Abstract
We inoculated an in vitro pharmacodynamic model simultaneously with clinical isolates of methicillin-resistant Staphylococcus aureus and an enterocin-producing enterococcus (vancomycin-resistant Enterococcus faecalis, ampicillin susceptible) at 7 log10 CFU/ml to examine enterocin effects and antimicrobial activity on staphylococci. The investigated antimicrobial regimens were 100 mg arbekacin every 12 h (q12h), 6 mg daptomycin per kg of body weight/day, 600 mg linezolid q12h, and 100 mg tigecycline q24h alone and in combination (daptomycin, linezolid, and tigecycline) with arbekacin. Simulations were performed in triplicate; bacterial quantification occurred over 48 h, and development of resistance was evaluated throughout. When we evaluated the impact of antimicrobial activity against S. aureus alone, daptomycin demonstrated bactericidal activity (≥3 log10 CFU/ml kill), whereas arbekacin, linezolid, and tigecycline displayed bacteriostatic activities (<3 log10 CFU/ml kill). In the mixed-pathogen model, early and distinctive stunting of S. aureus growth was noted (1.5 log CFU/ml difference) in the presence of enterocin-producing E. faecalis compared to growth controls run individually (P = 0.02). Most noteworthy was that in the presence of enterocin-producing E. faecalis, bactericidal activity was observed with arbekacin and tigecycline and with the addition of arbekacin to linezolid. Antagonism was noted for the combination of tigecycline and arbekacin against S. aureus in the presence of enterocin-producing E. faecalis. Our research demonstrates that the inhibitory effect of E. faecalis contributed significantly to its overall antimicrobial impact on S. aureus. This contribution was enhanced or improved compared to the activity of each antimicrobial alone. Further research is warranted to determine the impact of polymicrobial infections on antimicrobial activity.
Polymicrobial infections appear in a variety of clinical settings, including skin and soft tissue, abdominal, and catheter-related infections (7, 12). It can be difficult to discern each pathogen's contribution to the infection site, therefore rendering these infections difficult to treat and often times requiring broad-spectrum antimicrobial agents or combination therapy. Besides the impact of antimicrobial therapy, other influences contribute to the ability of bacteria to survive within a given infection site. These influences include environmental conditions which may favor one pathogen over another, quorum sensing via cell-to-cell communication, and the secretion of virulence factors or small peptides known as bacteriocins (called enterocins in enterococci), which inhibit the growth of competing bacteria (4). Frequently, bacteria that are identified together in clinical infections possess inhibitory properties over one another. These peptides, called enterocins, are heat stable, possess activity against staphylococci, and are produced by approximately 70% of enterococci (4).
A number of investigators have explored intraspecies bacterial interactions to determine whether the communication between bacterial pathogens can be used to discover new therapeutic targets and/or potential compounds. Certain gram-positive bacteria, including enterococci, have been commonly cultured in vivo in the presence of staphylococci and can frequently cohabit infection sites, especially in immune-compromised hosts (13).
We evaluated the impact of enterocin production from Enterococcus faecalis on Staphylococcus aureus alone and in combination with four antimicrobial agents possessing antistaphylococcus activity. These agents include arbekacin, an aminoglycoside commonly used in Japan for its methicillin-resistant Staphylococcus aureus (MRSA) activity; daptomycin, a novel lipopetide antibiotic; linezolid, a synthetic oxazolidinone; and tigecycline, a new glycylcycline antibiotic which was approved for use in 2005 for the treatment of complicated skin and soft tissue and interabdominal infections. Each agent has documented activity against MRSA and vancomycin-resistant enterococcus (6).
To date, there have been limited studies evaluating mixed pathogens in vitro or in vivo. Moreover, there are few studies evaluating antimicrobial efficacy or pharmacodynamic synergistic or inhibitory interactions of dual-pathogen infections. Utilizing an in vitro pharmacodynamic model, we investigated the impact of E. faecalis on the antimicrobial activities of four gram-positive, anti-infective agents against methicillin-resistant S. aureus.
(This work was presented in part at the 14th European Congress of Clinical Microbiology and Infectious Diseases, Prague, Czech Republic, 1 to 4 May 2004.)
MATERIALS AND METHODS
Detection of enterocin production.
