Abstract
Several studies have shown that phosphorothioate oligodeoxynucleotides (PS-ONs) have a sequence-independent antiviral activity against human immunodeficiency virus type 1 (HIV-1). It has also been suggested that PS-ONs inhibit HIV-1 by acting as attachment inhibitors that bind to the V3 loop of gp120 and prevent the gp120-CD4 interaction. Here we show that PS-ONs (and their fully 2′-O-methylated derivatives) are potent inhibitors of HIV-1-mediated membrane fusion and HIV-1 replication in a size-dependent, phosphorothioation-dependent manner. PS-ONs interact with a peptide derived from the N-terminal heptad repeat region of gp41, and the HIV-1 fusion-inhibitory activity of PS-ONs is closely correlated with their ability to block gp41 six-helix bundle formation, a critical step during the process of HIV-1 fusion with the target cell. These results suggest that the increased hydrophobicity of PS-ONs may contribute to their inhibitory activity against HIV-1 fusion and entry, because longer PS-ONs (≥30 bases) which have a greater hydrophobicity are more potent in blocking the hydrophobic interactions involved in the gp41 six-helix bundle formation and inhibiting the HIV-1-mediated cell-cell fusion than shorter PS-ONs (<30 bases). This novel antiviral mechanism of action of long PS-ONs has implications for therapy against infection by HIV-1 and other enveloped viruses with type I fusion proteins.
Several studies have suggested that the antiviral activity of phosphorothioated oligonucleotides (PS-ONs) with different sequences or different intended mechanisms of action (i.e., antisense or sequence-specific aptamers) all work as inhibitors of human immunodeficiency virus (HIV) viral entry by binding to the V3 loop of gp120 and preventing CD4 interaction and attachment to the host cell (13, 43). The general conservation of this mechanism of action with different oligonucleotides having different sequences suggested that the antiviral activity of oligonucleotides against HIV was sequence independent.
The sequence-independent nature of the antiviral activity of oligonucleotides (ONs) was more clearly demonstrated in several studies using degenerate or homopolymeric ONs (24, 30, 37). In these studies, the anti-HIV activities of 28-mer phosphorothioated polycytidylic, polyadenylic, and polythimidylic or fully degenerate ONs were studied in vitro, and in the case of polycytidylic ONs, it was also discovered that for ONs up to 28 bases in length increasingly larger ONs displayed more potent antiviral activity. The binding of 28-mer polycytidylic ONs to the V3 loop of gp120 was also described (38).
PS-ONs are also polyanionic, and the antiviral activities of a variety of other classes of polyanions, such as sulfated saccharides, polysulfonates, polyhydroxycarboxylates, and polyoxometalates, have also been well established (11, 28, 31, 34, 35). There is good evidence to suggest that these compounds also interfere with viral attachment by binding to the V3 loop of gp120 (2, 12, 33, 40).
To further explore the sequence-independent mechanisms of the antiviral activity of PS-ONs, we examined the antiviral activity against HIV type 1 (HIV-1) of completely degenerate (Randomer) PS-ONs of different sizes with their PS-2′-O-methylated and phosphodiester (PO)-2′-O-methylated counterparts. Both PS-ONs and their 2′-O-methylated derivates inhibited HIV-1-mediated membrane fusion and the fusion-active gp41 core formation in a well-correlated, size-dependent manner, whereas 2-O-methylated, nonphosphorothioated ONs were inactive. The optimal size for antiviral activity of PS-ONs was also much greater (∼50 to 60 bases) than previously reported. These results define a novel mechanism of action for longer (≥30 bases) PS-ONs which has important implications for its use as a therapy against infection by HIV and other viruses with type I fusion proteins.
MATERIALS AND METHODS
Reagents.
HIV-1IIIB, MT-2 cells, HIV-1IIIB chronically infected H9 cells (H9/HIV-1IIIB), GHOST-X4 cells, GHOST-R5 cells, MAGI-X4 cells, MAGI-R5 cells, HIV-1BaL, T-20-resistant HIV-1NL4-3 strains, V3 loop, and N36 and C34 peptides were obtained from the NIH AIDS Research and Reference Reagent Program. Additional preparations of peptides N36, C34, and C34-FITC (fluorescein isothiocyanate was added to the N terminus of C34) were synthesized by a standard solid-phase FMOC method in the MicroChemistry Laboratory of the New York Blood Center. The peptides were purified to homogeneity by high-performance liquid chromatography. The identity of the purified peptides was confirmed by laser desorption mass spectrometry (PerSeptive Biosystems). Rabbit antisera directed against the mixture of N36/C34 were prepared as previously described (21). Mouse monoclonal antibody NC-1 specific for the gp41 six-helix bundle (6-HB) was prepared and characterized as previously described (21). Rabbit and mouse immunoglobulin G (IgG) were purified using protein A/G beads (Pierce, Rockford, IL). Chicago Sky Blue (CSB) and zidovudine (AZT) were obtained from Sigma, and nevirapine (NVP) was obtained from the NIH AIDS Research and Reference Reagent Program.
Oligonucleotide synthesis.
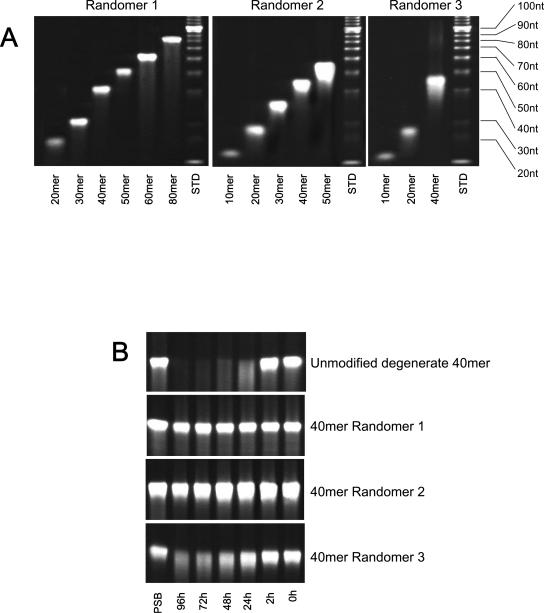
ONs were prepared at the University of Calgary DNA synthesis lab using standard phosphoramidite chemistry with commercially available reagents. Fully degenerate ONs (which we have termed Randomers) were prepared by mixing equimolar concentrations of all four amidites during coupling. ONs were deprotected and cleaved with NH4OH, desalted on a Sephadex 250 column, and lyophilized prior to quantification by absorbance at 260 nm using appropriate extinction coefficients where necessary (for homo- and heteropolymeric PS-ONs). All ONs were then prepared as 1 mM stocks in 10 mM Tris, pH 7.2, and diluted appropriately. Size analysis was performed on 10% urea-acrylamide gels using ethidium bromide to detect oligonucleotides (see Fig. 2A, below).
