Abstract
In our previous crystallographic studies of human immunodeficiency virus type 1 (HIV-1) protease-substrate complexes, we described a conserved “envelope” that appears to be important for substrate recognition and the selection of drug-resistant mutations. In this study, the complex of HIV-1 protease with the inhibitor RO1 was determined and comparison with the substrate envelope provides a rationale for mutational patterns.
Human immunodeficiency virus type 1 (HIV-1) protease is a key drug target for antiviral therapy (3). However, drug resistance occurs often, making many protease inhibitors (PIs) ineffective and allowing viral replication to occur (16-19). Previous studies from our laboratory on crystal structures of substrate complexes found that the protease substrates occupy an overlapping volume that we defined as the substrate envelope (15). We found that the location of primary resistance-associated mutation for existing inhibitors correlates with where that inhibitor protrudes beyond the substrate envelope (7, 8, 14) and therefore most active-site drug-resistant mutations occur at residues that are not predominantly contacted by the protease substrates. In this study, we determined the crystal structure of a new inhibitor, RO1, and compared its structure with our previously defined substrate envelope to predict likely drug resistance sites.
RO1 is a substrate-based HIV-1 PI (Fig. 1) with a 50% inhibitory concentration of 13 nM. RO1 has 5- to 10-fold greater inhibition than the 50% inhibitory concentration of several currently prescribed PIs (4). The crystal structure of the wild-type protease-RO1 complex has been solved and refined to 1.6 Å (Table 1). In addition the thermodynamics of binding confirm that the binding of RO1 is comparable to that of the first-generation drugs, with a dissociation constant (Kd) of 0.95 nM and a free-energy change (ΔG) of −12.1 kcal/mol. Protein sample preparation, isothermal titration calorimetry, and crystallography were carried out as described previously (8, 13). The protease dimer is in the asymmetric unit with two inhibitor orientations. The orientation-specific inhibitor-protease interactions involving the active-site residues Asp25-Asp30, Met46-Ile50, and Pro81-Ile84 were nonetheless identified during the crystallographic refinement.
FIG. 1.
Chemical structure of RO1.
TABLE 1.
Crystallographic statistics and thermodynamic parameters of RO1
| Parameter | Value (SD) |
|---|---|
| Data collection | |
| Space group | P212121 |
| Z | 4 |
| a (Å) | 50.87 |
| b (Å) | 58.17 |
| c (Å) | 61.65 |
| Resolution (Å) | 1.6 |
| Total no. of reflections | 141,598 |
| No. of unique reflections | 24814 |
| Rmerge (%) | 2.8 |
| Completeness (%) | 99.9 |
| I/σ1 | 19.2 |
| Refinement | |
| R value (%) | 16.3 |
| Rfree (%) | 19.8 |
| RMSDa | |
| Bond length (Å) | 0.006 |
| Bond angles (°) | 1.5 |
| No. of water molecules | 212 |
| Isothermal titration calorimetryb | |
| Kd (nM) | 0.95 (0.08) |
| ΔH (kcal/mol) | 2.41 (0.06) |
| −TΔS (kcal/mol) | −14.5 |
| ΔG (kcal/mol) | −12.1 |
RMSD, root mean squared deviation.
Mean values of experiments conducted in triplicate at 20°C.
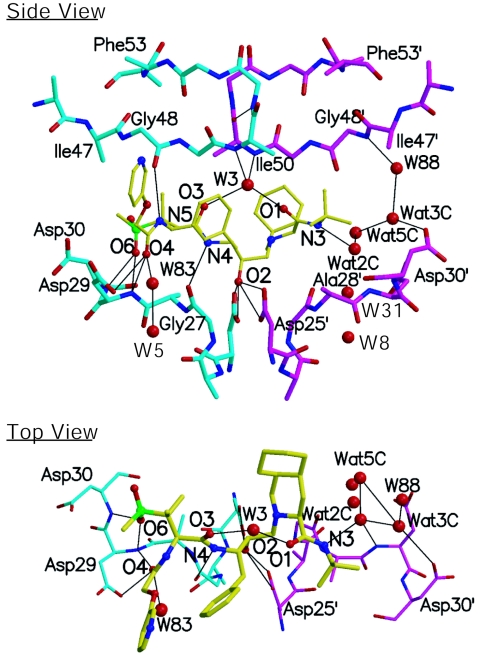
RO1-protease hydrogen bonds.
