Abstract
Two large conjugative resistance (R) plasmids from clinical strains of Salmonella enterica serovar Virchow carried a class 2 integron with the 5′ conserved sequence (5′CS)-dfrA1-sat1-aadA1-3′CS gene array, which is associated with defective Tn7 transposons. In addition, each contained a different class 1 integron (with 5′CS-aadA1-3′CS or 5′CS-sat-smr-aadA1-3′CS gene arrays) linked to Tn21-Tn9 sequences, and several non-integron-associated R determinants. An intact copy of Tn7 (including the class 2 integron) was present in the chromosome of each strain.
Integrons are gene expression elements that play an important role in the recruitment of antimicrobial drug resistance (R) determinants via site-specific recombination events catalyzed by the integron-encoded integrase (15). Based on the sequence of the integrase genes, several classes of integrons have been described (10, 12).
Integrons of classes 1 and 2 were simultaneously found in six isolates of Salmonella enterica serovar Virchow that were recovered as causal agents of acute gastroenteritis in a northern region of Spain. PCR amplifications (4, 20) and/or sequence analysis revealed 1,000-bp/aadA1 and 2,300-bp/sat-smr-aadA1 variable regions in the class 1 integrons of four and two isolates, respectively. The class 2 integrons from the six isolates were apparently identical (EMBL accession number AM055749) and carried the dfrA1-sat1-aadA1 gene array and the inactive integrase gene characteristic of Tn7 (5, 17, 19). Two serovar Virchow isolates (LSP 231/90 and 205/98), each containing one of the detected integron combinations, were selected for further characterization (Table 1). Both were resistant to six nonrelated antimicrobial drugs but displayed different R patterns. By conjugation experiments using Escherichia coli K-12 J53 as the recipient, transconjugants (Tc) with R phenotypes identical to those of the parental strains were obtained. The extraction of plasmid DNA using the S1-pulsed-field gel electrophoresis (PFGE) method (1) (Fig. 1A) revealed the presence of a large conjugative R plasmid (pUO-SvR1, ca. 275 kb) in LSP231/90 and its transconjugant (Tc-231), while a different conjugative plasmid (pUO-SvR2, of about the same size) was found in LSP205/98 and its transconjugant (Tc-205). PCR amplifications using Tc-231, Tc-205, and the recipient E. coli as the sources of template DNA confirmed the location of the expected R genes and integrons in the two identified plasmids. Of note, as far as we are aware, this is the first report on the presence of class 2 integrons in self-transferable plasmids of Salmonella.
TABLE 1.
Characteristics of serovar Virchow strains and their transconjugants
| Bacterial strain | R phenotypea | R genotypeb | VR size/R gene integron(s)
|
Transposonsd
|
|||
|---|---|---|---|---|---|---|---|
| Class 1 | Class 2 | Tn21 | Tn9 | Tn7 | |||
| LSP231/90 | Amp Chl Gen Sul Str Spt Tmp | blaTEM catA1 aacC2 suI1 strA strB aadA1 dfrA1 sat sat1 | 2,300/sat-smr-aadA1; qacEΔ1-sul1 | 2,300/dfrA1-sat1-aadA1 | + | + | + |
| Tc-231 | Amp Chl Gen Sul Str Spt Tmp (Rif)c | blaTEM catA1 aacC2 suI1 strA strB aadA1 dfrA1 sat sat | 2,300/sat-smr-aadA1; qacEΔ1-sul1 | 2,300/dfrA1-sat1-aadA1 | + | + | Δ |
| LSP205/98 | Chl Kan Sul Str Spt Tet Tmp | catA1 aphA1 suI1 strA strB aadA1 tetA dfrA1 sat sat1 | 1,000/aadA1; qacEΔ1-suI1e | 2,300/dfrA1-sat1-aadA1 | + | + | + |
| Tc-205 | Chl Kan Sul Str Spt Tet Tmp (Rif)c | catA1 aphA1 suI1 strA strB aadA1 tetA dfrA1 sat sat | 1,000/aadA1; qacEΔ1-sul1e | 2,300/dfrA1-sat1-aadA1 | + | + | Δ |
The strains were tested for susceptibility to antimicrobial drugs by disk diffusion according to the method of the CLSI (formerly NCCLS) (2).
Resistance determinants were identified by PCR amplification using previously described primers and conditions (4, 6). Resistance to streptothricin has not been tested, but the presence of sat genes was established by nucleotide sequencing.
Resistance from the E. coli recipient strain.