FIG. 2.
Synthesis and stability of modified ONs. (A) Size analogues of Randomer 1, 2, and 3 ONs were resolved on 10% urea-acrylamide gels to verify their size. In all cases, >90% of the oligo was observed to be full length. Size standards (in nucleotides [nt]) are identified on the right. (B) The temporal stabilities of 40-mer ONs with Randomer 1, 2, or 3 chemistries in fetal calf serum at 37°C were compared to the unmodified 40-mer degenerate ON. Following incubation for various times, ONs were resolved as for panel A. Degradation in both the 40-mer Randomer 1 and 2 ONs was undetectable after 96 h, the 40-mer Randomer 3 ON showed partial degradation beginning at 48 h, and the unmodified 40-mer degenerate ON was almost completely degraded by 48 h. ONs dissolved in PBS are provided as controls on the left.
Cell-cell fusion.
A dye transfer assay was used for detection of HIV-1-mediated cell-cell fusion as previously described (22). H9 cells chronically infected with HIV-1IIIB (H9/HIV-1IIIB) were labeled with a fluorescent reagent, calcein-AM (Molecular Probes, Inc., Eugene, Oreg.) and then incubated with MT-2 cells (ratio,1:5) in 96-well plates at 37°C for 2 h in the presence or absence of ONs. The fused and unfused calcein-labeled HIV-1-infected cells were counted under an inverted fluorescence microscope (Zeiss, Oberkochen, Germany) with an eyepiece micrometer disk. The percent inhibition of cell-cell fusion and the 50% inhibitory concentration (IC50) values were calculated as previously described (22).
Assessment of anti-HIV-1 infectivity.
The inhibitory activity of compounds on infection by laboratory-adapted HIV-1 strains was determined as previously described (18, 19). In brief, 104 MT-2 cells were infected with HIV-1 at 100 50% tissue culture infective doses in 200 μl of RPMI 1640 medium containing 10% fetal bovine serum in the presence or absence of compounds at graded concentrations overnight. After culture for another 2 h, the cells were washed to remove the free virus and compounds. Fresh medium without testing compounds was added. On the fourth day postinfection, 100 μl of culture supernatant was collected from each well, mixed with equal volumes of 5% Triton X-100, and assayed for p24 antigen, which was quantified by enzyme-linked immunosorbent assay (ELISA) as previously described (18). After incubation for two additional days, the indicator dye XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] tetrazolium dye (1 μg/ml; 50 μl/well; PolySciences, Inc., Warrington, PA) was added to the cells, followed by incubation at 37°C for 4 h. The soluble intracellular compound formazan was determined colorimetrically at 450 nm (19).
Time of addition assays.
HeLa CD4-LTR-β-gal cells (MAGI-X4) were plated 1 day prior to the experiment. On the day of the experiment, medium was removed from the wells, and 100 μl of the appropriate concentrations of control compounds and Randomer 1 were added to the time zero row of each plate, immediately followed by addition of an equal volume of diluted HIV-1IIIB at a high multiplicity of infection to all infected rows. Plates were incubated at 37°C in a humidified atmosphere containing approximately 5% CO2. After 2 h of infection, cells were washed to remove unbound virus and ON was added back to the 0-h postinfection rows as well as to the 2-h postinfection row. Medium was added back to all remaining wells. At the appropriate times postinfection (4, 6, 8, 12, and 24 h), ON was added to the remaining wells. Endpoints for antiviral efficacy for each experimental group were measured in triplicate. The chemiluminescent endpoint is used to determine the expression of β-galactosidase (β-Gal) as a function of HIV-1 infection of the cells. At 48 h postinfection, plates were aspirated and phosphate-buffered saline (PBS) was added to each well before freezing the plates at −20°C for subsequent chemiluminescence detection of β-galactosidase activity. β-Gal expression was measured using Gal screen (Applied Biosystems, Foster City, CA).
Inhibition of viral attachment with X4- and R5-tropic HIV-1 strains.
The viral attachment inhibition assay is designed to detect compounds that block virus attachment using MAGI-R5 or MAGI-X4 cells. Cells are routinely cultured with the required selection antibiotics. Twenty-four hours prior to initiation of the assay, the cells are trypsinized, counted, and plated in a 0.2-cm well in medium without selection antibiotics. At 24 h medium is removed and test compound in medium is placed on the cells and incubated for 15 min at 37°C. A known titer of HIV-1IIIB (X4) or BaL (R5) is then added to the wells (containing MAGI-X4 or MAGI R5 cells, respectively), and the incubation is continued for 1 to 2 h. At the end of the incubation the wells are washed two times with medium and the culture is continued for 40 to 48 h. At termination of the assay, medium is removed and β-galactosidase enzyme expression is determined by chemiluminescence per the manufacturer's instructions (Gal screen; Applied Biosystems). All determinations are performed in triplicate with serial log dilutions of ONs. The virus adsorption interval of 1 to 2 h is sufficiently short that AZT, which requires phosphorylation to achieve its active triphosphate form (AZT-TTP), is not active in this assay (data not shown).
ELISA for measuring inhibition of the gp41 six-helix bundle formation.
A sandwich ELISA with conformation-specific monoclonal antibody NC-1 (21) as previously described (18, 20) was used to screen for compounds that inhibit the gp41 six-helix bundle formation. Briefly, 2 μM peptide N36 was preincubated with a test compound at the indicated concentrations at 37°C for 30 min, followed by addition of 2 μM C34. After incubation at 37°C for 30 min, the mixture was added to wells of a 96-well polystyrene plate (Costar; Corning, Inc., Corning, N.Y.), which were precoated with IgG (2 μg/ml) purified from rabbit antiserum directed against the N36-C34 mixture. Then, monoclonal antibody NC-1, biotin-labeled goat anti-mouse IgG (Sigma Chemical Co., St. Louis, Mo.), streptavidin-labeled horseradish peroxidase (Zymed, South San Francisco, Calif.), and the substrate 3,3′,5,5′-tetramethylbenzidine (Sigma) were added sequentially. The A450 was measured by using a plate reader (Tecan Ultra 384). The percent inhibition by the ONs was calculated as previously described (19).