The inhibitor makes 10 backbone hydrogen bonds and a network of water bridges with the protease (Table 2 and Fig. 2). The RO1 atoms forming hydrogen bonds, except O-6, occupy substrate backbone positions of substrate-protease complexes. O-6 nonetheless overlies a substrate side chain atom which is usually Asp/AsnP2 OD1 or Glu/GlnP2 OE1 (Table 2). In addition, most RO1-protease hydrogen bonds resemble substrate-protease interactions. Another feature of RO1-protease hydrogen bonds is the involvement of both flaps (Gly48) and the floor of the active site (Asp25, Gly27, Asp29, and Asp30). In general, the flap residues do not participate in the inhibitor-protease hydrogen bonds as in indinavir (IDV), nelfinavir (NFV), TMC114, and amprenavir (APV) (Table 3). The substrates, in contrast, form hydrogen bonds with Gly48 and in certain cases even with Met46 (15).
TABLE 2.
Inhibitor-protease hydrogen bonds made by RO1 with the protease
| Protein | Atom | Inhibitor atom | Pattern found in substrate complexes (protein . . . substrate) | Distance (Å)
|
|
|---|---|---|---|---|---|
| Orientation 1 | Orientation 2 | ||||
| Asp29 | N | O-6 | Asp29/29′ N . . . OD1/OE1 P2/P2′ | 2.9 | 2.9 |
| Asp30 | N | O-6 | Asp30/30′ N . . . OD1/OE1 P2/P2′ | 2.9 | 2.8 |
| Gly48 | O | N-5 | Gly48 O . . . N P2 | 3.3 | 3.3 |
| Asp29 | N | O-4 | Asp29 N . . . O P3 | 3.0 | 2.9 |
| Asp29 | OD1 | O-4 | Asp29 OD2 . . . O P3 | 3.5 | 3.2 |
| Water31/83 | OH2 | O-4 | 3.1 | 2.9 | |
| Water3 | OH2 | O-3 | 3.0 | 2.9 | |
| Gly27 | O | N-4 | Gly27 O . . . N P1 | 3.4 | 3.3 |
| Asp25 | OD1 | O-2 | 2.6 | 2.7 | |
| Asp25 | OD2 | O-2 | 3.1 | 2.8 | |
| Asp25′ | OD1 | O-2 | 2.6 | 2.9 | |
| Asp25′ | OD2 | O-2 | 3.1 | 2.9 | |
| Water3 | OH2 | O-1 | 2.8 | 2.7 | |
| Water2C | OH2 | N-3 | 3.0 | 3.1 | |
FIG. 2.
Inhibitor protease hydrogen bonds: The two HIV-1 protease monomers are distinguished in cyan and magenta, while RO1 is shown in yellow. Nitrogen and oxygen atoms are illustrated in blue and red, respectively. The figure was created using Molscript (9).
TABLE 3.
Comparison of inhibitor protease hydrogen bondsa
| Protease position | Bond
|
||||||
|---|---|---|---|---|---|---|---|
| Inhibitor | RO1 | SQV | NFV | IDV | APV | TMC114 | |
| Asp30 OD2 | OH | No | No | Yes | No | No | No |
| Asp30 OD2 | NH2 | No | Yes | No | No | Yes | No |
| Gly27 O | NH | Yes | Yes | Yes | Yes | Yes | Yes |
| Asp29 N | O | Yes | Yes | No | No | Yes | Yes |
| Asp29 N | O | Yes | Yes | No | Yes | No | Yes |
| Asp29 OD1 | O | Yes | No | No | Yes | No | Yes |
| Asp30 N | O | Yes | Yes | No | No | Yes | Yes |
| Asp30 O | NH | No | No | No | No | Yes | Yes |
| Gly48 O | NH | Yes | Yes | No | No | No | No |
| Gly48 O | NH | No | Yes | No | No | No | No |
| Total | 6 | 7 | 2 | 3 | 5 | 6 | |
The four hydrogen bonds involving the Asp25/25′ side chain are omitted, as they are conserved in all the inhibitors. The hydrogen bond affected by the D30N NFV resistance mutation is highlighted in bold, while the conserved hydrogen bond found in all of the inhibitor complexes is highlighted with italics. The coordinates used are 1HXB (saquinavir [SQV]) (10), 1OHR (NFV) (5), 1HSG (IDV) (2), 1HPV (APV) (6), and 1T3R (TMC114) (8).