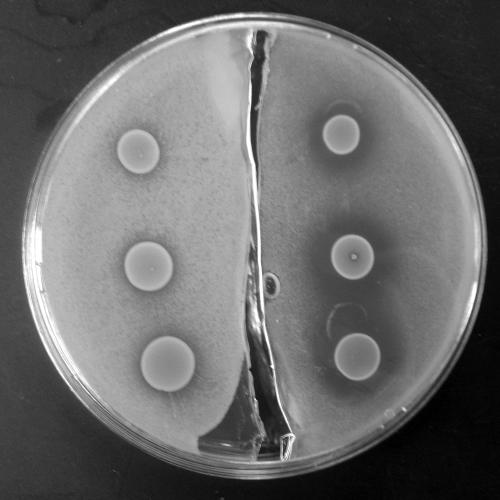
Three clinical isolates of Enterococcus faecalis (R585, R588, and R2526) were evaluated for enterocin production. Detection of the enterocin produced by E. faecalis was evaluated by a modified agar sandwich bioassay procedure as described by Navarro et al. (11) and modified as follows. Trypticase soy agar plus 0.5% yeast (TSA-Y; Becton Dickinson, Sparks, MD) was used for the foundation layer of the procedure. A 1.0 McFarland suspension of an overnight culture of E. faecalis was spotted onto the TSA-Y plates and incubated for 24 h at 37°C. Brain heart infusion supplemented with 0.8% agar (BHI-A; Difco Laboratories, Detroit, MI) was prepared and autoclaved to ensure sterility. Five milliliters of BHI-A seeded with 40 μl of 0.5 McFarland of the indicator organism, S. aureus, was poured onto the spotted foundation layer of each plate to form the second or top layer of the sandwich bioassay. Plates were then incubated for 24 h at 37°C. A clearing or opaque zone of inhibition around the E. faecalis spot indicated a positive test for enterocin production. The results demonstrated that the R2526 isolate (Fig. 1) had the most evident inhibition zones and was therefore selected for evaluation in the mixed-infection pharmacodynamic models and tested in combination with various antimicrobial agents.
FIG. 1.
Modified sandwich bioassay. On the right, three 20-μl spots of vancomycin-resistant E. faecalis R2526 are pipetted onto methicillin-resistant S. aureus (MRSA 494), with a resultant inhibition zone ± standard deviation of 8.07 ± 0.98 mm. The left side depicts the inhibitory effects of α-chymotrypsin and trypsin when added to the agar assay, with a resultant inhibition zone ± standard deviation of 2.23 ± 0.25 mm.
Inhibition of enterocin production.
Enterococcus faecalis R2526 was further evaluated for enterocin production. Using the previously described sandwich agar method, we modified the foundation layer to incorporate a hydrolytic enzyme that was suspected to have activity against enterocins. The foundation layer was divided to incorporate the enzyme into half of the foundation layer, whereas a comparison layer without enzyme was utilized in the second half as the control. Incorporated directly into the TSA-Y layer were 10 units/mg papain, 10 units/mg pepsin, 70 units/mg RNase, 592 units/mg DNase, 1,000 units/mg trypsin, 37 units/mg α-chymotrypsin, 0.6 units/mg α-chymotrypsin, or 150 units/mg heparin (Sigma Laboratories, St. Louis, MO). A 1.0 McFarland suspension of an overnight culture of E. faecalis was spotted onto the TSA-Y plates. Five milliliters of BHI-A seeded with S. aureus was poured onto the spotted foundation layer of each plate and incubated for 24 h at 37°C. Compared to the plain foundation layer, a decreased or absent zone of inhibition indicated an inhibition of the enterocin by the hydrolyzing enzyme and a positive result. The addition of α-chymotrypsin and trypsin (Fig. 1) had the most evident inhibition of the enterocin, as demonstrated by the reduction in inhibition zones of 5.83 and 2.37 mm, respectively, compared to that of the controls.
Bacterial strains.
Two clinical isolates were evaluated. MRSA 494 (mecA gene confirmed) was provided by Glenn Kaatz of the John D. Dingell Veterans Affairs Medical Center, Detroit, MI, and R2526 vancomycin-resistant Enterococcus faecalis (VRE) (DMC 83006, vanA gene confirmed) was provided by William Brown of the Detroit Medical Center.
Antimicrobials.
Arbekacin (Meiji Seika Kaisha, Ltd., Tokyo, Japan), daptomycin (Cubist Pharmaceuticals, Lexington, MA), and tigecycline (Wyeth Research, Pearl River, NY) analytical powders were obtained from their respective manufacturers. Linezolid (Pfizer Pharmaceuticals) was purchased commercially. Ampicillin and vancomycin analytical powders were purchased commercially from Sigma Chemical (St. Louis, MO). Stock solutions of each antibiotic were freshly prepared at the beginning of each week and kept frozen at −4°C.