FN-PAGE for measuring inhibition of the gp41 six-helix bundle formation.
Fluorescence native-polyacrylamide gel electrophoresis (FN-PAGE) was performed using precast Tris-glycine gels (18%) and a Novex X-Cell II Mini Cell (Invitrogen, Carlsbad, CA) as previously described (27). Briefly, the peptide N36 was incubated with PBS or ONs at the indicated concentrations at 37°C for 30 min before or after addition of FITC-labeled C34 (the final concentration of N36 and C34 was 40 μM in PBS). After further incubation at 37°C for 30 min, the sample was mixed with Tris-glycine native sample buffer (Invitrogen, Carlsbad, CA) at a ratio of 1:1 and was then loaded onto a 10-cm by 1-cm precast gel (25 μl each well). Gel electrophoresis was carried out with 125-V constant voltage at room temperature for 2 h. Immediately after electrophoresis, fluorescence bands in the gel were imaged with the FluorChem 8800 imaging system using a transillumination UV light source with excitation wavelength at 302 nm and a fluorescence filter with emission wavelength at 520 nm.
Fluorescence polarization for measuring ON-peptide interactions.
The binding affinities of HIV-1 peptides to various fluorescently labeled Randomer PS-ONs were monitored using fluorescence polarization. These peptides were serially diluted in assay buffer (10 mM Tris, pH 7.2, 80 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 0.1% Tween 20) and allowed to interact with 3 nM FITC-labeled Randomer PS-ONs for 30 seconds. Peptide binding was monitored by fluorescence polarization at 535 nm using a Tecan Ultra plate reader. The Kd was determined from the concentration of peptide that resulted in 50% of the maximal polarization observed (saturated peptide interaction). Averages and standard deviations were from at least three independent measurements.
RESULTS
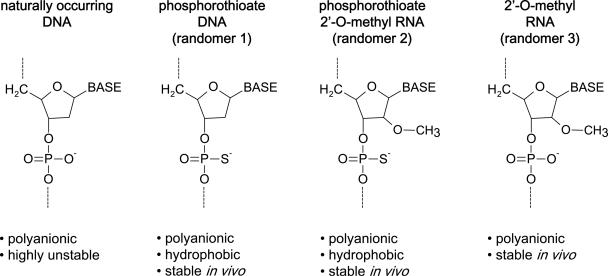
Since previous reports suggested that the polyanionic nature of PS-ONs was responsible for their anti-HIV activity, three different classes of fully degenerate ONs (Randomers) were synthesized: (i) ONs with phosphorothioation at each internucleotidic linkage, (ii) PS-ONs with 2′-O-methylation of each ribose moiety, and (iii) ONs with only 2′-O-methylation of each ribose moiety (without phosphorothioation). These different varieties of ON chemistries were designated as Randomers 1, 2, and 3, respectively (Fig. 1 and Table 1). Phosphorothioation stabilizes and increases the hydrophobicity of ONs (1). 2′-O-methylation also stabilizes ON structures (Fig. 2B) but does not increase their hydrophobicity, which allows the involvement of phosphorothioation in the antiviral activity of ONs to be assessed. While some degradation with Randomer 3 ONs was observed at 48 h and beyond (Fig. 2B), the stability of these compounds was sufficient to allow comparison of the activities of these ONs in tissue culture with Randomer 1 and 2 ONs and did not impact on the other assays performed here, which were either 2 h in tissue culture or in cell-free systems where nucleases were absent. Different-sized analogues (6 to 120 bases) of the three Randomer chemistries were synthesized (Fig. 2A). We chose to use fully degenerate oligonucleotides in this study to avoid any antisense or sequence-specific aptameric effects that could be associated with a specific sequence. The fully degenerate nature of these compounds was examined in a 40-base Randomer 1 PS-ON by quantifying with high-performance liquid chromatography the amount of each base incorporated into this PS-ON following oxidation and cleavage into individual nucleotides by S1 nuclease or snake venom phosphodiesterase (following the general methodologies of references 1 and 36), and the extent of phosphorothioation was assessed by 31P nuclear magnetic resonance (Table 2). This analysis demonstrated the approximately equivalent incorporation of all four nucleotides during synthesis and >99% efficiency of phosphorothioation.
FIG. 1.
Nucleic acid chemistry used in the experiments. Phosphorothioation (which stabilizes and increases hydrophobicity to ONs) and 2′-O-methylation (which only stabilizes ONs) were used to alter the chemical properties of Randomers used in this study.
TABLE 1.
Examples of oligonucleotide structure and chemistry used in this study
| Chemistry | Sequence (5′-3′)a | Chemical properties |
|---|---|---|
| Randomer 1 | NpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNp NpNpNpNpNpNpNpN (40-mer) | Polyanionic, hydrophobic |
| Randomer 2 | NpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNpNp NpNpNpNpNpNpNpN (40-mer) | Polyanionic, hydrophobic |
| Randomer 3 | NoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNoNo NoNoNoNoNoNoNoN (40-mer) | Polyanionic |
N, unmodified deoxyribonucleotide; N, 2′-O-methylated ribonucleotide; p, phosphorothioate linkage; o, phosphodiester linkage. Note that N also indicates the presence of any of the four naturally occurring nucleotides (adenosine, guanosine, cytosine, or thymidine/uridine).
TABLE 2.
Degeneracy and phosphorothioation content of a 40-base Randomer 1 PS-ON
| Batch | Phosphorothioate composition (%) | Base composition (avg no. of bases in a 40-mer) of S1 nuclease/SV phosphodiesterase
|
|||
|---|---|---|---|---|---|
| Cytosine | Thymidine | Adenosine | Guanosine | ||
| 1 | >99 | 8.15/8.39 | 11.28/12.11 | 8.45/7.84 | 12.12/11.66 |
| 2 | >99 | 9.3/9.1 | 11.7/11.2 | 8.8/9.1 | 10.2/10.6 |
| 3 | >99 | 10.7/10.5 | 10.7/10.9 | 7.81/7.57 | 11.3/11.0 |
PS-ONs inhibit HIV-1-mediated cell-cell fusion and HIV-1 replication in a size- and phosphorothioation-dependent manner.
Different sizes of Randomer 1, 2, and 3 ONs were tested for their inhibitory activity on HIV-1-induced cell-cell fusion in a dye transfer assay, and HIV-1 replication was determined by ELISA for p24 production and an XTT assay for cytopathic effect (CPE) (Table 3). With increasing length, the activities of Randomers 1 and 2 became more potent until they were ∼60 bases in length, after which we observed no further increase in anti-HIV activity with longer PS-ONs. None of the Randomer 3 size analogues, which were stable but not phosphorothioated, had any activity in inhibiting cell-cell fusion or HIV-1 replication.