+, positive for the indicated transposon; Δ, truncated Tn7.
Genes located in the 3′ conserved sequence region of the integron(s).
FIG. 1.
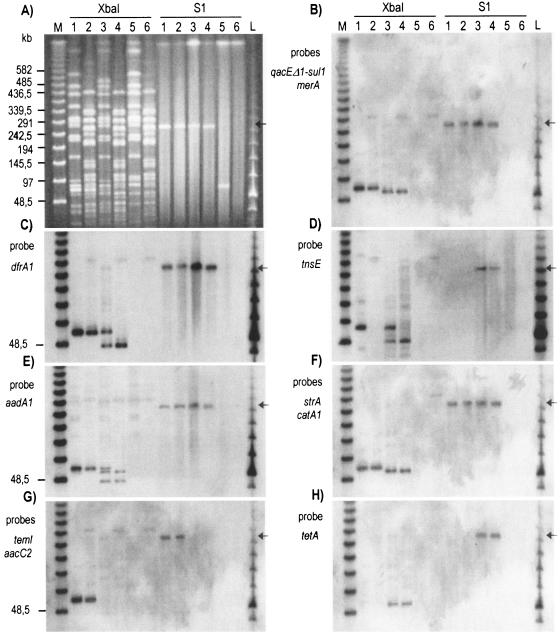
Mapping of integron, transposon, and R genes on the S. enterica serovar Virchow genome. (A) XbaI and S1 profiles of serovar Virchow strains and their transconjugants. (B through H) Hybridizations of panel A with the probes indicated at the left of each panel. Lanes M and L, PFGE marker I (New England BioLabs). Lanes 1 and 2, serovar Virchow LSP231/90 and Tc-231, both carrying pUO-SvR1. Lanes 3 and 4, serovar Virchow LSP205/98 and Tc-205, both carrying pUO-SvR2. Lane 5, serovar Virchow CECT 4154, susceptible to antimicrobial drugs. Lane 6, E. coli K-12 J53, plasmid free. The arrow indicates the ca. 275-kb plasmids.
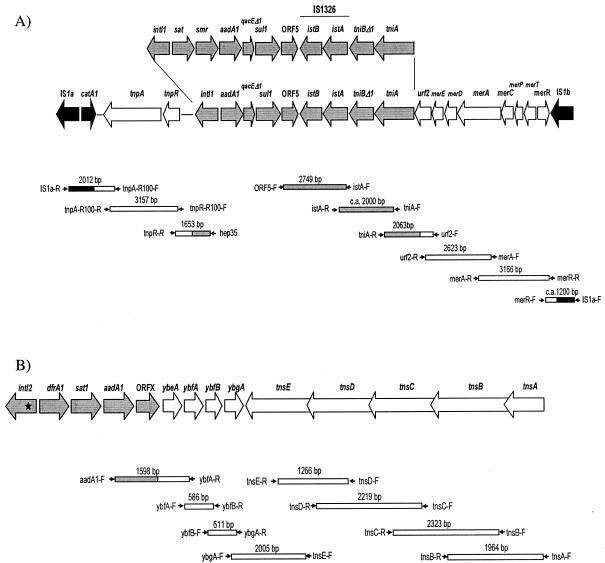
In order to investigate a possible association between integrons and transposons (7, 17, 19, 22), the presence of Tn21-, Tn9-, and Tn7-related sequences in the two serovar Virchow strains and their transconjugants was investigated by PCR amplifications of individual genes (using already-described primers and primers designed for the present work) (4) (Table 2). The four strains, but not E. coli K-12 J53, yielded right-sized amplicons with primers that were specific for the tnpA, tnpR, and merEDACPTR genes of Tn21. The same result was obtained for the catA1 gene, which was used as an indicator of Tn9. With regard to Tn7, all tested genes (ybfA, ybfB, ybgA, tnsE, tnsD, tnsC, tnsB, and tnsA) could be amplified from LSP231/90 and LSP205/98. However, genes downstream of ybfA and tnsD were not detected in Tc-231 and Tc-205, respectively. Additional amplifications of overlapping fragments (Fig. 2) demonstrated (i) the insertion of the two class 1 integrons within Tn21, which was in turn associated to Tn9, forming complex structures that differed from Tn2670 (7) by the absence of IS1353 (Fig. 2A), and (ii) the existence of an apparently intact copy of Tn7, including the class 2 integron, on the chromosome of each of the serovar Virchow strains and of a second copy of the integron, associated to a truncated Tn7, in each of the conjugative plasmids carried by these strains (Fig. 2B).
