Selection plates were prepared by adding an appropriate dilution of compound in 100% DMSO to 50 ml of molten agar at 55°C (ß-NAD and hemin for HTM [23] was added at this time for H. influenzae, fresh 5% defibrinated sheep blood [Bioreclamation, Inc., Hicksville, NY] was added to brain heart infusion agar for S. pneumoniae, and 5% laked horse blood [Quad Five, Ryegate, MT] was added to Mueller-Hinton broth agar for Enterococcus faecalis and Enterococcus faecium) and poured into sterile polystyrene petri plates (150 by 15 mm). The final concentration of compound was a multiple of the liquid MIC (2×, 4×, or 8×), and the final concentration of DMSO per plate was <0.1%. Plates were dried in a sterile hood for approximately 30 min prior to plating of bacteria. The late-log-phase inoculum was concentrated 10-fold by low-speed centrifugation, and approximately 1010 CFU were spread onto the surface of compound-containing agar plates. Plates were incubated at 35°C for a minimum of 3 days prior to counting of colonies. The resistance frequency was calculated as the number of compound-resistant colonies divided by the total number of CFU plated.
Bactericidal (time-kill) assays.
Five milliliters of appropriate growth medium was inoculated with approximately 106 log-phase cells, and cultures were grown in 15-ml sterile polypropylene conical tubes containing various dilutions of VRT-125853, VRT-752586, or control antibiotics. The final concentration of DMSO per culture was <0.5%. A sample of the inoculum was also serially diluted and plated to determine the starting number of CFU. Cultures were grown horizontally on a wheel (S. aureus ATCC 29213, S. pneumoniae ATCC BAA-255 [strain R6], and E. faecalis ATCC 29212) rotating rapidly (approximately 70 rpm) at 37°C or by shaking (H. influenzae ATCC 51907) in a tilted position in a floor shaker at 300 rpm. Samples (0.1 ml) were removed from cultures at timed intervals, serially diluted, and plated for CFU determinations on agar plates. Bactericidal activity was defined as a 3-log drop in the starting number of CFU in the culture within 24 h. Novobiocin, ciprofloxacin, and linezolid were run as controls.
RESULTS AND DISCUSSION
Dual targeting of DNA gyrase and topoIV enzymes by VRT-125853 and VRT-752586.
VRT-125853 and VRT-752586 were shown to be inhibitors of both the DNA gyrase and topoIV enzymes derived from E. coli and S. aureus (Table 1). For the E. coli enzymes, both compounds were significantly more potent against DNA gyrase than against topoIV. The lower Kis of both VRT-125853 and VRT-752586 against E. coli DNA gyrase versus topoIV suggested that DNA gyrase would be the primary target for both compounds in this organism in the absence of a cellular permeability barrier (see next section). In contrast to what was observed with the E. coli enzymes, the activities of VRT-125853 and VRT-752586 were more similar against both of the S. aureus enzymes, with topoIV/DNA gyrase Ki ratios of ∼5 and <0.4, respectively. The potent inhibition of both the gyrase and topoIV enzymes of E. coli and S. aureus demonstrated the in vitro dual-targeting properties of VRT-125853 and VRT-752586; however, the topoIV/DNA gyrase Ki ratios appeared to be both compound and organism specific. In contrast, novobiocin was found to be a more potent inhibitor of DNA gyrase than of topoIV in both sets of enzymes, consistent with findings in the literature that DNA gyrase is the major target of novobiocin in both E. coli and S. aureus (10, 21, 29). In a decatenation assay, neither aminobenzimidazole significantly inhibited human topoII (EC50, >25 μM; Table 1), a homologue with strong sequence similarity to both bacterial enzymes (5), indicating that the compounds are selective for the bacterial topoisomerases.
TABLE 1.
Dual inhibition of bacterial gyrase and topoIV by VRT-125853 and VRT-752586
| Compound |
Ki (nM)
|
E. coli topoIV/gyrase ratio |
Ki (nM)
|
S. aureus topoIV/gyrase ratio | EC50 (μM) human topoII decatenation assay | ||
|---|---|---|---|---|---|---|---|
| E. coli gyrase | E. coli topoIV | S. aureus gyrase | S. aureus topoIV | ||||
| VRT-125853 | 18 | 680 | 38 | 44 | 215 | 5 | >25 |
| VRT-752586 | <4 | 23 | >6 | 14 | <6 | <0.4 | >25 |
| Novobiocin | 13 | 160 | 12 | 19 | 900 | 47 | NDa |
ND, not determined.
Spectrum of activity of VRT-125853 and VRT-752586.
The antibacterial activities of VRT-125853 and VRT-752586 were determined against panels composed of recent drug-resistant and -susceptible aerobic, facultatively anaerobic, anaerobic, and atypical clinical isolates (Tables 2 and 3). Susceptibility results in Table 2 show that VRT-125853 and VRT-752586 had excellent activities against S. aureus, S. epidermidis, S. pneumoniae, S. pyogenes, E. faecalis, and E. faecium, suggesting their potential for use in a variety of hospital- and community-acquired infections. Although VRT-125853 and VRT-752586 were not active (MIC, >64 μg/ml) against Acinetobacter spp., E. coli, Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, Proteus mirabilis, Providencia spp., Pseudomonas aeruginosa, and Stenotrophomonas maltophilia (not shown), both compounds had significant activity against aerobic gram-negative respiratory pathogens H. influenzae and Moraxella catarrhalis (Table 2).
TABLE 2.