TABLE 3.
Randomer antiviral activity in HIV-1 is dependent on size and increased hydrophobicity
| Chemistry | Size (bases) | IC50 (μM, mean ± SD) for:
|
||
|---|---|---|---|---|
| Cell fusion | p24 (ELISA) | CPE (XTT) | ||
| Randomer 1 (phosphorothioate) | 6 | >2 | >2 | >2 |
| 10 | >2 | >2 | >2 | |
| 20 | >2 | 0.3699 ± 0.0359 | 0.7946 ± 0.1332 | |
| 30 | 0.465 ± 0.049 | 0.1439 ± 0.0046 | 0.3833 ± 0.0107 | |
| 40 | 0.285 ± 0.029 | 0.0909 ± 0.0001 | 0.1149 ± 0.0048 | |
| 50 | 0.148 ± 0.012 | 0.0207 ± 0.0037 | 0.0337 ± 0.0110 | |
| 60 | 0.064 ± 0.002 | 0.0158 ± 0.0016 | 0.0482 ± 0.0108 | |
| 80 | 0.069 ± 0.006 | 0.0084 ± 0.0014 | 0.0105 ± 0.0012 | |
| 120 | 0.052 ± 0.001 | 0.0089 ± 0.0015 | 0.0178 ± 0.0037 | |
| Randomer 2 (phosphorothioate + 2′-O-methyl) | 6 | >2 | >2 | >2 |
| 10 | >2 | >2 | >2 | |
| 20 | >2 | 0.2648 ± 0.0716 | 0.4428 ± 0.0540 | |
| 30 | 0.209 ± 0.009 | 0.0454 ± 0.0048 | 0.0414 ± 0.0047 | |
| 40 | 0.150 ± 0.027 | 0.0353 ± 0.0048 | 0.0402 ± 0.0078 | |
| 50 | 0.136 ± 0.013 | 0.0253 ± 0.0031 | 0.0321 ± 0.0070 | |
| Randomer 3 (2′-O-methyl) | 6 | >2 | >2 | >2 |
| 10 | >2 | >2 | >2 | |
| 20 | >2 | >2 | >2 | |
| 40 | >2 | >2 | >2 | |
Inhibition of HIV-1-mediated cell-cell fusion by PS-ONs is sequence independent.
A series of analogues of Randomer 1 consisting of specific nucleotides, including poly(A), -(C), -(T), -(G), -(A · C), -(A · G), -(T · C), and -(T · G) were synthesized and tested for their inhibitory activity on HIV-1-mediated cell-cell fusion at identical concentrations (individual ON concentrations were established using the specific extinction coefficients for each nucleotide). Except for poly(G), which has the propensity for forming complicated tertiary structures (23), and poly(A), which as a polypurine may also form similar secondary structure, all other ONs had similar inhibitory activities as Randomer 1 (Table 4). We noted that the poly(C) compound was significantly more active than the other hetero- or homopolymers tested, and we attribute this to the increased nuclease resistance (using purified nucleases) observed with this compound (data not shown).
TABLE 4.
HIV fusion inhibition activity of various 40-base hetero- and homopolymer PS-ONs
| PS-ON sequence | IC50 of cell fusion (μM) (mean ± SD) |
|---|---|
| G40 | >4.000 |
| A40 | 1.272 ± 0.042 |
| T40 | 0.244 ± 0.007 |
| C40 | 0.094 ± 0.001 |
| (TG)20 | 0.330 ± 0.012 |
| (AC)20 | 0.225 ± 0.009 |
| (TC)20 | 0.479 ± 0.007 |
| (AG)20 | 0.386 ± 0.005 |
PS-ONs block HIV-1 entry.
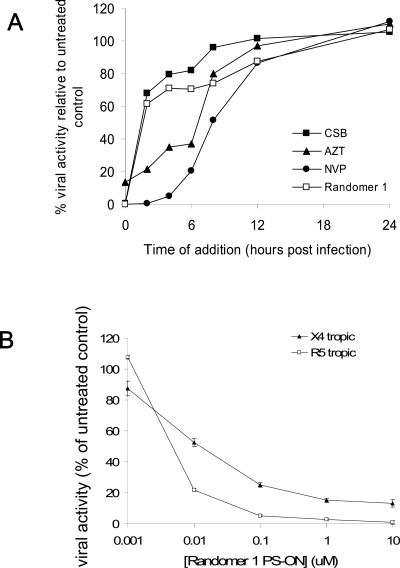
To determine whether the PS-ONs were HIV-1 entry inhibitors, we tested the inhibitory activity of a 40-base Randomer 1 PS-ON on HIV-1 replication in a time-of-addition assay by adding the compound at different time intervals of inoculation of HIV-1. Three anti-HIV-1 compounds, CSB (a polyanionic dye known be an attachment inhibitor [9]), AZT (a nucleotide reverse transcriptase [RT] inhibitor), and NVP (a nonnucleotide RT inhibitor), were used as controls. As shown in Fig. 3A, CSB only showed potent antiviral activity when present during infection, while both AZT and NVP retained activity when added 6 to 8 h after infection, consistent with their postinfective mechanisms of action. The 40-base Randomer 1 PS-ON displayed the same timeframe of activity as CSB, only having potent activity when present during infection. This compound was also tested for its ability to prevent the attachment of X4- or R5-tropic HIV-1 strains to host cells (Fig. 3B). This PS-ON prevented both X4- and R5-tropic strains of HIV-1 from attaching to the host cell. This demonstrates that PS-ONs are HIV-1 entry inhibitors that can block X4- or R5-tropic HIV-1 attachment and fusion.
FIG. 3.
(A) Effect of time of addition on the antiviral activity of a 40-base Randomer 1. Various control compounds (see Results) and Randomer 1 were added to cultured cells at various times during (<2 h) or after (>2 h) infection with HIV-1. All compounds were used at IC99. (B) Inhibitory activity of PS-ONs in attachment and fusion. A 40-base Randomer 1 PS-ON was tested in an attachment-fusion assay against both X4- and R5-tropic HIV-1 strains.
PS-ONs block the gp41 six-helix bundle formation by targeting the gp41 NHR region.