TABLE 2.
Primers designed for this work
| Region or gene | Name | Sequence (5′ to 3′) | Temp (°C) | Accession no. |
|---|---|---|---|---|
| IS1 from Tn9 | IS1a-F | CTGACGGGGTGGTGCGTAACGGC/ | 65 | NC_002134 |
| IS1a-R | GTAAAACAGCCAGCGCTGGCGCG | |||
| tnpA | tnpA-R100-F | GATCGGCGCGGGGAAGTTCAGGC | 65 | NC_002134 |
| tnpA-R100-R | AGCCGGTTGCAGAGGCCGTAGC | |||
| tnpR | tnpR-R100-F | GAGCGTCAGCGCGAGGGTATTG | —a | AF_071413 |
| istA | istA-F | GACAGCGTTGGCAAGTTAAGTCC | 60 | AF_071413 |
| istA-R | GGCATGCCAGTGGGCATCAAACA | |||
| tniA | tniA-F | GATCCTCCGGCGCACGCTGACCC | 65 | AF_071413 |
| tniA-R | AATCTGGTCGAACGGTTTGGCGG | |||
| urf2 | urf2-F | GGAGATCGAACTGACCGAATCGG | 65 | AF_071413 |
| urf2-R | CAGGAGCTGGCTGCACAACAGC | |||
| merE | merE-F | CGCCCCTGACAAACTGCCGCCC | 60 | AF_071413 |
| merE-R | CGAGCGCGGCAACACCCCAATG | |||
| merD | merD-F | TACGGCCGGTGGCCTGCACCACG | 60 | AF_071413 |
| merD-R | CGCTCGGCTGGCATGGAGGCCAG | |||
| merA | merA-F2 | AAGGCGCACGTGTCACGCTGATC | 70 | AF_071413 |
| merA-R2 | CCGCTGCCTTCTTCAACCACCAG | |||
| merC | merC-F | AGCGTCGTTTCCGCGATGGGCTG | 60 | AF_071413 |
| merC-R | CCGGTCCGCAACGGCGATGCGCC | |||
| merP | merP-F | CTCCCTTGCCCTCGCCGCCGCTG | 65 | AF_071413 |
| merP-R | GGACGGATAGCCGGCGTCTGCGG | |||
| merT | merT-F | GCGCCACGCGAGAAGCGGCCGC | 60 | AF_071413 |
| merT-R | GAAATCCAAGCGCGACCAGGAC | |||
| merR | merR-F | GGGGTCAACGTGGAGACAATCCG | 65 | AF_071413 |
| merR-R | CCTTGAGCTTGTGTTCGGCCAGG | |||
| ybfA | ybfA-F | CGCCCAAGTAAGTATCCAGCTGTG/ | 60 | NC_002525 |
| ybfA-R | CTTGACATCTCATCAATACCACC | |||
| ybfB | ybfB-F | CGATGTGGCTACGAGTCTGATC/ | 55 | NC_002525 |
| ybfB-R | CTTCTCTCTAGATAGCAGGG | |||
| ybgA | ybgA-F | CTATGCACTTTGTGACAAGTTGG/ | 60 | NC_002525 |
| ybgA-R | CTTGAAGTGCAGGCGAGCACG | |||
| tnsA | tnsA-F | GCAGCAGCCTTACAAGACGAGCG | 60 | NC_002525 |
| tnsA-R | GGCTAATACAGAGATCTGCACAC | |||
| tnsB | tnsB-F | TGTAGGTTTGCCAGATGTGTTGC | 60 | NC_002525 |
| tnsB-R | TGACCCCGAGTAATACAAACCCC | |||
| tnsC | tnsC-F | GGCAATTACAGCTTTTACAACGC | 60 | NC_002525 |
| tnsC-R | CTGCGATATCTAGCTGAAGTTGG | |||
| tnsD | tnsD-F | ACAGGGATTGGCTAGTTCACTGG/ | 60 | NC_002525 |
| tnsD-R | CTTTGTGCTTCCTCAGTTATCCG | |||
| tnsE | tnsE-F | GGCGGATCGTGTGATTGAGTTTG/ | 60 | NC_002525 |
| tnsE-R | GCGTACTACCATCACCTGCCTCC |
tnpR-R100-F was used together with tnpA-R100-R at an annealing temperature of 65°C.
FIG. 2.