Media.
Mueller-Hinton broth (Difco Laboratories, Sparks, MD) was supplemented with 25 mg/liter calcium and 12.5 mg/liter magnesium (SMHB). This medium was used for in vitro pharmacodynamic models. For the models evaluating daptomycin, physiological concentrations equivalent to 50 mg/liter of ionized calcium (1.02 ± 0.38 mmol/liter of ionized calcium) were supplemented according to CLSI (formerly NCCLS) guidelines due to daptomycin's dependency on physiological calcium for its activity (2, 9, 10, 14). Colony counts were determined using TSA (Difco Laboratories, Sparks, MD). For the isolation of VRE, Mueller-Hinton agar antibiotic plates containing vancomycin at eight times the MIC of the MRSA isolate were used. For the isolation of MRSA, Mueller-Hinton agar antibiotic plates containing ampicillin at eight times the MIC of the VRE were used, since the chosen strain of VRE was susceptible to ampicillin. To determine the impact, if any, of ampicillin or vancomycin incorporated into the agar on organism growth, an inoculum check versus the control (no antibiotic) was performed for both the enterococcus and staphylococcus test organisms.
Susceptibility testing.
MICs and minimum bactericidal concentrations (MBCs) of the antimicrobial agents were determined by broth microdilution with an inoculum of 5 × 105 and freshly prepared SMHB supplemented as described above, according to the CLSI (9, 10).
In vitro pharmacodynamic model.
An in vitro pharmacodynamic model consisting of a 250-ml one-compartment glass chamber with multiple ports for the removal of SMHB, delivery of antibiotics, and collection of bacterial and antimicrobial samples was utilized. The apparatus was prefilled with medium, and antibiotics were administered as boluses into the central compartment via an injection port. All model simulations were conducted over 48 h and were performed in triplicate to ensure reproducibility. Prior to each experiment, bacterial colonies from overnight growth on TSA were made to obtain a 5 × 108 CFU/ml suspension (0.5 McFarland of MRSA and 1.0 McFarland of VRE, verified with a calorimeter) of each organism. Then, 2.5 ml of diluted organism suspension (5 × 108) was added to each of the pharmacodynamic models to produce a starting inoculum of 5 × 106 CFU/ml for each organism. Each model was placed in a 37°C water bath for the duration of the experiment, with a magnetic stir bar in each model to produce continuous mixing of the medium. A peristaltic pump (Masterflex; Cole-Parmer Instrument Company, Chicago, Ill.) was used to continually replace antibiotic-containing medium with fresh SMHB (at a rate to simulate the half-lives [t1/2] of respective antibiotics). All antimicrobials were infused into the dosing port over approximately 1 min. Daptomycin was given, with corresponding exposures reflecting approximate free daptomycin concentrations. Free concentrations were based on daptomycin's high affinity for protein (93% protein bound) (15). Total concentrations were simulated for arbekacin, linezolid, and tigecycline. Regimen simulations (estimated peak concentration and t1/2 are given in parentheses, respectively) were as follows: 100 mg arbekacin every 12 h (8 mg/liter; 4 h), 6 mg daptomycin per kg of body weight every 24 h (6.65 mg/liter; 8 h), 600 mg linezolid every 12 h (18 mg/liter; 5 h), and 100 mg tigecycline every 24 (1.17 mg/liter; 31 h).
Pharmacodynamic analysis.