Activities of VRT-125853 and VRT-752586 against aerobic and facultatively anaerobic bacteria
| Organism (no. of isolates) | Antimicrobial | MIC (μg/ml)
|
No. of isolatesc
|
|||
|---|---|---|---|---|---|---|
| Range | 90%d | S | I | R | ||
| Enterococcus faecalis (35) | VRT-125853 | 0.5-1 | 1 | —a | — | — |
| VRT-752586 | 0.03-0.06 | 0.06 | — | — | — | |
| Ampicillin | 0.5-4 | 2 | 35 | — | — | |
| Levofloxacinb | 1->4 | >4 | 22 | 0 | 13 | |
| Linezolidb | 2->8 | 2 | 32 | 1 | 2 | |
| Novobiocin | 4-16 | 8 | — | — | — | |
| Oxacillin | 4->32 | 32 | — | — | — | |
| Quinupristin-dalfopristinb | 2->8 | >8 | 0 | 2 | 33 | |
| Telithromycin | 0.06->4 | 4 | — | — | — | |
| Vancomycinb | 1->32 | 16 | 31 | 1 | 3 | |
| Enterococcus faeclum (34) | VRT-125853 | 0.25-4 | 2 | — | — | — |
| VRT-752586 | 0.015-0.25 | 0.12 | — | — | — | |
| Ampicillinb | 0.5->16 | >16 | 6 | 0 | 28 | |
| Levofloxacinb | 1->4 | >4 | 3 | 2 | 29 | |
| Linezolidb | 1->8 | >8 | 28 | 1 | 5 | |
| Novobiocin | 1-8 | 2 | — | — | — | |
| Oxacillin | 8->32 | >32 | — | — | — | |
| Quinupristin-dalfopristinb | 0.5->8 | 4 | 19 | 11 | 4 | |
| Telithromycin | 0.03->4 | >4 | — | — | — | |
| Vancomycinb | 1->32 | >32 | 17 | 0 | 17 | |
| Staphylococcus aureus (54) | VRT-125853 | 1-4 | 4 | — | — | — |
| VRT-752586 | 0.06-0.25 | 0.12 | — | — | — | |
| Ampicillinb | ≤0.25->16 | >16 | 2 | 0 | 52 | |
| Levofloxacinb | 0.12->4 | >4 | 15 | 1 | 38 | |
| Linezolid | 1->8 | 4 | 53 | — | — | |
| Novobiocin | ≤0.25-1 | 0.5 | — | — | — | |
| Oxacillinb | 0.25->32 | >32 | 18 | 0 | 36 | |
| Quinupristin-dalfopristin | 0.25-2 | 2 | 46 | 8 | 0 | |
| Telithromycinb | 0.06->4 | >4 | 25 | 0 | 29 | |
| Vancomycinb | 1->32 | 4 | 50 | 2 | 2 | |
| Staphylococcus epidermidis (24) | VRT-125853 | 1-4 | 2 | — | — | — |
| VRT-752586 | 0.03-0.12 | 0.12 | — | — | — | |
| Ampicillinb | ≤0.25->16 | >16 | 2 | 0 | 22 | |
| Levofloxacinb | 0.12->4 | >4 | 11 | 0 | 13 | |
| Linezolid | 1-2 | 2 | 24 | — | — | |
| Novobiocin | ≤0.25-1 | 0.5 | — | — | — | |
| Oxacillinb | 0.25->32 | >32 | 5 | 0 | 19 | |
| Quinupristin-dalfopristin | 0.25-2 | 1 | 23 | 1 | 0 | |
| Telithromycinb | 0.03->4 | >4 | 16 | 0 | 8 | |
| Vancomycin | 1-4 | 4 | 24 | 0 | 0 | |
| Streptococcus pneumoniae (64) | VRT-125853 | 0.12-0.5 | 0.5 | — | — | — |
| VRT-752586 | 0.004-0.03 | 0.03 | — | — | — | |
| Amoxicillinb | ≤0.015-8 | 8 | 53 | 3 | 8 | |
| Azithromycinb | 0.06->2 | >2 | 36 | 1 | 27 | |
| Ceftriaxoneb | 0.03->4 | 2 | 53 | 7 | 4 | |
| Levofloxacinb | 0.5->8 | >8 | 53 | 0 | 11 | |
| Linezolid | 0.5-2 | 2 | 64 | — | — | |
| Novobiocin | ≤0.5-4 | 2 | — | — | — | |
| Quinupristin-dalfopristin | ≤1-≤1 | ≤1 | 64 | 0 | 0 | |
| Telithromycin | 0.015-1 | 0.5 | 64 | 0 | 0 | |
| Streptococcus pyogenes (22) | VRT-125853 | 0.015-8 | 4 | — | — | — |
| VRT-752586 | 0.004-0.12 | 0.12 | — | — | — | |
| Amoxicillin | ≤0.015-0.06 | 0.06 | 22 | — | — | |
| Azithromycinb | 0.06->2 | >2 | 7 | 0 | 15 | |
| Ceftriaxone | 0.03-0.12 | 0.12 | 22 | — | — | |
| Levofloxacin | ≤0.25-1 | 1 | 22 | 0 | 0 | |
| Linezolid | ≤0.25-2 | 1 | 22 | — | — | |
| Novobiocin | ≤0.5-4 | 4 | — | — | — | |
| Quinupristin-dalfopristin | ≤1-≤1 | ≤1 | 22 | 0 | 0 | |
| Telithromycin | 0.015->4 | >4 | — | — | — | |
| Haemophilus influenzae (36) | VRT-125853 | 2->16 | >16 | — | — | — |
| VRT-752586 | 0.5->16 | >16 | — | — | — | |
| Amoxicillin-clavulanateb | ≤1->8 | 8 | 32 | 0 | 4 | |
| Ampicillinb | ≤1->4 | >4 | 21 | 0 | 15 | |
| Azithromycin | ≤0.5->8 | 2 | 35 | — | — | |
| Ceftriaxone | ≤0.03-0.5 | 0.06 | 36 | — | — | |
| Levofloxacin | 0.015->4 | >4 | 31 | — | — | |
| Novobiocin | ≤1-4 | ≤1 | — | — | — | |
| Telithromycinb | ≤0.5->16 | 4 | 34 | 1 | 1 | |
| Moraxella catarrhalis (23) | VRT-125853 | 0.25-2 | 2 | — | — | — |
| VRT-752586 | 0.03-0.25 | 0.25 | — | — | — | |
| Amoxicillin-clavulanate | ≤1-≤1 | ≤1 | — | — | — | |
| Ampicillin | ≤1->4 | >4 | — | — | — | |
| Azithromycin | ≤0.5-≤0.5 | ≤0.5 | — | — | — | |
| Ceftriaxone | ≤0.03-2 | 2 | — | — | — | |
| Levofloxacin | 0.03-0.12 | 0.06 | — | — | — | |
| Novobiocin | ≤1-≤1 | ≤1 | — | — | — | |
| Telithromycin | ≤0.5-1 | ≤0.5 | — | — | — | |
—, CLSI interpretive criteria for susceptibility and resistance have not been defined.