The RO1 inhibitor is also stabilized by a network of water molecules, in addition to the five water molecules preserved in most HIV-1 protease complexes (Fig. 2, water molecules labeled W3, W5, W8, W31, and W83), four additional water molecules (W88, Wat2C, Wat3C, and Wat5C) form a network of bridges mediating the inhibitor (N-3 atom of P2′) and the protease (Gly48′ of N and Asp30′ of OD2). This water network compensates for lack of direct hydrogen bonds on the P1′-P2′ side of RO1.
The substrate-like interactions and the minimal involvement of protease side chains could allow RO1 to circumvent specific drug-resistant mutations. For instance, a specific interaction between the Asp30 side chain and NFV results in a D30N resistance mutation (Table 3) (19) while this interaction is absent in RO1. Preliminary viral studies (4) indicate that viral samples with D30N protease are inhibited by RO1 but not NFV, which is consistent with this structural analysis.
van der Waals (VDW) interactions and comparison of RO1 with the substrate envelope.
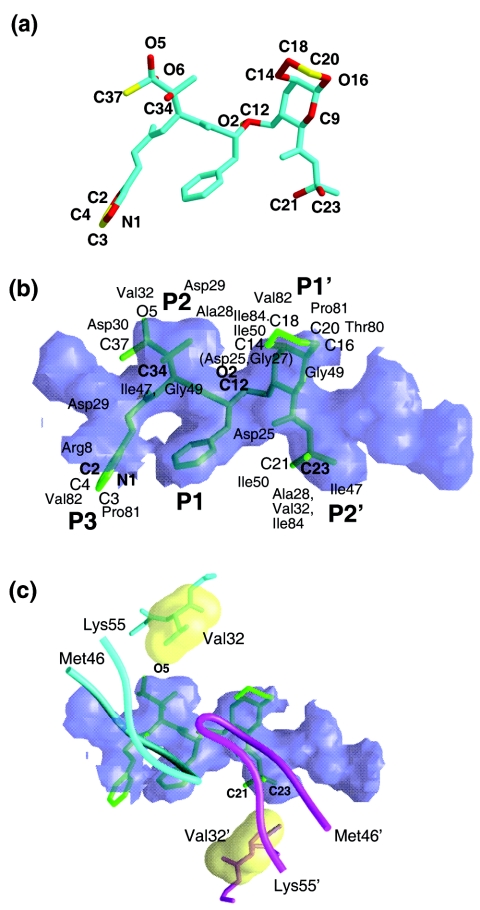
The RO1 atoms protruding outside the substrate envelope were visually evaluated (Fig. 3a) and their distances from the nearest substrate positions were also computed. An inhibitor atom is then implicated as “outlying” when it protrudes outside the substrate envelope and is more than 1.4 Å away from the nearest substrate atom (averaged over six substrate complexes). Seventeen outlying atoms were identified (12 deviate by between 1.4 and 2.0 Å and five deviate by >2.0 Å) (Fig. 3a and b). These outliers form VDW interactions with Arg8, Asp25, Ala28, Asp29, Asp30, Val32, Ile47, Ile50, Thr80, Pro81, Val82, and Ile84 (Fig. 3b). Many of these residues (Arg8, Asp25, Ala28, Asp29, Thr80, and Pro81) are highly conserved, mutating less than 1 percent of the time in the patient population (18).
FIG. 3.
(a) Outlying atoms of RO1 identified as protruding outside the substrate envelope. Atoms deviating by 1.4 to 2.0 Å and greater than 2.0 Å are distinguished in red and yellow, respectively. (b) Inhibitor RO1 superimposed on the substrate envelope: The substrate envelope is shown as a purple translucent model, while the inhibitor region embedded inside the substrate envelope and protruding atoms are shown in dark and light shades of green, respectively (also refer to the previous panel). The protruding atoms are labeled except for O-6 and C-9, which are protruding in the opposite direction. (c) Possible rationale for V32I mutation. The VDW packing interaction between Val32/32′ (illustrated as yellow surfaces) and the atoms protruding outside the P2 (O-5) and P2′ (C-21 and C-23) sites of the substrate envelope are shown. The flaps are displayed in tube representation for reference. The figures were created using Grasp (11).
Structural interpretation of mutational patterns.