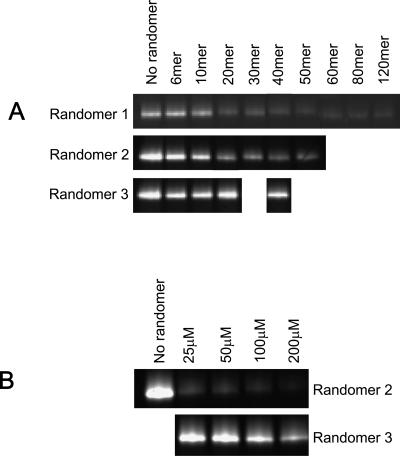
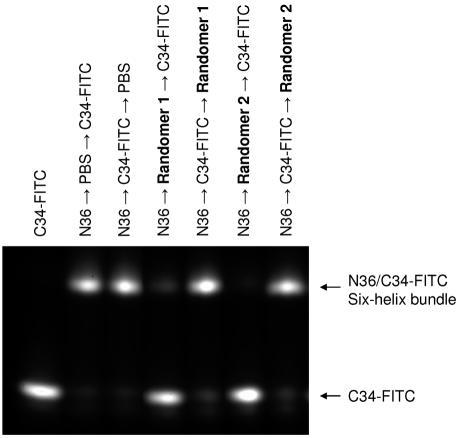
The ability of PS-ONs to block gp41 6-HB formation, a critical step of HIV-1-mediated membrane fusion (6), was examined using a sandwich ELISA (20) with monoclonal antibody NC-1, which specifically recognizes the 6-HB formed by the peptides N36 and C34 (21), and by FN-PAGE, which showed different bands of the 6-HB formed by the N36/C34 complex and the individual peptides N36 and C34 (27). Strikingly, PS-ONs (whether 2′-O-methylated or not) inhibited 6-HB formation in a size-dependent manner, whereas ONs that were only 2-O-methylated had no inhibitory activity (Table 5 and Fig. 4). These data are consistent with the size-dependent, phosphorothioation-dependent antiviral activity of PS-ONs (Table 3). We noted that this inhibition was noncompetitive, as 40-base Randomer 1 or 2 PS-ONs added to the N36 peptide after the C34 peptide resulted in no inhibition of 6-HB formation (Fig. 5). We also noted that the absolute activities of Randomers in the ELISA were lower than those shown in Table 3, but we consider this a function of the differences between these two assay systems (direct antiviral activity measurement in Table 3 versus inhibition of the N36-C34 peptide interaction in Table 5). What is consistent between these assays is a well-correlated size-dependent, phosphorothioation-dependent activity of oligonucleotides in inhibiting antiviral activity in vitro (Table 3), inhibiting 6-HB formation measured by FN-PAGE (Fig. 4) and by ELISA (Table 5).
TABLE 5.
Size- and chemistry-dependent ON inhibition of 6-HB formation by ELISA and FN-PAGE
| Chemistry | Size (bases) | 6-HB (ELISA) IC50 (μM, mean ± SD) | 6-HB inhibition (FN-PAGE)a |
|---|---|---|---|
| Randomer 1 | 6 | >100 | − |
| 10 | >100 | − | |
| 20 | >100 | ++ | |
| 30 | 9.80 ± 2.65 | ++ | |
| 40 | 8.37 ± 0.67 | ++ | |
| 50 | 0.18 ± 0.09 | +++ | |
| 60 | 0.39 ± 0.12 | +++ | |
| 80 | 0.83 ± 0.07 | +++ | |
| 120 | 0.23 ± 0.02 | +++ | |
| Randomer 2 | 6 | >100 | − |
| 10 | >100 | − | |
| 20 | 24.94 ± 4.13 | ++ | |
| 30 | 1.20 ± 0.13 | ++ | |
| 40 | 0.47 ± 0.001 | +++ | |
| 50 | 0.48 ± 0.07 | +++ | |
| Randomer 3 | 6 | >100 | − |
| 10 | >100 | − | |
| 20 | >100 | − | |
| 40 | >100 | − |
With 200 μM Randomer. −, no inhibition; ++, moderate inhibition; +++, >90% inhibition.
FIG. 4.
PS-ON size and phosphorothioation dependence on inhibition of 6-HB formation by FN-PAGE. (A) Different-size analogues of Randomer 1, 2, and 3 ONs (200 μM) were added to the N36 peptide (40 μM) before the addition of the FITC-C34 peptide (40 μM). A size-dependent inhibition was observed with Randomer 1 and 2 PS-ONs, but no significant inhibition was observed with Randomer 3 ONs of any size. (B) Dose response of the inhibition of the N36/FITC-C34 complex (both at 40 μM) by increasing concentrations of 40-base Randomer 2 or 3 ONs. For simplicity only the region of the gel displaying the 6-HB complex is shown in panels A and B.
FIG. 5.
FN-PAGE analysis of inhibition of 6-HB formation by PS-ONs. The 40-base Randomer 1 or 2 ONs (200 μM) were added to the N36 peptide (40 μM) before or after the addition of FITC-labeled C34 peptide (40 μM). The N36/FITC-C34 6-HB complex has a retarded mobility (as indicated on the right of the gel). Randomers 1 and 2 were active in preventing 6-HB formation when they were incubated with N36 prior to the addition of FITC-C34.
PS-ONs are highly potent at inhibiting infection by T-20-resistant HIV-1 strains.
Because the T-20 peptide is partially derived from the CHR region of gp41, the sensitivity of T-20-resistant HIV-1 strains to the action of a 40-base Randomer 1 PS-ON was assessed in MAGI-X4 cells (Table 6). The T-20-resistant strains, which showed significantly reduced susceptibility to T-20, were equivalently sensitive to the 40-base Randomer 1 PS-ON compared to wild-type strains. We noted that the activities of this compound in this assay were significantly better than those in Table 3, and we attribute these differences to different cell lines, assay conditions, and methods of detecting viral activity.
TABLE 6.
Activity of a 40-base Randomer 1 PS-ON against T-20-resistant HIV-1 strains
| HIV-1NL4-3 gp41 variant | Resistance to T-20 | IC50 (nM)
|
|
|---|---|---|---|
| T-20 | 40-base PS-ON (Randomer 1) | ||
| D36G (parental NL4-3) | None | 2.02 | 13.2 |
| D36G N42S | None | 1.58 | 16.1 |
| D36G V38A | 16-fold | 22.6 | 5.21 |
| D36G V38A, N42D | 140-fold | 114 | 4.86 |
| D36G V38A, N42T | 149-fold | 177 | 5.36 |
| D36G V38E, N42S | 513-fold | 678 | 4.86 |
| D36G N42T, N43K | 32-fold | 52.6 | 7.61 |
Randomer interaction with N36, C34, and the V3 loop.