Isolates resistant to this antibiotic were present in the panel.
Relevant resistance phenotypes were confirmed by using CLSI breakpoints for susceptibility (S), intermediate susceptibility (I), and resistance (R).
MIC for 90% of strains tested.
TABLE 3.
Activities of VRT-125853 and VRT-752586 against anaerobic and atypical bacteria
| Organism (no. of isolates) | Antimicrobial | MIC (μg/ml)
|
No. of isolatesc
|
|||
|---|---|---|---|---|---|---|
| Range | 90%e | S | I | R | ||
| Bacteroides distasonis (11) | VRT-125853 | >16->16 | >16 | —a | — | — |
| VRT-752586 | 2-4 | 4 | — | — | — | |
| Amoxicillin-clavulanate | 2-8 | 8 | 9 | 2 | 0 | |
| Imipenem | 0.5-1 | 1 | 11 | 0 | 0 | |
| Clindamycinb | 4->8 | >8 | 0 | 1 | 10 | |
| Metronidazole | 0.5-1 | 1 | 11 | 0 | 0 | |
| Penicillinb | 4->32 | >32 | 0 | 0 | 11 | |
| Bacteroides fragilis (11) | VRT-125853 | >16->16 | >16 | — | — | — |
| VRT-752586 | 2-4 | 4 | — | — | — | |
| Amoxicillin-clavulanate | 0.5-8 | 1 | 10 | 1 | 0 | |
| Imipenem | 0.25-1 | 0.5 | 11 | 0 | 0 | |
| Clindamycinb | 0.5-8 | 4 | 8 | 2 | 1 | |
| Metronidazole | 0.5-1 | 1 | 11 | 0 | 0 | |
| Penicillinb | 1-32 | 16 | 0 | 1 | 10 | |
| Bacteroides thetaiotaomicron (11) | VRT-125853 | >16->16 | >16 | — | — | — |
| VRT-752586 | 2-2 | 2 | — | — | — | |
| Amoxicillin-clavulanateb | 1->16 | 4 | 10 | 0 | 1 | |
| Imipenem | 0.25-1 | 1 | 11 | 0 | 0 | |
| Clindamycinb | 4-8 | 8 | 0 | 3 | 8 | |
| Metronidazole | 0.5-2 | 2 | 11 | 0 | 0 | |
| Penicillinb | 8->32 | >32 | 0 | 0 | 11 | |
| Bacteroides vulgatus (11) | VRT-125853 | >16->16 | >16 | — | — | — |
| VRT-752586 | 2-4 | 4 | — | — | — | |
| Amoxicillin-clavulanateb | 1-16 | 8 | 9 | 1 | 1 | |
| Imipenem | 0.12-1 | 1 | 11 | 0 | 0 | |
| Clindamycinb | ≤0.25->8 | >8 | 8 | 0 | 3 | |
| Metronidazole | 0.25-2 | 2 | 11 | 0 | 0 | |
| Penicillinb | 1->32 | >32 | 0 | 1 | 10 | |
| Clostridium difficile (11) | VRT-125853 | 1->16 | >16 | — | — | — |
| VRT-752586 | 0.06-4 | 4 | — | — | — | |
| Amoxicillin-clavulanate | 0.25-4 | 1 | 11 | 0 | 0 | |
| Imipenemb | 0.5-16 | 4 | 10 | 0 | 1 | |
| Clindamycinb | 0.5->8 | >8 | 6 | 1 | 4 | |
| Metronidazole | 0.25-2 | 0.5 | 11 | 0 | 0 | |
| Penicillinb | ≤0.5->32 | 4 | 5 | 3 | 3 | |
| Clostridium perfringens (11) | VRT-125853 | 8-16 | 16 | — | — | — |
| VRT-752586 | 0.5-2 | 2 | — | — | — | |
| Amoxicillin-clavulanate | ≤0.12-0.25 | 0.25 | 11 | 0 | 0 | |
| Imipenem | 0.06-0.25 | 0.25 | 11 | 0 | 0 | |
| Clindamycinb | ≤0.25-8 | 8 | 5 | 4 | 2 | |
| Metronidazole | 0.5-4 | 4 | 11 | 0 | 0 | |
| Penicillin | ≤0.5-≤0.5 | ≤0.5 | 11 | 0 | 0 | |
| Fusobacterium sp. (11) | VRT-125853 | 4->16 | >16 | — | — | — |
| VRT-752586 | 2->16 | >16 | — | — | — | |
| Amoxicillin-clavulanateb | ≤0.12-16 | 4 | 10 | 0 | 1 | |
| Imipenem | 0.03-0.5 | 0.5 | 11 | 0 | 0 | |
| Clindamycinb | ≤0.25->8 | 0.5 | 10 | 0 | 1 | |
| Metronidazoleb | ≤0.12->32 | 0.25 | 10 | 0 | 1 | |
| Penicillinb | ≤0.5-32 | ≤1 | 9 | 1 | 1 | |
| Peptostreptococcus sp. (11) | VRT-125853 | 16->16 | >16 | — | — | — |
| VRT-752586 | 0.5->16 | >16 | — | — | — | |
| Amoxicillin-clavulanate | ≤0.12-4 | 0.5 | 11 | 0 | 0 | |
| Imipenem | ≤0.015-1 | 0.12 | 11 | 0 | 0 | |
| Clindamycinb | ≤0.25->8 | 4 | 9 | 1 | 1 | |
| Metronidazoleb | ≤0.12->32 | 32 | 9 | 0 | 2 | |
| Penicillinb | ≤0.5->32 | ≤0.5 | 10 | 0 | 1 | |
| Prevotella sp. (11) | VRT-125853 | >16->16 | >16 | — | — | — |
| VRT-752586 | 4-16 | 8 | — | — | — | |
| Amoxicillin-clavulanate | ≤0.12-4 | 4 | 11 | 0 | 0 | |
| Imipenem | 0.06-0.5 | 0.5 | 11 | 0 | 0 | |
| Clindamycinb | ≤0.25-8 | 8 | 4 | 4 | 3 | |
| Metronidazole | 0.5-16 | 2 | 10 | 1 | 0 | |
| Penicillinb | ≤0.5->32 | >32 | 3 | 0 | 8 | |
| Legionella pneumophila (11) | VRT-125853 | 4-8 | 8 | — | — | — |