Integron-transposon associations. (A) Physical linkage between the class 1 integrons, Tn21, and Tn9. The scheme is based on sequencing data from R100 Shigella flexneri plasmid (NC_002134). The class 1 integron, Tn21, and Tn9 sequences are indicated by gray, white, and black arrows, respectively. All of the overlapping fragments that are depicted below the scheme could be amplified from the two serovar Virchow strains and their transconjugants. ORF5, open reading frame 5. (B) Physical linkage between class 2 integrons and Tn7. The schematic representation is based on sequencing data from the R721 plasmid of E. coli (NC_002525). Integron and transposon genes are indicated by gray and white arrows, respectively. All of the overlapping amplicons were generated from the two serovar Virchow strains, except the last three from Tc-205 and the first two from Tc-231. Primers used in PCR amplifications are compiled in Table 2.
LSP231/90, LSP205/98, and their transconjugants were also analyzed by macrorestriction using XbaI-PFGE (11). The two serovar Virchow strains generated distinctive profiles (Jaccard's coefficient of similarity, 0.79), which included 16 matching and 7 nonmatching fragments (Fig. 1A). Selected probes for classes 1 (qacEΔ1, sul1, and aadA1) and 2 (aadA1, dfrA1) integrons, for Tn21 (merA), Tn9 (catA1), and Tn7 (tnsE and tnsA) transposons, and for R genes that were not associated to integrons or transposons [aacC2, strA, strB, blaTEM, and tetA(A)] were mapped on the XbaI and S1-PFGE profiles of the four strains (8, 13).
With LSP231/90 and Tc-231, all probes except tnsE mapped on a ca. 75-kb fragment of the XbaI profiles and on the ca. 275-kb band corresponding to pUO-SvR1 (Fig. 1B, C, and E through G). The tnsE probe mapped on a 75-kb fragment from LSP231/90 but failed to hybridize with the XbaI profile of Tc-231 and with pUO-SvR1 (Fig. 1D). These results located the class 1 integron that was linked to Tn21-Tn9 sequences and the class 2 integron inserted into a defective Tn7, as well as all independent R genes within a ca. 75-kb XbaI fragment that was generated from pUO-SvR1. The hybridization of tnsE on a fragment of about the same size suggests the existence of two comigrating bands on the macrorestriction profile of LSP231/90, one from the chromosome and one from pUO-SvR1. In the former, the intact copy of Tn7 (including the class 2 integron) would be located. The high intensity of the relevant fragment on the agarose gel (Fig. 1A) supports this possibility.
In LSP205/98 and its transconjugant, the qacEΔ1-sul1, merA, and catA1 probes (and hence the Tn2670-like transposon with the class 1 integron), as well as the probes for independent R genes, mapped on the ca. 275-kb pUO-SvR2 and on a ca. 65-kb XbaI-fragment that was common to the macrorestriction profiles of the two strains and therefore of plasmid origin (Fig. 1B, F, and H). The dfrA1 probe hybridized with the 275-kb plasmid, a ca. 40-kb XbaI fragment from the transconjugant, and two XbaI fragments (of ca. 75 and 40 kb) from the donor strain (Fig. 1C). These results are consistent with the presence of the class 2 integron in both the plasmid and the chromosome of LSP205/98. This was confirmed by hybridizations with probes for aadA1 (Fig. 1E), a gene cassette shared by the classes 1 and 2 integrons of LSP205/98, and tnsE (Fig. 1D). The former gene mapped on the 275-kb plasmid and on two XbaI fragments (65 and 40 kb) from Tc-205. By comparison with results that were obtained with the qacEΔ1-sul1 and dfrA1 probes, the 65-kb fragment would contain the aadA1 cassette of the class 1 integron and the 40-kb fragment would contain the aadA1 cassette of the class 2 integron. In the XbaI profile of LSP205/98, an additional fragment of ca. 75 kb (where dfrA1 was previously located) hybridized with aadA1 and tnsE, hence verifying the existence of the chromosomal copy of the Tn7 integron (Fig. 1D and E). The association of the extrachromosomal copies of the class 2 integron with truncated Tn7 transposons was finally corroborated by the absence of hybridization of a tnsA probe with the two plasmids and with the XbaI profiles of the transconjugants (not shown).
With regard to public health, serovar Virchow has emerged as the third or fourth most common Salmonella serovar that causes human gastroenteritis in Europe (3, 18) and its incidence is even higher in countries like Australia and Israel, where it has been reported as an important cause of bacteremia in children (9, 14, 16, 21). Taking this into account, the emergence of multiple drug-resistant strains, such as those reported here, is an obvious cause of concern, as it is this wide range of mobile genetic elements that can efficiently contribute to the acquisition, maintenance, and spread of resistance determinants.