Previous studies have described the interaction of PS-ONs with the V3 loop of gp120 (38). In the present study, the direct interaction of 40-base Randomer 1, 2, and 3 ONs with N36 (HR1) and C34 (HR2) peptides from the HIV-1 gp41 and a V3 loop peptide from the HIV-1 gp120 was examined. Binding studies using fluorescence polarization (Table 7) showed that all 40-base Randomers (1, 2, and 3) bound the N36 peptide, none bound to the C34 peptide, and only the 40-base Randomer 1 and Randomer 2 PS-ONs bound to the V3 loop peptide. These data suggest that PS-ONs can interact with both gp41 and gp120, consistent with previously reported interactions of PS-ONs with the V3 loop of gp120, but also that increased hydrophobicity (phosphorothioation) is required for the interaction with gp41 and the V3 loop of gp120.
TABLE 7.
Binding of 40-base Randomers to N36, C34, and V3 loop peptides
| 40-base ON chemistry |
Kd (μM, mean ± SD)
|
||
|---|---|---|---|
| N36 | C34 | V3 loop | |
| Randomer 1 | 4.03 ± 0.26 | No binding | 6.97 ± 2.88 |
| Randomer 2 | 5.34 ± 1.74 | No binding | 6.30 ± 3.44 |
| Randomer 3 | 7.54 ± 0.72 | No binding | No binding |
DISCUSSION
The data presented here suggest that in addition to the previously established ability of PS-ONs to inhibit the attachment of HIV virions with the host cell, longer (≥30-base) PS-ONs are more potent HIV-1 entry inhibitors that act by interfering with HIV-1-mediated membrane fusion. This novel fusion inhibition activity by ONs in HIV-1 is size and phosphorothioation dependent and sequence independent, consistent with the activity of degenerate (Randomer) PS-ONs and also the comparable activity of most homo- and heteropolymeric PS-ON sequences (Tables 3 and 4). A few notable exceptions to this rule were the inactivity of the 40-mer poly(G) sequence, which can be attributed to significant tertiary structure formed as previously described for polyguanosine tracts (23) and suggests a requirement for a relaxed ON conformation for efficient binding to its target. The weaker activity of the 40-mer poly(A) sequence may be attributable to a similar but less stable secondary structure, as it is also a polypurine like poly(G). We also noted that the 40-mer poly(C) sequence was the most potent of all the sequences tested, and this is consistent with the increased nuclease resistance we observed with poly(C) ONs compared to degenerate and other hetero- and homopolymeric sequences (data not shown).
Degenerate ONs must exhibit some secondary structure in solution due to the vast number of different sequences that are present, and it could be argued that the increased antiviral activity of degenerate PS-ONs was a result of increased secondary structure (i.e., G-quartet structures) present in longer PS-ONs. However, it has been clearly demonstrated that 2′-O-methyl-modified RNA has an equal propensity for forming G-quartet and other structures compared to phosphorothioated DNA (10). Thus, if any G-quartet-based or secondary structure-based activity were involved in the antiviral activity of Randomers, this would have been seen with the Randomer 3 ONs. The absence of activity with these compounds shows that secondary structure (including G-quartet structures) is a negligible component of the antiviral activity of these compounds. This control also serves to show that there is a real length dependence for antiviral activity.
Longer PS-ONs also have a potent size- and phosphorothioation-dependent inhibitory activity on the formation of the gp41 six-helix bundle (Fig. 4 and Table 5), a critical step during the process of HIV-1 fusion (7) that is well-correlated with their inhibition of HIV-1-mediated membrane fusion. The direct interaction of PS-ONs with the heptad repeats of gp41 is further supported by the fact that fully degenerate PS-ONs can bind to N36, a peptide derived from the gp41 NHR region. The binding of the 40-mer Randomer 3 ON to N36 (Table 7) at first presented a paradox, as this compound had no antiviral activity, but since the 40-mer Randomer 3 ON was inactive in preventing the formation of the 6-HB (Fig. 4b and Table 5), it seems likely that the 40-mer Randomer 3 ON binds to the hydrophilic face of the N36 α-helix (consistent with its polyanionic character) and does not prevent the interaction of the hydrophobic face of the N36 α-helix with its C34 counterpart (Fig. 5). We noted that none of the 40-mer Randomer ONs interacted with the C34 peptide; however, this is consistent with the fact that this peptide in solution does not form any ordered structure (S. Jiang, unpublished results) and the requirement for an amphipathic α-helical structure for PS-ON interaction.
The requirement of phosphorothioation of ONs for their antiviral activity can be explained by the effect this modification has on the chemical makeup of ONs. It is known that phosphorothioation increases the hydrophobicity of ONs (1). Therefore, the increased hydrophobicity of PS-ONs may contribute to the increased amphipathic and fusion-inhibitory activity of PS-ONs; longer PS-ONs may have greater hydrophobic character or may be able to interact more completely with the entire α-helix. In fact, the interaction between the N36 peptide and 40-base PS-ONs is apparently noncompetitive (Fig. 5), suggesting the presence of a very-high-affinity interaction. The strong binding affinity with the gp41 NHR α-helices and potent inhibitory activity on the gp41 six-helix bundle formation are consistent with potent inhibition of HIV-1 fusion and entry. Thus, while shorter PS-ONs (<30 bases) may be good attachment inhibitors, longer PS-ONs (≥30 bases) inhibit both the attachment and fusion of HIV-1 virions with the host cell.
In this study, we also observed that 40-mer Randomer 1 and 2 PS-ONs could bind to the V3 loop, consistent with reports that suggested PS-ONs may inhibit HIV-1 attachment through their polyanionic features (38, 43). However, we noted that Randomer 3 ONs did not display any antiviral activity, although they also possess similar polyanionic properties. These observations suggested that the interaction of PS-ONs with the V3 loop was not dependent on their polyanionic nature but rather on their increased hydrophobic nature. While historically the V3 loop-CD4 interaction has been considered to be primarily charge based, there is substantial evidence to suggest that this may not entirely be the case. First, the positive charge of the V3 loop is highly variable (16), suggesting that the receptor interactions may not be only charge based. Furthermore, the V3 loop contains a conserved α-helix in several different strains of HIV (5, 32), suggesting that amphipathic interactions similar to those seen for gp41 are involved as well. Thus, PS-ONs may inhibit both HIV-1 attachment and HIV-1-mediated membrane fusion by a mechanism of action dependent on their amphipathic as opposed to polyanionic character. While it is possible that other polyanionic molecules exert their antiviral activity via a different mechanism, the results reported here suggest that the antiviral activity of polyanions may also be related in part to their hydrophobic properties.