| VRT-752586 | 0.12-0.25 | 0.25 | — | — | — | |
| Azithromycin | 0.06-2 | 1 | — | — | — | |
| Doxycycline | 1-8 | 4 | — | — | — | |
| Levofloxacin | 0.015-0.03 | 0.03 | — | — | — | |
| Telithromycin | 0.03-0.06 | 0.06 | — | — | — | |
| Mycoplasma pneumoniae (6) | VRT-125853 | 2-8 | NAd | — | — | — |
| VRT-752586 | 0.12-0.5 | NA | — | — | — | |
| Azithromycin | ≤0.008->8 | NA | — | — | — | |
| Doxycycline | 0.06-0.12 | NA | — | — | — | |
| Levofloxacin | 0.5-1 | NA | — | — | — | |
| Telithromycin | ≤0.015->16 | NA | — | — | — | |
—, CLSI interpretive criteria for susceptibility and resistance have not been defined.
Isolates resistant to this antibiotic were present in the panel.
Relevant resistance phenotypes were confirmed by using CLSI breakpoints for susceptibility (S), intermediate susceptibility (I), and resistance (R).
NA, too few isolates to determine MIC90.
MIC for 90% of isolates tested.
Confirmation of a gyrase-mediated mechanism of action of both compounds in bacteria was demonstrated by an upward shift in the MIC with an S. aureus strain containing a GyrB T173I mutation compared to wild-type parental strain ATCC 29213. Mutations in threonine 173 in S. aureus GyrB have been previously implicated in conferring novobiocin resistance (29); the MIC of novobiocin (0.063 μg/ml) was increased by eightfold for the GyrB T173I mutant strain; however, the MICs of other antibiotics (ciprofloxacin, vancomycin, and linezolid) were unaffected (data not shown). The GyrB T173I mutation increased the MICs of VRT-125853 (2 μg/ml) and VRT-752586 (0.032 μg/ml) by 8-fold and 16-fold, respectively, indicating that the MICs of these compounds for wild-type S. aureus are dependent on an interaction with GyrB and that their site of interaction with GyrB overlaps the binding site of novobiocin.
Increased susceptibilities of E. coli mutant strains with either improved permeability (impA mutant) or decreased efflux (tolC mutant), and an H. influenzae mutant with decreased efflux (acrA::kan), suggested that permeability and efflux reduce the antibacterial potency of both compounds in these, and perhaps other, gram-negative organisms (Table 4). The improved potency of VRT-752586 versus VRT-125853 against gram-positive bacteria such as staphylococci, enterococci, and streptococci (Table 2) could be attributed, at least in part, to the superior enzymatic potency and dual-targeting properties of VRT-752586 (Table 1). It is also possible that VRT-752586 is better able to permeate or is less prone to efflux in gram-positive bacteria. Susceptibility testing against an S. aureus NorA-overproducing strain, SA-1199B, and a MepA-overproducing strain, SA-K2068, indicated that these particular efflux pumps, implicated in fluoroquinolone resistance (13, 14), do not appreciably recognize VRT-125853 and VRT-752586; the susceptibility of both efflux pump mutant strains was similar to that of the corresponding wild-type parental strains (data not shown). These results do not rule out the possibility that other gram-positive efflux systems are involved in the efflux of our aminobenzimidazole compounds to different degrees. Interestingly, we found that the naturally novobiocin-resistant organism Staphylococcus saprophyticus was susceptible to both VRT-125853 and VRT-752586 (MICs equal to 2 and <0.008 μg/ml, respectively), comparable to what was observed with the other staphylococci tested. The novobiocin MIC for the S. saprophyticus isolate tested was >8 μg/ml. This result indicated that the mechanism of novobiocin resistance in this organism has no effect on its susceptibility to aminobenzimidazole inhibitors.