Samples (approximately 1 ml each) from each model were collected at 0, 1, 2, 4, 6, 8, 24, 28, 32, and 48 h and serially diluted in cold 0.9% sodium chloride. Bacterial counts were determined by plating 100-μl aliquots of each diluted sample on TSA, using an automated spiral dispenser (Whitley Automatic Spiral Plater; Don Whitley Scientific Limited, West Yorkshire, England). Plated samples were incubated at 37°C for 24 h, and colony counts (log10 CFU per milliliter) were determined using a laser colony counter (ProtoCOL [version 2.05.02]; Synbiosis, Cambridge, England). The limit of detection for this method of colony count determination is 2.0 log10 CFU/ml. Growth curves were conducted for each pathogen alone and in combination to determine the impact of E. faecalis on the growth characteristics of the S. aureus isolate. For the mixed-pathogen models, samples were removed and plated (as described above) on TSA plates containing antibiotic at eight times the MIC of either ampicillin to isolate colonies of S. aureus or vancomycin to isolate colonies of E. faecalis. For all samples, antimicrobial carryover was accounted for by serial dilution of the plated samples. If the anticipated dilution was near the MIC, then vacuum filtration was also used. When vacuum filtration was used, samples were washed through a 0.45-μm filter with normal saline to remove the antimicrobial agent. These filters were plated onto TSA and incubated at 37°C for 24 h. Model time-kill curves were determined by plotting mean colony counts (log10 CFU per milliliter) from each model versus time. Bactericidal activity (99.9% kill) was defined as a ≥3 log10 CFU/ml reduction in colony count from that of the initial inoculum. Bacteriostatic activity was defined as a <3 log10 CFU/ml reduction in colony count from that of the initial inoculum, while inactivity was defined as no observed reductions in colony counts of initial inoculums. The impact of E. faecalis with or without arbekacin on the activity of daptomycin, linezolid, or tigecycline was determined by comparing the change in the number of CFU/ml of S. aureus at 48 h. Enhancement of activity by the presence of E. faecalis or by the addition of arbekacin was defined as an increase in kill of ≥2 log10 CFU/ml versus that of the most active single agent of that combination. Improvement was defined as a 1 to 2 log10 CFU/ml increase in kill in comparison to that of the most active single agent, while combinations that resulted in ≤1 log10 CFU/ml bacterial growth in comparison to the least-active single agent were considered to represent antagonism. The terms “improvement” and “enhancement” were used because our simulations used therapeutically obtained serum concentration, and this did not permit the mathematical modeling necessary to consider the standard terms “additivity” and “synergy” (1). Reductions in colony counts were determined over a 48-h period and compared between regimens. The time required to achieve 99.9% killing was determined by linear regression (if r2 was ≥0.95) or by visual inspection.
Pharmacokinetic analysis.
Samples were obtained, through the dosing port, at 0.5, 1, 2, 4, 8, 24, 32, and 48 h for verification of target antimicrobial agent concentrations. All samples were stored at −70°C until analysis of each antibiotic was conducted. Concentrations of arbekacin and vancomycin were determined using fluorescence polarization immunoassays (TDX assay; Abbott Diagnostics, Tokyo, Japan, and North Chicago, Ill., respectively). The arbekacin assay had a limit of detection of 1.93 μg/ml and a between-day coefficient of variation (CV) of <6%. The vancomycin assay had a limit of detection of 2.0 μg/ml and a between-day CV of ≤4.8%. Linezolid samples were determined by high-pressure liquid chromatography by the Division of Infectious Diseases at the National Jewish Medical and Research Center (Denver, Colo.) as previously described (6a). The standard curves for linezolid ranged from 2.06 to 19.51, and the between-day CV was ≤9.43%.
Concentrations of all other agents were determined using standard agar diffusion bioassay procedures. Daptomycin concentrations were determined using antibiotic assay medium 5 supplemented with 50 mg/liter calcium and Micrococcus luteus ATCC 9341. Blank 1/4-mm disks were spotted with 20 μl of 1, 2, 5, 7.5, and 10 μg/ml standards (standard curve) or hourly samples. We tested each standard in triplicate by placing the disk onto the appropriate agar plates that were preswabbed with a 0.5 McFarland suspension of the test organism. Plates were incubated for 18 to 24 h at 37°C before zone sizes were measured. The daptomycin bioassay had a lower limit of detection of 1 μg/ml and a between-day CV of ≤11.1%. Correlation coefficients for all assays were >0.93. For the tigecycline assay, agar medium was prepared by adding 8 g nutrient broth (BBL; Becton Dickinson) and 11 g agarose (Sigma Chemical) (1.1% vol/vol) per liter of distilled water. We prepared a stock solution of tigecycline at a concentration of 1,000 μg/ml by weighing and dissolving the standard powder in 0.06 M phosphate-buffered saline (pH 7.8). The solution was then diluted for a concentration range of 4, 2, 1, 0.5, 0.25, and 0.12 μg/ml for the standard curve. Wells (approximately 4-mm diameter) were cut into the agar assay plate. The standard curve and the samples were placed into the wells in a random array. The plates were placed at 4°C for 2 hours and then incubated at 30°C for an additional 18 h. The between-day CV for this assay was 8.4%. Peak, trough, and t1/2 were calculated for each antimicrobial agent using concentration-time plots of the model samples. The area under the concentration-time curve from 0 to 24 h (AUC0-24) was calculated using linear trapezoid methods and the PKAnalyst program (version 1.10; MicroMath Scientific Software, Salt Lake City, UT).