The novel fusion inhibition activity of long (≥30-base) PS-ONs has important therapeutic implications. If the increased hydrophobicity of PS-ONs is a major source for its antiviral activity, PS-ONs may also nonspecifically interact with other hydrophobic molecules (such as serum proteins), which could affect the in vivo efficacy of PS-ONs. However, it is important to note that while it is widely accepted that PS-ONs interact with serum proteins in the blood, this does not apparently prevent PS-ONs from reaching their therapeutic target: first-generation (PS-ON) antisense compounds like Genasense (17) and Alicaforsen (41) have recently been shown to be therapeutically effective at well-tolerated doses. Moreover, the requirement for these antisense compounds to penetrate inside the cell, a barrier which does not exist for the antiviral activity of PS-ONs described here, bodes well for the potential antiviral therapeutic activity of antiviral PS-ONs, which act outside the cell.
The mechanism of action of these long PS-ONs suggests that their anti-HIV activity may be derived from their hydrophobic interactions with multiple target sites along the gp41 amphipathic α-helices, rather than a specific active site that is sensitive to point mutation. Therefore, individual point mutations may be insufficient to alter the interaction of PS-ONs with the gp41 coiled-coil domain. Moreover, any significant mutation in the conserved hydrophobic regions of the gp41 core, the potential targets of PS-ONs, may result in an attenuated activity of the virus due to the reduced capacity of the gp41 NHR and CHR to form 6-HBs and allow membrane fusion with target cells. Therefore, by targeting the entire α-helical surface, chronic treatment with PS-ONs may be unlikely to result in the development of significant resistance and will also have broad activity against different HIV-1 strains. This potential broad-spectrum anti-HIV activity is supported by the fact of the unaltered antiviral activity of PS-ONs against several T-20-resistant strains (Table 6) which also target gp41 and also by the fact that PS-ONs have significant antiviral activity in multiple HIV-1 subtypes and strains resistant to RT and/or protease inhibitors (data not shown). Moreover, the unaltered activity of the 40-base Randomer 1 PS-ON in wild-type versus T-20-resistant strains suggests their mechanism of action is dissimilar to that of T-20. In fact, it has been recently reported that the T-20 peptide does not interfere with the α-helical regions in gp41 (26), which is consistent with the results reported here.
The interaction of Randomers with the α-helices of gp41, the prototypical type I viral fusion protein, has important implications for the potential broad-spectrum antiviral activities of longer PS-ONs. The α-helical domains found in HIV-1 gp41 are common structures present in all type I fusion proteins that have been characterized to date. The conservation of these domains is a good example of convergent evolution, in which these structures (the α-helices) are highly similar in viruses from several different families but are encoded by amino acid sequences that share no homology. To date, type I fusion proteins (α-helices) have been identified in the following viral families: Retroviridae (e.g., HIV-1 gp41) (7, 39, 42), Orthomyxoviridae (e.g., hemagglutinin of influenza virus) (4, 8), Paramyxoviridae (e.g., F protein of respiratory syncytial virus and parainfluenza virus) (3, 44), Filoviridae (e.g., glycoprotein 2 of Ebola virus) (29), and most recently Coronaviridae (e.g., spike protein of severe acute respiratory syndrome coronavirus) (25) and Herepesviridae (glycoprotein H of herpes simplex virus) (14, 15). The properties of PS-ONs that target the α-helices in the HIV-1 fusion protein gp41 may also allow them to function as antiviral agents against other viruses with type I fusion proteins. Consistent with this hypothesis, we have evidence that long PS-ONs are also effective antiviral agents against other viruses with type I fusion proteins in the families of Herpesviridae, Orthomyxoviridae, Paramyxoviridae, and Filoviridae (data not shown). We also have evidence that long PS-ONs have potent in vivo activity against the above-mentioned viral families and are well tolerated when using aerosol, parenteral, and topical administration of the compounds (A. Vaillant and J.-M. Juteau, unpublished data). Thus, longer PS-ONs have the potential to be effective antiviral drugs active against HIV infection/AIDS. Furthermore, the mechanism of action described herein shows the potential of α-helix bundle formation as a target for therapeutic activity.
Acknowledgments
We thank Jinkui Niu and James Farmer at the MicroChemistry Laboratory, New York Blood Center, for peptide synthesis. AZT, NVP, C34, N34, and V3 loop peptides (from DAIDS, NIAID) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Agrawal, S., J. Y. Tang, and D. M. Brown. 1990. Analytical study of phosphorothioate analogues of oligodeoxynucleotides using high performance liquid chromatography. J. Chromatogr. 509:396-399. [DOI] [PubMed] [Google Scholar]
- 2.Baba, M., R. Pauwels, J. Balzarini, J. Arnout, J. Desmyter, and E. De Clercq. 1988. Mechanism of inhibitory effect of dextran sulfate and heparin on replication of human immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 85:6132-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, A. C. Treharne, R. W. Ruigrok, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Catasti, P., E. M. Bradbury, and G. Gupta. 1996. Structure and polymorphisms of HIV third variable loops. J. Biol. Chem. 271:8236-8242. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D., C. Chutlowski, and P. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D., and P. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., J. J. Skehel, and D. C. Wiley. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 96:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clanton, D. J., R. A. Moran, J. B. McMahon, O. S. Weislow, R. W. Buckheit, Jr., M. G. Hollingshead, V. Ciminale, B. K. Felber, G. N. Pavlakis, and J. P. Bader. 1992. Sulfonic acid dyes: inhibition of the human immunodeficiency virus and mechanism of action. J. Acquir. Immune Defic. Syndr. 5:771-781. [PubMed] [Google Scholar]
- 10.Dapic, V., P. J. Bates, J. O. Trent, A. Rodger, S. D. Thomas, and D. M. Miller. 2002. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry 11:3676-3685. [DOI] [PubMed] [Google Scholar]
- 11.De Clercq, E. 2001. Molecular targets for antiviral agents. J. Pharmacol. Exp. Ther. 297:1-10. [PubMed] [Google Scholar]
- 12.Este, J. A., D. Schols, K. De Vreese, K. Van Laethem, A.-M. Vandamme, J. Desmyter, and E. De Clercq. 1997. Development of resistance of human immunodeficiency virus type 1 to dextran sulfate associated with the emergence of specific mutations in the envelope gp120 glycoprotein. Mol. Pharmacol. 52:98-104. [DOI] [PubMed] [Google Scholar]
- 13.Este, J., C. Cabrera, D. Schols, P. Cherepanov, A. Gutierrez, M. Witvrouw, C. Pannecouque, Z. Debyser, R. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (Zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 14.Gianni, T., P. L. Martelli, R. Casadio, and G. Campidelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianni, T., L. Menotti, and G. Campidelli-Fiume. 2005. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha helix with attributes of a fusion peptide, is critical for virus entry and inhibition. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung, C.-S., N. Vander Hayden, and L. Ratner. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 73:8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itri, L. 2005. Genesense (BCL-2 antisense): efficacy and safety from randomized phase 3 clinical trials in patients with melanoma and chronic lymphocytic leukemia (CLL). Oligonucleotide Therapeutics Society, New York, N.Y.