TABLE 4.
Activities of VRT-125853 and VRT-752586 against gram-negative bacteria
| Bacterium | Isolate | MIC (μg/ml)a
|
|||
|---|---|---|---|---|---|
| VRT-125853 | VRT-752586 | Novobiocin | Ciprofloxacin | ||
| H. influenzae | ATCC 51907 (Rd) | 4 | 1 | 0.13 | 0.004 |
| H. influenzae | Rd acrA::kan mutant | 1 | 0.13 | 0.016 | 0.008 |
| E. coli | ATCC 25922 | >64 | >64 | 64 | 0.016 |
| E. coli | impA mutantb | 8 | 0.5 | 1 | 0.008 |
| E. coli | tolC mutantb | 2 | 0.13 | 1 | 0.004 |
All MICs were determined in duplicate.
E. coli K-12 strain.
Improved activity of VRT-752586 versus VRT-125853 was also observed for most of the anaerobic and atypical organisms tested (Table 3). Presumably, this finding is also due to the greater enzymatic potency and dual-targeting properties of VRT-752586. Additionally, neither compound had any measurable effect on the growth of Candida albicans up to 64 μg/ml (highest tested concentration), which was used as a convenient indicator of nonspecific eukaryotic toxicity (data not shown).
Activity of aminobenzimidazole compounds against drug-resistant pathogens.
Consistent with the novel mechanism of action of VRT-125853 and VRT-752586, susceptibility was unaffected by commonly encountered resistance phenotypes (Tables 2 and 3). Oxacillin-, levofloxacin-, telithromycin-, and vancomycin-resistant S. aureus strains were as susceptible to both compounds as were nonresistant isolates. Amoxicillin, ceftriaxone, azithromycin, and levofloxacin resistance similarly had no effect on the susceptibility of S. pneumoniae. Levofloxacin, linezolid, and vancomycin resistance had no effect on the susceptibility of E. faecalis and E. faecium to VRT-125853 and VRT-752586. In H. influenzae, where for 13 out of 36 isolates the MIC of VRT-752586 was >2 μg/ml, no correlation of ß-lactam resistance with reduced aminobenzimidazole susceptibility was observed. Unlike the multigenerational members of the ß-lactam, macrolide, glycopeptide, and fluoroquinolone antibiotic families, it is anticipated that this new class of aminobenzimidazole antibiotics would not be vulnerable to most of the resistance mechanisms present in current clinical isolates. The finding that the antibacterial activities of VRT-125853 and VRT-752586 were unaffected by fluoroquinolone resistance (Table 2) suggests that fluoroquinolone resistance mutations in GyrA and ParC are far enough removed in the heterotetramer and do not impact the ATP-binding site in GyrB and ParE.
Antimycobacterial activities of VRT-125853 and VRT-752586.
VRT-125853 and VRT-752586 were active in vitro against multiple strains of drug-susceptible and -resistant M. tuberculosis (Table 5), suggesting that, similar to fluoroquinolones (8), these aminobenzimidazoles would be effective against current and emerging multidrug-resistant strains. VRT-752586 exhibited limited activity against MAC isolates; VRT-125853 was inactive against all of the MAC isolates tested (Table 5). Similar to other observed antibacterial activities, VRT-752586 had greater potency against M. tuberculosis than did VRT-125853. That the activities of both compounds against M. tuberculosis and MAC isolates (VRT-752586 only) are mediated through inhibition of gyrase remains to be confirmed. Mycobacterial DNA gyrase has been shown to possess both efficient negative supercoiling and decatenase activities in vitro under physiological conditions (18). Given that M. tuberculosis has no distinct topoIV enzyme (18), the improved potency of VRT-752586 cannot be attributed to dual targeting and may result from an ability to permeate, reduced recognition by efflux pumps, or potent inhibition of the bifunctional enzyme in this organism.
TABLE 5.
Activities of VRT-125853 and VRT-752586 against M. tuberculosis and the MAC
| Organism and strain | Phenotypea | MIC (μg/ml)
|
|
|---|---|---|---|
| VRT-125853 | VRT-752586 | ||
| M. tuberculosis | |||
| H37Rv (ATCC 27294) | SM/INH/RIF/EMB-S | 2 | 0.12 |
| MSI 04-294 | SM-R/INH-R | 2 | 0.25 |
| MSI 04-295 | SM-R/INH-R | 2 | 0.25 |
| MSI 04-3543 | RIF-R | 2 | 0.12 |
| MSI 04-7770 | INH-R | 2 | 0.12 |
| MAC | |||
| ATCC 700898 | NAb | >4 | 4 |
| MSI 04-7929 | NAb | >4 | 2 |
| MSI 04-7766 | NAb | >4 | 2 |
| MSI 04-6714 | NAb | >4 | >4 |
| MSI 04-7744 | NAb | >4 | 4 |
Confirmed phenotypes for M. tuberculosis: SM, streptomycin; INH, isoniazid; RIF, rifampin; EMB, ethambutol; S, susceptible; R, resistant. All isolates were fluoroquinolone sensitive.