Resistance.
Samples (100 μl) from each time point were plated on TSA plates containing four- and eightfold the MIC of each antimicrobial to assess the development of resistance. Plates were visually inspected for growth of resistant subpopulations after 24, 32, and 48 h of incubation at 37°C.
Statistical analysis.
Bacterial densities (number of CFU/ml) at 48 h and the time to 99.9% kill were compared by two-way analysis of variance with Tukey's posthoc test. Relationships between bacterial densities at 48 h were evaluated by nonlinear regression. A P value of ≤0.05 was considered significant. All data were plotted and graphed using SigmaPlot software (version 6.1; SPSS, Inc., Chicago, Ill.). All statistical analyses were performed using SPSS statistical software (release 11.1; SPSS, Inc., Chicago, Ill.).
RESULTS
Susceptibility testing.
Microdilution MICs and MBCs for S. aureus and E. faecalis are listed in Table 1. S. aureus was resistant to ampicillin (>512 μg/ml) and susceptible to arbekacin, daptomycin, linezolid, tigecycline, and vancomycin. E. faecalis was resistant to vancomycin (>256 μg/ml) and arbekacin (64 μg/ml) and was susceptible to ampicillin, daptomycin, linezolid, and tigecycline.
TABLE 1.
Susceptibility testing results of MRSA and VRE
| Antimicrobial agent | MIC (μg/ml) (MBC [μg/ml]) for:
|
|
|---|---|---|
| MRSA 494 | VRE R2526 | |
| Ampicillin | 512 (>512) | 1 (2) |
| Arbekacin | 0.25 (1) | 64 (>64) |
| Daptomycin | 0.25 (0.5) | 0.5 (1) |
| Linezolid | 2 (8) | 2 (8) |
| Tigecycline | 0.125 (0.5) | 0.125 (0.5) |
| Vancomycin | 0.25 (0.5) | >256 (>256) |
Pharmacokinetic analysis.
Achieved t1/2, maximum (peak) concentrations, and AUC0-24 for each antibiotic are demonstrated in Table 2. For all model simulations, pH ranged between 6.88 and 7.05, and the temperature was maintained at 37°C throughout the 48-h experiments.
TABLE 2.
Values of pharmacokinetic parameters obtained with mixed-pathogen modelsa
| Regimen | Peak concn (μg/ml) | Half-life (h) | AUC0-24 (μg · h/ml) |
|---|---|---|---|
| 100 mg arbekacin q12h | 7.85 ± 0.40 | 3.26 ± 0.19 | 78.3 ± 0.38 |
| 600 mg linezolid q12h | 21.90 ± 1.57 | 5.5 ± 0.64 | 306.6 ± 5.88 |
| 100 mg tigecycline q24h | 1.19 ± 0.11 | 32 ± 1.22 | 22.3 ± 0.53 |
| 6 mg/kg daptomycin q24* | 6.67 ± 0.56 | 8 ± 0.22 | 68.12 ± 2.2 |
Values are means ± standard deviations. q12h, every 12 h. *, pharmacokinetic calculations of free drug.
Growth control.
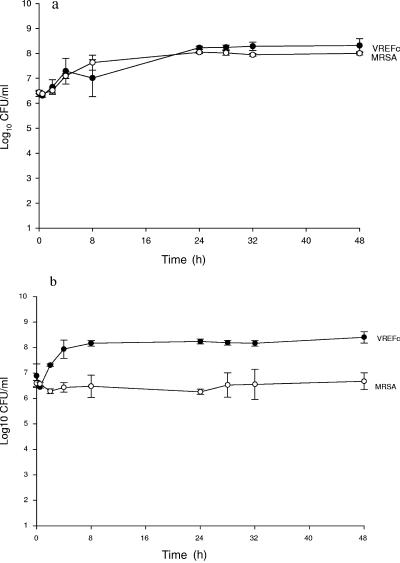
Results of the population analysis comparing the organism concentration in antibiotic-incorporated medium to that in the control medium demonstrated no appreciable effect on the growth of the target organism (target versus control [number of CFU/ml ± standard deviation]: E. faecalis, 5.80 ± 0.01 versus 5.88 ± 0.03; S. aureus, 5.23 ± 0.40 versus 5.56 ± 0.01) via the antibiotic used to separate the organisms for the purpose of colony count determination. The growth controls of S. aureus and E. faecalis in the single-pathogen model and together in a mixed-pathogen model are demonstrated in Fig. 2a and b. When S. aureus and E. faecalis were grown individually, the growth controls of both organisms demonstrated equal logarithmic-phase growth during the first 24 h and then remained in stationary phase throughout the 48-hour experiment. In contrast, S. aureus demonstrated a significantly different growth curve in the presence of E. faecalis. A 1.5 log decrease in growth as early as 4 h was observed for S. aureus compared to the growth curve of this isolate in the absence of E. faecalis (P < 0.01).