RO1 appears to be a fairly robust inhibitor of resistant variants. One protrusion beyond the substrate envelope does not appear in and of itself to allow the development of resistance, as isolates with I50V, V82A, and I84V are inhibited (4). However, in a background of existing resistant mutations (M46L, G48V, I62V, L63P, T74S, V77I, and V82A) after 21 viral passages, V32I and I54V emerged and allowed growth under higher levels of RO1 (4). In a structural context, Ile54 is located in the flap and does not make contact with either the inhibitor or the substrates, and thus rationalization of its mutational pattern is not predictable from this analysis. However, the RO1 atoms protruding outside the P2 and P2′ regions make extensive interactions with Val32 from both monomers (Fig. 3c). This interaction may be perturbed by any modification of Val32 leading to drug resistance, and a V32I mutation may lead to a VDW clash between residue 32 and RO1, possibly accounting for the selection of this particular mutation.
The V32I mutation has also been observed with lopinavir (1, 12), and a comparison of lopinavir with the substrate envelope revealed that certain outlying atoms make VDW interactions with Val32 (7). For the inhibitor RO1 resistance appears to occur when mutations arise at multiple positions that protrude beyond the substrate envelope. Therefore utilization of the substrate envelope in future inhibitor design is likely to help assess and reduce the occurrence of drug resistance.
Acknowledgments
We thank Christina Ng for collecting ITC data, Claire Baldwin for editorial assistance, Balaji Bhyravbhatla and Luca Leone for technical help, and Mohan Somasundharan for discussion.
This research was supported by the National Institutes of Health (grants R01-GM64347-05 and P01-GM66524-04).
REFERENCES
- 1.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Z., Y. Li, E. Chen, D. L. Hall, P. L. Darke, C. Culberson, J. A. Shafer, and L. C. Kuo. 1994. Crystal structure at 1.9 angstrom resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J. Biol. Chem. 269:26344-26348. [PubMed] [Google Scholar]
- 3.Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retroviruses 8:153-164. [DOI] [PubMed] [Google Scholar]
- 4.Heilek-Snyder, G., A. Kohli, J. Ezpeleta-Brilliant, N. Cammack, and N. Parkin. 2003. Presented at the XII International HIV Drug Resistance workshop, Los Cabos, Mexico, 10 to 14 June 2003.
- 5.Kaldor, S., V. Kalish, J. N. Davies, B. Shetty, J. Fritz, K. Appelt, J. Burgess, K. Campanale, N. Chirgadze, D. K. Clawson, B. A. Dressman, S. D. Hatch, D. A. Khalil, M. B. Kosa, P. P. Lubbehusen, M. A. Muesing, A. K. Patick, S. H. Reich, K. S. Su, and J. H. Tatlock. 1997. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 40:3979-3985. [DOI] [PubMed] [Google Scholar]
- 6.Kim, E. E., C. T. Baker, M. D. Dwyer, M. A. Murcko, et al. 1995. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 117:1181-1182. [Google Scholar]
- 7.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, and C. A. Schiffer. 2004. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 11:1333-1338. [DOI] [PubMed] [Google Scholar]
- 8.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, P. Wigerinck, M.-P. de Bethune, and C. A. Schiffer. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012-12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraulis, P. J. 1991. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 10.Krohn, A., S. Redshaw, J. C. Ritchie, B. J. Graves, and M. H. Hatada. 1991. Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere. J. Med. Chem. 34:3340-3342. [DOI] [PubMed] [Google Scholar]
- 11.Nicholls, A., K. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Gen. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 12.Parkin, N. T., C. Chappey, and C. J. Petropoulos. 2003. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS 17:955-961. [DOI] [PubMed] [Google Scholar]
- 13.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2004. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 78:12446-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2003. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy. J. Virol. 77:1306-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prabu-Jeyabalan, M., E. A. Nalivaika, and C. A. Schiffer. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: Analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 16.Ridky, T., and J. Leis. 1995. Development of drug resistance to HIV-1 protease inhibitors. J. Biol. Chem. 270:29621-29623. [DOI] [PubMed] [Google Scholar]
- 17.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1997. Mutations in retroviral genes associated with drug resistance. Int. Antiviral News 5:129-142. [Google Scholar]
- 18.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner, D., J. M. Schapiro, B. G. Brenner, and M. A. Wainberg. 2004. The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients. Antiviral Ther. 9:301-314. [PubMed] [Google Scholar]