Members of the MAC are intrinsically resistant to all of the first-line antimycobacterial drugs.
Effects of human serum on aminobenzimidazole activity.
As an indicator of relative serum-binding potential, compounds were tested in an MIC assay with S. aureus ATCC 29213 in the presence and absence of 50% human serum, similar to the actual serum content of blood. Compound activity in the presence of serum provides an estimation of the amount of unbound compound available to inhibit bacterial growth in human blood. A twofold shift in the VRT-125853 MIC and a 16-fold shift in the VRT-752586 MIC suggested that the latter compound was relatively more highly protein bound by human serum. Regardless of the greater degree of serum binding, the serum-shifted MIC of VRT-752586 still remained below 1 μg/ml. Consistent with reports in the literature (16, 28), in this assay the MIC of novobiocin was shifted greater than 60-fold, confirming previous results showing that novobiocin is highly protein bound in human serum. The clinical relevance of these serum-binding results for VRT-125853 and VRT-752586 remains to be explored by animal efficacy studies.
Spontaneous-resistance frequencies and mutant prevention concentrations (MPCs) of VRT-125853 and VRT-752586.
It has been estimated that the bacterial population at a given infection site can reach on the order of 1010 cells (7). If spontaneous antibiotic-resistant mutants arise during the expansion of an infection, then antibiotic treatment will likely fail to eradicate the infection. To estimate the probability that resistant mutants would arise in a naive population of approximately 1010 cells, the in vitro frequencies of spontaneous resistance of several relevant pathogens to VRT-125853, VRT-752586, and comparators were measured (Table 6). In general, across the organism panel, the spontaneous-resistance frequencies observed for VRT-125853 and VRT-752586 were comparable to, or in some cases better than, those obtained with novobiocin, ciprofloxacin, and linezolid at similar multiples of their respective MICs (Table 6).
TABLE 6.
Frequencies of spontaneous resistance to VRT-125853 and VRT-752586a
| Compound and concn (fold over MIC)b | Frequency of spontaneous resistance
|
||||
|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. pneumoniae ATCC BAA255 (R6) | E. faecalis ATCC 29212 | E. faecium ATCC 49624 | H. influenzae ATCC 51907 (Rd) | |
| VRT-125853 | |||||
| 2 | 1.4 × 10−8 | >1.0 × 10−6 | <1.3 × 10−9 | 1.6 × 10−8 | 1 × 10−7 |
| 4 | <1.3 × 10−10 | 2.8 × 10−8 | <1.3 × 10−9 | 3.6 × 10−9 | 3.2 × 10−9 |
| 8 | <1.3 × 10−10 | 9.0 × 10−9 | <1.3 × 10−9 | <4.5 × 10−10 | 7.0 × 10−10 |
| VRT-752586 | |||||
| 2 | 2.4 × 10−8 | 6.0 × 10−7 | <2.1 × 10−10 | <1.3 × 10−10 | >1 × 10−6 |
| 4 | 7.4 × 10−9 | 4.3 × 10−8 | <2.1 × 10−10 | <1.3 × 10−10 | 5.5 × 10−8 |
| 8 | <5.7 × 10−10 | <5.5 × 10−10 | <2.1 × 10−10 | <1.3 × 10−10 | 2.8 × 10−9 |
| Novobiocin | |||||
| 2 | 3.5 × 10−8 | 5.4 × 10−7 | <3.8 × 10−10 | <3.6 × 10−10 | >1.0 × 10−6 |
| 4 | 1.8 × 10−8 | 6.5 × 10−8 | <3.8 × 10−10 | <3.6 × 10−10 | 3.1 × 10−8 |
| 8 | 1.2 × 10−8 | 3.3 × 10−8 | <3.8 × 10−10 | <3.6 × 10−10 | 7.0 × 10−9 |
| Ciprofloxacin | |||||
| 2 | >1.0 × 10−6 | >1.0 × 10−6 | >1.0 × 10−6 | >1.0 × 10−6 | >1.0 × 10−6 |
| 4 | 3.3 × 10−8 | 1.4 × 10−6 | >1.0 × 10−6 | >1.0 × 10−6 | 3.5 × 10−9 |
| 8 | 1.1 × 10−9 | <8.2 × 10−10 | <3.8 × 10−10 | <3.6 × 10−10 | 5.8 × 10−10 |
| Linezolid | |||||
| 2 | <3.7 × 10−10 | <8.2 × 10−10 | 5.4 × 10−9 | 2.4 × 10−8 | <2.3 × 10−10 |
| 4 | <3.7 × 10−10 | <8.2 × 10−10 | 1.9 × 10−9 | <4.0 × 10−10 | <2.3 × 10−10 |
| 8 | <3.7 × 10−10 | <8.2 × 10−10 | <3.8 × 10−10 | <4.0 × 10−10 | <2.3 × 10−10 |
Representative values for duplicate determinations are presented.
MIC (micrograms per milliliter) of VRT-125853 and VRT-752586, respectively, are as follows: S. aureus ATCC 29213, 2 and 0.032; S. pneumoniae ATCC BAA255, 0.13 and 0.004; E. faecalis ATCC 29212, 0.5 and 0.016; E. faecium ATCC 49624, 2 and 0.031; H. influenzae ATCC 51907, 4 and 1.