FIG. 2.
Growth control of (a) single-pathogen model and (b) mixed-pathogen model.
Daptomycin.
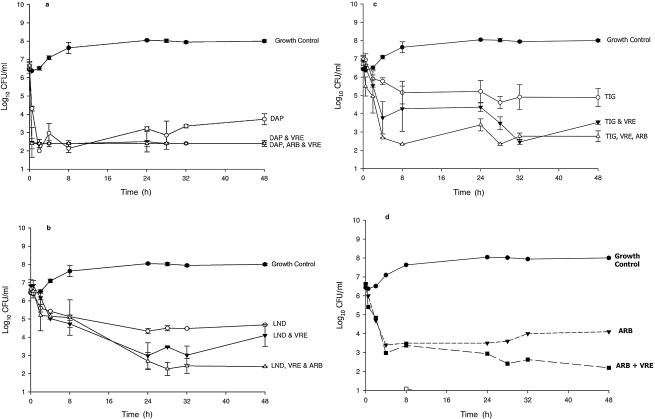
Daptomycin's activity against MRSA, alone and in combination with VRE and arbekacin, is demonstrated in Fig. 3a. Daptomycin demonstrated similar bactericidal kill against both the E. faecalis (data not shown) and the S. aureus isolates in the single-pathogen model throughout the 48-h experiment. However, in the presence of E. faecalis, a significant enhancement of kill was noted for daptomycin against S. aureus compared to the activity of daptomycin against S. aureus alone. Specifically, at the 0.5-h time point, activity against S. aureus was enhanced by 2.2 log kill in the presence of daptomycin and E. faecalis compared to that of daptomycin alone. When arbekacin was added to the mixed-pathogen model, no additional kill was noted, as the combination of daptomycin in the presence of E. faecalis was already at the limit of detection for evaluating kill.
FIG. 3.
Daptomycin (DAP) (a), linezolid (LND) (b), tigecycline (TIG) (c), and arbekacin (ARB) (d) activities against MRSA alone and in combination with VRE and arbekacin (a, b, and c).
No development of resistance was observed against S. aureus or E. faecalis at any time point throughout the 48-h model with daptomycin alone or in combination with E. faecalis or arbekacin. The free AUC/MIC ratio for daptomycin against S. aureus was 273.
Linezolid.
The activity of linezolid against MRSA, alone and in combination with VRE and arbekacin, is demonstrated in Fig. 3b. Against S. aureus, linezolid demonstrated bacteriostatic activity throughout the 48-h study. However, enhanced kill of S. aureus was observed at the 24-, 28-, and 32-h time points in the presence of E. faecalis. Most interesting is the significant bactericidal killing (99.9% kill) of S. aureus when linezolid and arbekacin were combined in the presence of E. faecalis.
The AUC/MIC ratio ± standard deviation for linezolid against S. aureus was 153 ± 3.32; the time above the MIC was 100% throughout the experiments, and no development of resistance was noted against either isolate throughout the 48-h model.
Tigecycline.
The activity of tigecycline against MRSA, alone and in combination with VRE and arbekacin, is demonstrated in Fig. 3c. Against S. aureus, tigecycline demonstrated bacteriostatic activity throughout the 48-h study. However, improvement of tigecycline's activity killing occurred in the presence of E. faecalis. The addition of arbekacin to tigecycline in the mixed-pathogen model resulted in enhancement, with significant rapid bactericidal activity (99.9% kill) as early as 4 h against the S. aureus isolate. Conversely, antagonism was noted when tigecycline was used in combination with arbekacin against the E. faecalis organism in the mixed-pathogen model (Table 3). We found this antagonistic observation interesting, most notably because the enterococcus isolate was resistant to arbekacin (MIC, 64 mg/liter). Tigecycline significantly killed the S. aureus and E. faecalis isolates alone, but when arbakacin was added in the mixed-pathogen model, antagonism was noted only against E. faecalis. The AUC/MIC ratio for tigecycline against S. aureus was 178.4, and the time above the MIC was 100% against both organisms throughout the 48-hour experiment.