- 18.Jiang, S., H. Lu, S., Lui, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 48:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, S., K. Lin, and A. R. Neurath. 1991. Enhancement of human immunodeficiency virus type-1 (HIV-1) infection by antisera to peptides from the envelope glycoproteins gp120/gp41. J. Exp. Med. 174:1557-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, S., K. Lin, L. Zhang, and A. K. Debnath. 1999. A screening assay for antiviral compounds targeted to the HIV-1 gp41 core structure using a conformation-specific monoclonal antibody. J. Virol. Methods 80:85-96. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the HIV-1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J., C. Cheong, and P. Moore. 1991. Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature 351:331-332. [DOI] [PubMed] [Google Scholar]
- 24.Lavigne, C., J. Yelle, G. Sauve, and A. Thierry. 2002. Is antisense an appropriate nomenclature or design for oligodeoxynucleotides aimed at the inhibition of HIV-1 replication? AAPS Pharmacol. Sci. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between the heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implication for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S., H. Lu, J. Niu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259-11273. [DOI] [PubMed] [Google Scholar]
- 27.Liu, S., Q. Zhao, and S. Jiang. 2003. Determination of the HIV-1 gp41 fusogenic core conformation modeled by synthetic peptides: applicable for identification of HIV-1 fusion inhibitors. Peptides 24:1303-1313. [DOI] [PubMed] [Google Scholar]
- 28.Lynch, G., L. Low, S. Li, A. Sloane, S. Adams, C. Parish, B. Kemp, and A. L. Cunningham. 1994. Sulfated polyanions prevent HIV infection of lymphocytes by disruption of the CD4-gp120 interaction, but do not inhibit monocyte infection. J. Leukoc. Biol. 56:266-272. [DOI] [PubMed] [Google Scholar]
- 29.Malashkevitch, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein gp2 from Ebola virus at 1.9 Å resolution. Proc. Natl. Acad. Sci. USA 96:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsukura, M., K. Shinozuka, G. Zon, H. Mitsuya, M. Reitz, J. Cohen, and S. Broder. 1987. Phosphorothioate analogs of oligodeoxynucleotides: inhibitors of replication and cytopathics of human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 84:7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meerbach, A., J. Neyts, J. Balzarini, B. Helbig, E. De Clercq, and P. Wutzler. 2002. In vitro activity of polyhydroxycarboxylates against herpesviruses and HIV. Antivir. Chem. Chemother. 12:337-345. [DOI] [PubMed] [Google Scholar]
- 32.Meyers, G., B. T. Korber, B. T. Foley, K.-T. Jeang, J. W. Mellor, and S. Wain-Hobson (ed.). 1996. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 33.Moulard, M., H. Lortat-Jacob, I. Mondor, G. Roca, R. Wyatt, J. Sodroski, L. Zhao, W. Olsen, P. Kwong, and Q. Sattentau. 2000. Selective interactions of polyanions with basic surfaces of human immunodeficiency virus type 1 gp120. J. Virol. 74:1948-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neurath, A. R., N. Strick, Y.-Y. Li, and A. K. Debnath. 2001. Cellulose acetate phthalate, a common pharmaceutical excipient, inactivates HIV-1 and blocks the coreceptor binding site on the virus envelope glycoprotein gp120. BMC Infect. Dis. 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neurath, A. R., N. Strick, and Y.-Y. Li. 2002. Anti-HIV activity of anionic polymers: a comparative study of candidate microbiocides. BMC Infect. Dis. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuette, J., D. Cole, and G. Srivatsa. 1994. Development and validation of a method for routine base composition analysis of phosphorothioate oligonucleotides. J. Pharmacol. Biomed. Anal. 12:1345-1353. [DOI] [PubMed] [Google Scholar]
- 37.Stein, C. A., M. Matsukura, C. Subasinghe, S. Broder, and J. Cohen. 1989. Phosphorothioate oligodeoxynucleotides are potent sequence nonspecific inhibitors of de novo infection by HIV. AIDS Res. Hum. Retrovir. 5:639-646. [DOI] [PubMed] [Google Scholar]
- 38.Stein, C. A., L. Neekers, B. Nair, S. Mumbauer, G. Hoke, and R. Pal. 1991. Phosphorothioate oligodeoxycytidine interferes with the binding of HIV-1 gp120 to CD4. J. Aquir. Immune Defic. Syndr. 4:686-693. [PubMed] [Google Scholar]
- 39.Tan, K., J. Lie, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vives, R. R., A. Imberty, Q. J. Sattentau, and H. Lorat-Jacob. 2005. Heparan sulfate targets the HIV-1 envelope glycoprotein gp120 coreceptor binding site. J. Biol. Chem. 280:21353-21357. [DOI] [PubMed] [Google Scholar]
- 41.Wedel, M. 2005. Evaluation of 1st and 2nd generation antisense drugs in clinical trials for the treatment of inflammatory and metabolic disorders. Oligonucleotide Therapeutics Society, New York, N.Y.
- 42.Weissenhorn, W., A. Dessen, S. Harrison, J. Skehel, and D. Wiley. 1997. Atomic strucutre of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi, K., B. Papp, D. Zhang, A. Ali, S. Agrawal, and R. Byrn. 1997. The multiple inhibitory mechanisms of GEM 91, a gag antisense phosphorothioate oligonucleotides, for human immunodeficiency virus type-1. AIDS Res. Hum. Retrovir. 13:545-554. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, X., M. Singh, V. N. Malashkevitch, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]