The MPC has recently been suggested as a way to rank antibiotics with regard to the ability to suppress the emergence of resistance during infection (7). The MPC reflects the ability of an antibiotic to block the growth of first-step mutants without regard for whether the mutations cause alterations in the molecular target, drug efflux, drug inactivation, or drug exclusion. Theoretically, if the MPC for an infecting organism is maintained at the infection site, the growth of resistant mutants will be suppressed. In most cases, except for H. influenzae (VRT-125853 and VRT-752586) and S. pneumoniae (VRT-125853 only), the spontaneous-resistance frequencies were approximately <10−10 at concentrations within eight times the MIC, illustrating favorable MPCs of these compounds for each organism (Table 6). Lower spontaneous-resistance frequencies are consistent with the inhibition of more than one essential target at a given compound concentration. The spontaneous-resistance frequencies of E. faecalis (both compounds) and E. faecium (VRT-752586 only) were below the level of detection at two times the MIC, suggesting the inhibition of more than one target in these organisms. For other organisms, complete suppression of spontaneous emergence of resistance was achieved at higher multiples of the MIC (Table 6). Additional experiments with animal models of infection are necessary to determine whether the MPC for each compound with regard to each organism is achieved at a given site of infection.
Bactericidal activities of VRT-125853 and VRT-752586.
All antibiotics are bacteriostatic (i.e., they stop bacterial cell growth); however, some antibiotics also kill bacteria (i.e., they are bactericidal). Bacteriostatic antibiotics require the host's natural defense mechanisms to play a role in completely eradicating the infection, while bactericidal antibiotics are able to clear infections independently of the host's defenses. In the case of immunocompromised host defenses, antibiotics with bactericidal activity are therefore preferred.
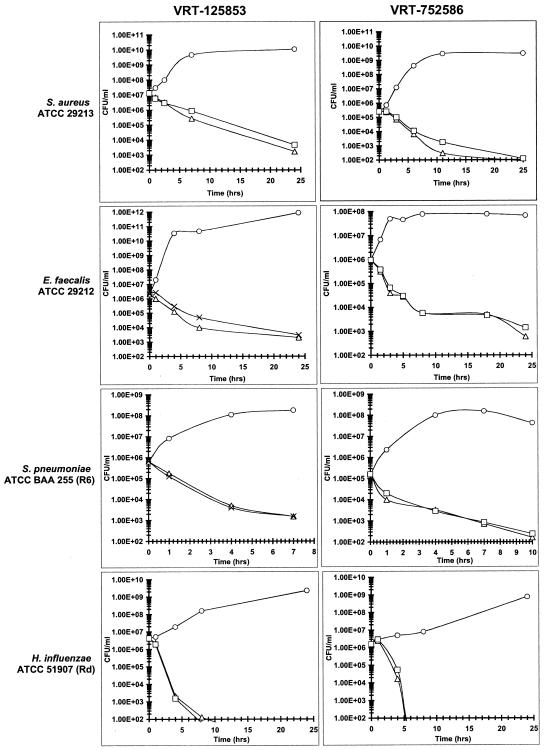
The in vitro bactericidal activities of VRT-125853 and VRT-752586 against S. aureus, E. faecalis, S. pneumoniae, and H. influenzae were evaluated in standard time-kill assays. Representative time-kill curves are shown in Fig. 2. Both VRT-125853 and VRT-752586 were found to be bactericidal against all of the strains tested, as defined by a 3-log reduction in the starting inoculum by 24 h (Fig. 2). Interestingly, the rate of killing was fastest for H. influenzae, where a 3-log reduction was observed in 3 to 5 h. While the explanation for this finding is unclear, the rapid bactericidal action against H. influenzae seems to be a property of the aminobenzimidazole class since a similar phenomenon has been observed with other members of the class as well (data not shown). Because the mechanism of action of VRT-125853 and VRT-752586 is inhibition of the ATPase activity of DNA gyrase and topoIV, the rate of killing was not as rapid as that seen with ciprofloxacin (data not shown), a drug which promotes DNA breakage, triggering a rapid cascade of lethal events in the cell (9). Linezolid, as expected, was bacteriostatic against all bacteria except H. influenzae in this assay (data not shown). Consistent with the related mechanism of action, the rate of killing for both VRT-125853 and VRT-752586 was similar to, or slightly faster than, the rate of killing by novobiocin for all of the organisms tested (data not shown).
FIG. 2.
Bactericidal activities of VRT-125853 and VRT-752586. Cells were cultured as described in Materials and Methods. Cells were grown in the presence of the indicated compound concentrations: ○, no drug; ▵, 4 times the MIC; ×, 8 times the MIC; □, 10 times the MIC. MICs (μg/ml) for VRT-125853 and VRT-752586, respectively, were as follows: S. aureus ATCC 29213, 2 and 0.032; S. pneumoniae ATCC BAA255, 0.13 and 0.004; E. faecalis ATCC 29212, 0.5 and 0.016; H. influenzae ATCC 51907, 4 and 1.
Summary.
VRT-125853 and VRT-752586 are representatives of a novel aminobenzimidazole class of bacterial topoisomerase inhibitors. Their potent activity against resistant and susceptible staphylococci, enterococci, streptococci, and respiratory gram-negative bacteria (H. influenzae and M. catarrhalis) suggests their potential utility against a variety of both nosocomial and community infections. VRT-125853 and VRT-752586 also showed favorable pharmacokinetic properties in animals and demonstrated efficacy in animal models of infection, supporting their use for skin and skin structure infections, as well as respiratory infections caused by drug-susceptible and -resistant pathogens (unpublished data), thereby making them a promising new class of antibacterials for combating the problem of drug resistance.
Acknowledgments
We thank John Thomson, Eric Olson, Ann Kwong, and Michael Briggs for support and helpful suggestions. We are grateful to Focus Technologies for help in designing and performing the MIC90 studies.