TABLE 3.
Inoculum changes in single- and mixed-pathogen models of MRSA and VRE
| Antimicrobial agent(s) | Change in bacterial density (log10 CFU/ml) over 48 h in model of:
|
|||
|---|---|---|---|---|
| MRSA alone | VRE alone | MRSA in presence of VRE | VRE in presence of MRSA | |
| None (control) | +1.57 | +1.91 | +0.08a | +1.51 |
| Arbekacin | −2.54b | +1.05 | −4.43b,c | −3.06b |
| Daptomycin | −2.99b | −3.06b | −4.20b | −2.05b |
| Linezolid | −1.79b | −1.87b | −2.59b | −2.05b |
| Tigecycline | −1.82b | −1.06b | −3.38b,c | −2.35b |
| Daptomycin + arbekacin | −4.26b,c | −2.49b | ||
| Linezolid + arbekacin | −4.27b,c | −1.35b | ||
| Tigecycline + arbekacin | −4.24b,c | +0.06d | ||
P = 0.02 compared to VRE growth control in a mixed-pathogen model.
P ≤ 0.05 compared to respective growth control.
P ≤ 0.05 compared to antimicrobial and MRSA alone.
Antagonism observed.
Arbekacin.
In the single-pathogen model, arbekacin demonstrated bactericidal activity against S. aureus, whereas arbekacin demonstrated no significant activity against E. faecalis (Fig. 3d). The combination of arbekacin and E. faecalis demonstrated significant bactericidal activity and improvement of kill against S. aureus in the mixed-pathogen model. No resistance was noted with S. aureus against arbekacin throughout the 48-h model. The maximum concentration of arbekacin in serum/MIC ratio was 31 against S. aureus.
DISCUSSION
Mixed or multiple gram-positive bacterial infections commonly appear in both hospital and community settings (7, 12). There are several influences on the growth and killing potential of bacteria in these types of infections. These include both the external (antimicrobial agents) and internal (virulence factors, peptide secretion, and cell-to-cell communication) environmental influences of the bacterial infection site (8). It was our main objective to evaluate the impact of enterocin-producing vancomycin-resistant Enterococcus faecalis on the activity of several antimicrobial agents against methicillin-resistant Staphylococcus aureus in a mixed-pathogen model.
It is well documented that organisms both cooperate and compete in ecosystems (4). The ability of a single pathogen to become the predominant organism within a mixed infection is a competitive and complex effort. The inhibition of competing organisms can be accomplished by a variety of means, including alteration of pH, the production of hydrogen peroxide, differences in the ability or inability to adapt (adhesion fitness, eliciting immune system response, etc.), and cell-to-cell communication via quorum sensing (4, 8, 11).
Enterocins are small, heat-stable peptides created by a variety of organisms, including E. faecalis and Enterococcus faecium, that possess activities against several species of bacteria, including staphylococci (4). These peptides have been demonstrated to inhibit certain food-borne pathogens, and there are continuing attempts to find useful applications of these peptides in veterinary and human medicine (3).
We have previously demonstrated the activities of daptomycin, arbekacin, tigecycline, and linezolid against MRSA (6). However, we are not aware of any time-kill evaluation of these antimicrobials in the presence of enterocins for the evaluation of improved, enhanced, or antagonistic killing between intraspecies bacteria. The simulated therapeutic regimens for the various antimicrobials tested were well within targeted concentrations, half-lives, and targeted pharmacokinetic/pharmacodynamic values that are consistent with the literature (1, 5, 16). Despite the achievement of these targeted values for optimal activity, we were able to demonstrate a further consistent enhancement or improvement of antimicrobial activity against S. aureus in the presence of E. faecalis. While we did not measure directly for enterocin peptide production in our E. faecalis isolate, it seems likely that these peptides are produced by this isolate based upon the bioassay and the inhibition of this activity via proteolytic enzymes. There was a significant stunting of growth seen in our MRSA isolate in the presence of enterocin-producing E. faecalis. The addition of E. faecalis contributed significantly to the overall kill against S. aureus for all antibiotics tested. Although no emergence of resistance was observed over the 48-h model experiments, the enhanced antibiotic killing observed in the presence of E. faecalis may be important in deterring resistance development. A limitation of our study is that we tested only three E. faecalis isolates for inhibitory effects on MRSA, and only one isolate was examined in antimicrobial combination experiments. However, it has been reported that over 70% of all enterococci produce the cationic peptides that have inhibitory effects on bacteria, including staphylococci (4).