Nucleotide sequence accession number.
The sequence for sat-smr-aadA was deposited in the EMBL database under accession number AM055748.
Acknowledgments
We thank M. A. González-Hevia (Laboratorio de Salud Pública, Asturias, Oviedo) and CECT (Colección Española de Cultivos Tipo) for clinical and reference strains.
I. Rodríguez is the recipient of a grant from the Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología (FICYT-BP04-086). The work was supported by projects FIS PI020172 (Ministerio de Sanidad y Consumo, Spain) and SAF2005-04212 (Ministerio de Educación y Ciencia, Spain).
REFERENCES
- 1.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2005. Performance standards 7 for antimicrobial susceptibility testing—15th informational supplement. Approved 8 standard M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 3.Echeita, M. A., A. M. Aladueña, R. Díez, M. Arroyo, F. Cerdán, R. Gutiérrez, M. de la Fuente, R. González-Sanz, S. Herrera-León, and M. A. Usera. 2005. Distribución de los serotipos y fagotipos de Salmonella de origen humano aislados en España en 1997-2001. Enferm. Infecc. Microbiol. Clin. 23:127-134. [DOI] [PubMed] [Google Scholar]
- 4.Guerra, B., E. Junker, A. Miko, R. Helmuth, and M. C. Mendoza. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83-91. [DOI] [PubMed] [Google Scholar]
- 5.Hansson, K., L. Sundström, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levèsque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez, N., M. C. Mendoza, B. Guerra, M. A. Gonzalez-Hevia, and M. R. Rodicio. 2005. Genetic basis of antimicrobial drug resistance in clinical isolates of Salmonella enterica serotype Hadar from a Spanish region. Microb. Drug Resist. 11:185-193. [DOI] [PubMed] [Google Scholar]
- 9.Messer, R. D., T. H. Warnock, R. J. Heazlewood, and J. N. Hanna. 1997. Salmonella meningitis in children in far north Queensland. J. Paedriatr. Child Health 33:535-538. [DOI] [PubMed] [Google Scholar]
- 10.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 11.Peters, T. M., C. Maguire, E. J. Threlfall, I. S. T. Fisher, N. Gill, and A. J. Gatto. 2003. The Salm-gene project—a European collaboration for DNA fingerprinting for food-related salmonellosis. Eurosurveillance 8:46-50. [DOI] [PubMed] [Google Scholar]
- 12.Rowe-Magnus, D. A., and D. Mazel. 2002. The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 292:115-125. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Shimoni, Z., S. Pitlik, L. Leibovici, Z. Samra, H. Konigsverger, M. Drucker, V. Agmon, S. Ashkenazi, and M. Weinberger. 1999. Nontyphoid Salmonella bacteremia: age-related differences in clinical presentation, bacteriology, and outcome. Clin. Infect. Dis. 28:822-827. [DOI] [PubMed] [Google Scholar]
- 15.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan, A. M., L. R. Ward, B. Rowe, J. B. Woolcock, and J. M. Cox. 1998. Phages types of Australian isolates of Salmonella enterica subsp. enterica serovar Virchow. Lett. Appl. Microbiol. 27:216-218. [DOI] [PubMed] [Google Scholar]
- 17.Sundström, L., P. H. Roy, and O. Skold. 1991. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Threlfall, E. J., I. S. T. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschäpe, M. Cormican, I. Luzzi, F. Schnieder, W. Wannet, J. Machado, and G. Edwards. 2003. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Eurosurveillance 8:41-45. [DOI] [PubMed] [Google Scholar]
- 19.Tietze, E., J. Brevet, and H. Tschäpe. 1987. Relationships among the streptothricin resistance transposons Tn1825 and Tn1826 and the trimethoprim resistance transposon Tn7. Plasmid 18:246-249. [DOI] [PubMed] [Google Scholar]
- 20.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagupsky, P., N. Maimon, and R. Dagan. 2002. Increasing incidence of nontyphi Salmonella bacteremia among children living in southern Israel. Int. J. Infect. Dis. 6:94-97. [DOI] [PubMed] [Google Scholar]
- 22.Young, H. K., M. J. Qumsieh, and M. L. McIntosh. 1994. Nucleotide sequence and genetic analysis of the type Ib trimethoprim-resistant, Tn4132-encoded dihydrofolate reductase. J. Antimicrob. Chemother. 34:715-725. [DOI] [PubMed] [Google Scholar]