REFERENCES
- 1.Aspa, J., O. Rajas, F. Rodriguez de Castro, J. Blanquer, R. Zalacain, A. Fenoll, R. de Celes, A. Vargas, F. R. Salvanes, P. P. Espana, J. Rello, and A. Torres. 2004. Drug-resistant pneumococcal pneumonia: clinical relevance and related factors. Clin. Infect. Dis. 38:787-798. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, C. T., and J. F. Barrett. 2003. Antibacterials: are the new entries enough to deal with the emerging resistance problems? Curr. Opin. Biotech. 14:621-626. [DOI] [PubMed] [Google Scholar]
- 3.Bellon, S., J. D. Parsons, Y. Wei, K. Haykawa, L. L. Swenson, P. S. Charifson, J. A. Lippke, R. Aldape, and C. H. Gross. 2004. Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob. Agents Chemother. 48:1856-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2004. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004, issued October 2004. www.cdc.gov/ncidod/hip/NNIS/2004NNISreport.pdf. [DOI] [PubMed]
- 5.Champoux, J. J. 2001. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 6.Deibler, R. W., S. Rahmati, and E. L. Zechiedrich. 2001. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 15:748-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K. 2001. A strategy for fighting antibiotic resistance. ASM News 67:27-33. [Google Scholar]
- 8.Ginsburg, A. S., J. H. Grosset, and W. R. Bishai. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet 3:432-442. [DOI] [PubMed] [Google Scholar]
- 9.Gootz, T. D., and K. E. Brighty. 1998. Chemistry and mechanism of action of the quinolone antibacterials, p. 29-80. In V. T. Andriole (ed.), The quinolones, second edition. Academic Press, Inc., New York, N.Y.
- 10.Gross, C. H., J. D. Parsons, T. H. Grossman, P. S. Charifson, S. Bellon, J. Jernee, M. Dwyer, S. P. Chambers, W. Markland, M. Botfield, and S. A. Raybuck. 2003. Active-site residues of Escherichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob. Agents Chemother. 47:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heisig, P. 2001. Inhibitors of bacterial topoisomerases: mechanisms of action and resistance and clinical aspects. Planta Med. 67:3-12. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, M. R. 2003. Worldwide trends in antimicrobial resistance among common respiratory tract pathogens. Pediatr. Infect. Dis. J. 22:S109-S119. [DOI] [PubMed] [Google Scholar]
- 13.Kaatz, G. W., V. V. Moudgal, S. M. Seo, and J. E. Kristiansen. 2003. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaatz, G. W., F. McAleese, and S. M. Seo. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49:1857-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, J.-I., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Susuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 16.Kucers, A., S. M. Crowe, M. L. Grayson, and J. F. Hoy (ed.). 1997. The use of antibiotics, 5th edition, p. 663-666. Butterworth-Heinemann, Linacre House, Jordan Hill, Oxford, United Kingdom.
- 17.Livermore, D. M. 2004. The need for new antibiotics. Clin. Microbiol. Infect. 10(Suppl. 4):1-9. [DOI] [PubMed] [Google Scholar]
- 18.Manjunatha, U. H., M. Dalal, M. Chatterji, D. R. Radha, S. S. Visweswariah, and V. Nagaraja. 2002. Functional characterization of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 30:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 20.Morrison, J. F. 1982. The slow-binding, tight-binding inhibition of enzyme catalyzed reactions. Trends Biochem. Sci. 7:102-105. [Google Scholar]
- 21.Munoz, R., M. Bustamante, and A. G. de la Campa. 1995. Ser-127-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 177:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutnick, A. H., V. Enne, and R. N. Jones. 2003. Linezolid resistance since 2001: SENTRY Antimicrobial Surveillance Program. Ann. Pharmacother. 37:769-774. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th edition, volume 23, number 2. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.National Committee for Clinical Laboratory Standards. 2004. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 6th edition, volume 24, number 2. Approved standard M11-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes, volume 23, number 18. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 26.Pottumarthy, S., T. R. Fritsche, and R. N. Jones. 2005. Comparative activity of oral and parenteral cephalosporins tested against multidrug-resistant Streptococcus pneumoniae: report from the SENTRY Antimicrobial Surveillance Program (1997-2003). Diagn. Microbiol. Infect. Dis. 51:147-150. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schio, L., F. Chatreaux, V. Loyau, M. Murer, A. Ferreira, P. Mauvais, A. Bonnefoy, and M. Klich. 2001. Fine tuning of physico-chemical parameters to optimise a new series of novobiocin analogues. Bioorg. Med. Chem. Lett. 11:1461-1464. [DOI] [PubMed] [Google Scholar]
- 29.Stieger, M., P. Angehrn, B. Wohlgensinger, and H. Gmunder. 1996. GyrB mutations in Staphylococcus aureus strains resistant to cyclothialidine, coumermycin, and novobiocin. Antimicrob. Agents Chemother. 40:1060-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waites, K. B., L. B. Duffy, T. Schmid, D. Crabb, M. S. Pate, and G. H. Cassell. 1991. In vitro susceptibilities of Mycoplasma pneumoniae, Mycoplasma hominis, and Ureaplasma urealyticum to sparfloxacin and PD127391. Antimicrob. Agents Chemother. 35:1181-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zechiedrich, E. L., A. B. Khodrusky, S. Bachellier, R. Schneider, D. Chen, D. M. J. Lilley, and N. R. Cozzarelli. 2000. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 11:8103-8113. [DOI] [PubMed] [Google Scholar]