In addition, we chose to study the impact of enterococci on S. aureus using a fixed inoculum of 5 × 106 CFU/ml. It is possible that at a lower inoculum, the impact of the enterocin would be much greater on the enhancement of these antibiotics. Although we did not study the impact of various inocula here in this initial evaluation, we plan to do so in future studies.
In conclusion, we demonstrated enhanced or improved activities of arbekacin, daptomycin, linezolid, and tigecycline against MRSA in the presence of an enterocin-producing enterococcus in a mixed pharmacodynamic model. Further research is warranted to delineate the benefits of enterocins and antimicrobial combinations against multi-drug-resistant bacterial pathogens.
REFERENCES
- 1.Andes, D., M. L. van Ogtrop, J. Peng, and W. A. Craig. 2002. In vivo pharmacodynamics of a new oxazolidinone (linezolid). Antimicrob. Agents Chemother. 46:3484-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, A. L., P. C. Fuchs, and S. D. Brown. 2001. In vitro activities of daptomycin against 2,789 clinical isolates from 11 North American medical centers. Antimicrob. Agents Chemother. 45:1919-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 4.Giraffa, G. 2003. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 88:215-222. [DOI] [PubMed] [Google Scholar]
- 5.Hanberger, H., L. E. Nilsson, R. Maller, and B. Isaksson. 1991. Pharmacodynamics of daptomycin and vancomycin on Enterococcus faecalis and Staphylococcus aureus demonstrated by studies of initial killing and postantibiotic effect and influence of Ca2+ and albumin on these drugs. Antimicrob. Agents Chemother. 35:1710-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaPlante, K. L., and M. J. Rybak. 2004. Clinical glycopeptide-intermediate staphylococci tested against arbekacin, daptomycin, and tigecycline. Diagn. Microbiol. Infect. Dis. 50:125-130. [DOI] [PubMed] [Google Scholar]
- 6a.LaPlante, K. L., and M. J. Rybak. 2004. Impact of high-inoculum Staphylococcus aureus on the activities of nafcillin, vancomycin, linezolid, and daptomycin, alone and in combination with gentamicin, in an in vitro pharmacodynamic model Antimicrob. Agents Chemother. 48:4665-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantey, I., R. L. Hill, A. V. Foster, S. Wilson, J. J. Wade, and M. E. Edmonds. 2000. Infection of foot ulcers with Staphylococcus aureus associated with increased mortality in diabetic patients. Commun. Dis. Public Health 3:288-290. [PubMed] [Google Scholar]
- 8.Moise-Broder, P. A., G. Sakoulas, G. M. Eliopoulos, J. J. Schentag, A. Forrest, and R. C. Moellering, Jr. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700-1705. [DOI] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2003. Approved standard M7-A6. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 10.National Committee for Clinical Laboratory Standards. 2003. MIC testing supplemental tables. M100-S13. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Navarro, L., M. Zarazaga, J. Saenz, F. Ruiz-Larrea, and C. Torres. 2000. Bacteriocin production by lactic acid bacteria isolated from Rioja red wines. J. Appl. Microbiol. 88:44-51. [DOI] [PubMed] [Google Scholar]
- 12.Rocha, H., M. Barros, and M. Magnavita. 1965. [Clinical study of mixed infections of the urinary tract.] Hospital (Rio de Janeiro) 67:789-793. [PubMed] [Google Scholar]
- 13.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise, R., J. M. Andrews, and J. P. Ashby. 2001. Activity of daptomycin against gram-positive pathogens: a comparison with other agents and the determination of a tentative breakpoint. J. Antimicrob. Chemother. 48:563-567. [DOI] [PubMed] [Google Scholar]
- 15.Wise, R., T. Gee, J. M. Andrews, B. Dvorchik, and G. Marshall. 2002. Pharmacokinetics and inflammatory fluid penetration of intravenous daptomycin in volunteers. Antimicrob. Agents Chemother. 46:31-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhanel, G. G., K. Homenuik, K. Nichol, A. Noreddin, L. Vercaigne, J. Embil, A. Gin, J. A. Karlowsky, and D. J. Hoban. 2004. The glycylcyclines: a comparative review with the tetracyclines. Drugs 64:63-88. [DOI] [PubMed] [Google Scholar]